Abstract
Histone modifications that regulate chromatin-dependent processes are catalyzed by multisubunit complexes. These can function in both targeting activities to specific genes and in regulating genomewide levels of modifications. In Saccharomyces cerevisiae, Esa1 and Rpd3 have opposing enzymatic activities and are catalytic subunits of multiple chromatin modifying complexes with key roles in processes such as transcriptional regulation and DNA repair. Esa1 is an essential histone acetyltransferase that belongs to the highly conserved MYST family. This study presents evidence that the yeast histone deacetylase gene, RPD3, when deleted, suppressed esa1 conditional mutant phenotypes. Deletion of RPD3 reversed rDNA and telomeric silencing defects and restored global H4 acetylation levels, in addition to rescuing the growth defect of a temperature-sensitive esa1 mutant. This functional genetic interaction between ESA1 and RPD3 was mediated through the Rpd3L complex. The suppression of esa1's growth defect by disruption of Rpd3L was dependent on lysine 12 of histone H4. We propose a model whereby Esa1 and Rpd3L act coordinately to control the acetylation of H4 lysine 12 to regulate transcription, thereby emphasizing the importance of dynamic acetylation and deacetylation of this particular histone residue in maintaining cell viability.
THE genome of eukaryotic cells is packaged in chromatin, where the DNA is organized into a nucleosomal subunit structure. Nucleosomes consist of DNA wrapped around a histone octamer that contains two copies of each of the four core histones (H2A, H2B, H3, and H4), each of which can be post-translationally modified with multiple types of chemical and protein additions. The addition and removal of these modifications are catalyzed by histone modifying enzymes that function in a wide range of nuclear processes.
One dynamic histone modification is the acetylation and deacetylation of lysine residues. Enzymes that add an acetyl group to a lysine residue are known as histone acetyltransferases (HATs), and the enzymes that remove acetyl groups are called histone deacetylases (HDACs). The opposing activities of these two types of enzymes control the status of histone acetylation in the cell. For example, in the budding yeast Saccharomyces cerevisiae, regulation of H4 lysine 16 (H4K16) acetylation is critical in maintaining transcriptionally silent chromatin at the telomeres and regulating replicative life span, via activities of the HAT Sas2 and the HDAC Sir2 (Kimura et al. 2002; Suka et al. 2002; Dang et al. 2009).
Roles for the other acetylated lysines on H4 are less clearly defined. Some information has come from studying these modifications on a genomewide level. Through one microarray expression study, it became apparent that H4K5, H4K8, and H4K12 might contribute nonspecifically but jointly to transcription. Each lysine, when individually mutated to an unacetylatable amino acid, results in minimal changes in genomewide transcription. However, when combined to make double or triple lysine mutants, they display additive effects on transcription (Dion et al. 2005). In addition to participating in transcriptional control, H4 acetylation is critical for other nuclear processes, including DNA replication and repair (Megee et al. 1995; Bird et al. 2002; Choy and Kron 2002). Esa1 and Rpd3 are yeast enzymes with opposing activities toward H4 lysine acetylation, and are also members of two highly conserved families of histone modifying enzymes (reviewed in Doyon and Côté 2004; Yang and Seto 2008).
Rpd3 is one of the founding members of class I HDACs, which include the human proteins HDAC1, 2, 3, and 8 that are often overexpressed in human cancer cells. Indeed, HDAC inhibitors are being actively used and studied as therapeutic agents for multiple types of cancer (reviewed in Yang and Seto 2008). In yeast, Rpd3 deacetylates lysines on both H3 and H4 (Rundlett et al. 1996) and is involved in a wide range of nuclear processes. On a global scale, Rpd3 is responsible for transcriptional regulation of a large number of genes (Bernstein et al. 2000; Sabet et al. 2004; Alejandro-Osorio et al. 2009). For many, but not all of these genes, transcriptional regulation occurs through modification of the lysines on the H3 and H4 N termini (Sabet et al. 2004). When examined at specific loci, Rpd3 represses transcription of INO1 and IME2 by deacetylating histones at the promoters of these genes (Kadosh and Struhl 1998b; Rundlett et al. 1998). In contrast, there are several transcripts that require Rpd3 for their activation (de Nadal et al. 2004; Sertil et al. 2007). For example, Rpd3 is required for expression of the DNA damage inducible genes HUG1 and RNR3 (Sharma et al. 2007). In line with Rpd3 having a role in DNA repair, rpd3 mutants are defective in nonhomologous end joining (Jazayeri et al. 2004). Mutants of rpd3 increase silencing at the telomeres, ribosomal DNA (rDNA) repeats, and HM cryptic mating-type loci (de Rubertis et al. 1996; Rundlett et al. 1996; Vannier et al. 1996; Sun and Hampsey 1999), although the mechanism for this is unknown. Rpd3 also contributes to cell cycle control, in that rpd3 mutants undergo early DNA replication origin firing (Vogelauer et al. 2002; Aparicio et al. 2004).
Rpd3 exists in two biochemically defined complexes, named Rpd3S (small) and Rpd3L (large) to reflect their relative sizes (Carrozza et al. 2005b; Keogh et al. 2005). The identification of the subunits in each of the two Rpd3-containing complexes has begun to reveal a separation in Rpd3 complex functions. Rpd3L is likely responsible for Rpd3's role at gene promoters, as Rpd3 is recruited to chromatin via the Rpd3L-specific subunit Ume6, which recognizes specific upstream promoter sequences (Kadosh and Struhl 1997; Carrozza et al. 2005a). Rpd3L-specific mutants also display increased rDNA, telomeric, and HM loci silencing (Vannier et al. 1996; Zhang et al. 1998; Sun and Hampsey 1999; Loewith et al. 2001; Carrozza et al. 2005a; Keogh et al. 2005), and replication timing defects (Knott et al. 2009) similar to rpd3Δ itself, indicating that Rpd3L is responsible for Rpd3's role in silencing and regulation of replication initiation. The smaller Rpd3S complex is recruited to methylated H3K36 within coding sequences to repress intragenic transcription initiation (Carrozza et al. 2005b; Keogh et al. 2005), a role not shared by Rpd3L.
Whereas Rpd3 is a class I HDAC, Esa1 is part of the evolutionarily conserved MYST family of HATs. Tip60, the human homolog of Esa1, is associated with many human diseases, including HIV, Alzheimer's and multiple cancers. Tip60 acetylates the tumor suppressor p53 and acts as a transcriptional coactivator for c-Myc and NF-κB (reviewed in Avvakumov and Côté 2007). In yeast, Esa1 is essential for viability (Smith et al. 1998; Clarke et al. 1999), although Esa1's essential function may not be limited to its catalytic activity (Decker et al. 2008). Esa1 acetylates specific lysine residues on histones H2A, H3, H4 (Smith et al. 1998; Clarke et al. 1999), and the histone variant H2A.Z (Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006). Similar to Rpd3, Esa1 is the catalytic subunit of two chromatin modifying complexes that have shared subunits. The larger of these is known as NuA4, whereas the smaller is piccolo (Allard et al. 1999; Boudreault et al. 2003). In vitro activity assays indicate that piccolo is the more active acetyltransferase complex on chromatin, but separate roles for the two complexes have not been established in vivo (Boudreault et al. 2003).
Through the study of conditional esa1 mutants, Esa1 has been discovered to play an important role in many nuclear processes. Esa1 functions in cell cycle progression (Clarke et al. 1999) and DNA repair (Bird et al. 2002). Esa1 also contributes both to transcriptional activation and repression. Esa1 binding has been observed at the promoters of many transcriptionally active genes (Robert et al. 2004), and specifically at ribosomal protein genes, where Esa1 is needed for their transcriptional activation (Reid et al. 2000). In a somewhat opposing role, Esa1 is required for transcriptional silencing at the telomeres and the rDNA. Specifically, Pol II reporter genes that are normally repressed when inserted at the telomeres and the rDNA display increased expression in esa1 mutants (Clarke et al. 2006).
In this study, we defined an in vivo collaboration between the histone acetyltransferase Esa1 and the histone deacetylase Rpd3. Genetic dissection of the functional interactions revealed that the collaboration is mediated specifically through the Rpd3L complex. Deletion of RPD3 suppressed multiple phenotypes of esa1 mutants, including temperature sensitivity, rDNA and telomeric silencing defects, and restored global H4 acetylation defects. Deletion of genes encoding Rpd3L-specific subunits Pho23 or Sds3 likewise promoted suppression of esa1 phenotypes, suggesting that Esa1 coordinates acetylation specifically with Rpd3L. Consistent with this interpretation, deletion of the Rpd3S subunit encoded by RCO1 did not suppress esa1 phenotypes. Finally, suppression of the growth defect in the esa1 mutant by deletion of Rpd3L subunits was specifically dependent on acetylation of lysine 12 on histone H4, thereby pointing to a crucial yet previously unsuspected role for this specific residue.
MATERIALS AND METHODS
Yeast strains and plasmids:
All strains and plasmids are listed in supporting information, Table S1 and Table S2. Except where noted with specific allele designations, all mutations used are null alleles constructed using standard molecular methods. The rpd3∷kanMX (LPY12154), hda1∷kanMX (LPY13472), hos1∷kanMX (LPY13706), and hos2∷kanMX (LPY13583) mutants were constructed using marker swap plasmids M3926, M3929, and M3925 as described in Voth et al. (2003) on rpd3∷LEU2 (DY1539) (Kasten et al. 1997), hda1∷TRP1 (DY4891), hos1∷HIS3 (DY6073), hos2∷TRP1 (DY4549) (all generous gifts from D. Stillman) and then backcrossed prior to use. The pho23Δ∷kanMX (LPY12732), sds3Δ∷kanMX (LPY12958), and rco1Δ∷kanMX (LPY12645) mutants were constructed by amplification (oligonucleotides listed in Table S3) of the kanMX cassettes from Saccharomyces Genome Deletion Project strains, transformed into W303-1a (LPY5) and backcrossed prior to use. All double mutants and the silencing markers rDNA∷ADE2-CAN1 (Fritze et al. 1997) and TELVR∷URA3 (Renauld et al. 1993) were introduced through standard genetic crosses. Construction of the RPD3 catalytic mutant plasmid, rpd3-H150A-H151A, is described in Ruault and Pillus (2006). Histone mutant strains, derived from MSY1905 (a generous gift from M. M. Smith) (Ruault and Pillus 2006) are chromosomally deleted for both HHF-HHT loci and initially contained wild-type histones on the plasmid pJH33 (CEN URA3 HTA1 HTB1 HHF2 HHT2) (Ahn et al. 2005). For mutant construction, strains were transformed with a TRP1 plasmid containing the relevant H4 (HHF2) mutation, and then subjected to a plasmid shuffle by counterselection on 5-FOA to remove pJH33. Histone mutant plasmids were constructed by site-specific mutagenesis using oligonucleotides listed in Table S3.
Growth dilution assays and silencing assays:
Unless otherwise noted, all dilution assays represent fivefold serial dilutions, starting from an A600 of 1.0 after growth to saturation in 3 ml of liquid synthetic complete (SC) medium. Growth and silencing plates were incubated at 30°. Suppression of esa1's growth defect on SC plates was assayed at the restrictive temperature of 35°. The rDNA silencing assays were performed with strains containing the rDNA∷ADE2-CAN1 reporter as described (van Leeuwen and Gottschling 2002). Strains were grown in SC−Ade−Arg medium to saturation, normalized as above, and plated on SC−Ade−Arg (growth control) and SC−Ade−Arg plates containing 32 μg/ml of canavanine (to assay rDNA silencing). Telomeric silencing assays were performed with the TELVR∷URA3 reporter as described (Renauld et al. 1993; van Leeuwen and Gottschling 2002). Strains were grown in SC medium and plated on both SC (growth control) and SC with 0.1% 5-FOA (to assay telomeric silencing). Camptothecin (CPT) sensitivity was assayed using CPT dissolved in DMSO added to plates buffered with 100 mm potassium phosphate (pH 7.5) to maintain maximal drug activity (Nitiss and Wang 1988). Growth control plates contained equal concentrations of DMSO and phosphate buffer. All images were captured after 2–4 days of growth.
Protein immunoblots:
Whole cell extracts were prepared from cells grown to A600 of 0.6–1.0 at 30°. In the temperature-shift experiment, cells were grown first at 30° and then shifted to 34° for six hours. Extracts were prepared by bead beating as described previously (Clarke et al. 1999). Briefly, cells were resuspended in phosphate buffered saline with protease inhibitors and lysed by vortexing with glass beads. Whole cell extracts were then denatured by boiling in sample loading buffer and separated from the insoluble pellet by centrifugation. Proteins were separated on 18% SDS-polyacrylamide gels and transferred to nitrocellulose (0.2 μm). Primary antisera used were anti-H4K5Ac (1:5000 dilution, Serotec), anti-H4K8Ac (1:2500 dilution, Serotec), anti-H4K12Ac (1:2500 dilution, Serotec), anti-H4K16Ac (1:2500 dilution, Upstate), and anti-H3 CT (1:10,000 dilution, Upstate). Goat anti-rabbit conjugated to horseradish peroxidase (Promega) was used as a secondary antibody, and signal was detected with Western Lightning Chemiluminescence Reagent (Perkin Elmer) on Kodak X-Omat film.
RESULTS
Deletion of the histone deacetylase gene RPD3 suppressed the growth defect of esa1:
Histone acetylation and deacetylation are opposing chemical modifications that must be balanced for transcriptional regulation. The interplay between HATs and HDACs is complicated by the presence of numerous enzymes, some of which have very specific substrates, whereas others share overlapping histone targets. In attempting to dissect these relationships, large-scale studies have uncovered an intricate network of genetic interactions between multiple HATs and HDACs (Collins et al. 2007; Lin et al. 2008; Mitchell et al. 2008).
In a study directly examining Esa1's functions in rDNA silencing, a surprising relationship was discovered between Esa1 and the class III HDAC Sir2 (Clarke et al. 2006). When either ESA1 or SIR2 is overexpressed, each suppresses the rDNA silencing defects of the other mutant. For example, overexpression of ESA1 rescues the rDNA silencing defect of the sir2 mutant (Clarke et al. 2006).
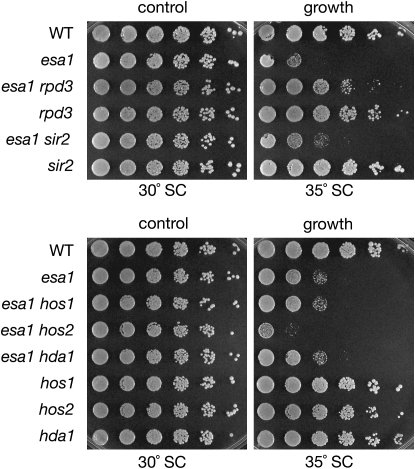
Neither increased dosage nor deletion of SIR2, however, had any effect on the growth defect of the esa1-414 temperature-sensitive mutant at elevated temperatures. In searching for other genes encoding chromatin modifying enzymes that might functionally interact with esa1 mutants, deletion of RPD3 was discovered to specifically suppress this growth defect (Figure 1).
Figure 1.—
Deletion of the histone deacetylase gene RPD3 suppresses the growth defect of esa1. Deletion of RPD3 suppressed the temperature sensitivity of the esa1-414 allele, whereas deletion of other genes encoding histone deacetylases did not. Top panel: serial dilutions of wild type (LPY5), esa1 (LPY4774), esa1 rpd3 (LPY12156), rpd3 (LPY12154), esa1 sir2 (LPY11160), and sir2 (LPY11) are shown on SC at the restrictive temperature for esa1 (35°), compared to growth at the permissive temperature (30°). Bottom panel: serial dilutions of wild type (LPY5), esa1 (LPY4774), esa1 hos1 (LPY13712), esa1 hos2 (LPY13585), esa1 hda1 (LPY13478), hos1 (LPY13706), hos2 (LPY13583), and hda1 (LPY13472) on SC at restrictive and permissive temperatures. Note that some of these interactions overlap with published results from a genomewide study (Lin et al. 2008), yet others are distinct. The differences may be due to strain background- or allele-specific effects (Y. Lin and J. Boeke, personal communication).
To ask whether deletion of genes encoding other HDACs could also support viability of the temperature-sensitive esa1-414 allele at restrictive temperatures, double mutants of esa1-414 were constructed in combination with a series of HDAC mutants. Along with the class III family deacetylase Sir2, deletion of genes encoding HDACs of classes I and II were tested. Rpd3, Hos1, Hos2, and Hda1 are all yeast HDACs of classes I and II that share 25–50% protein sequence identity. Of these, only RPD3 deletion supported growth of esa1 mutants at elevated temperatures (Figure 1). Mutation of the other genes either had no effect or in the case of hos2, exacerbated the severity of the esa1 growth defect. Some of these results are parallel to interactions reported in a genomewide study (Lin et al. 2008).
The esa1-414 temperature-sensitive mutant contains a frameshift mutation that results in an early truncation of the protein and displays reduced HAT activity both in vitro and in vivo (Clarke et al. 1999). To test the allele specificity of the suppression, RPD3 was deleted in another esa1 temperature-sensitive allele, esa1-L254P, and was also found to suppress the esa1 growth defect at restrictive temperature (Figure S1, A). The esa1-L254P allele contains a point mutation that resides near the HAT domain, and similar to esa1-414, is temperature sensitive and lacks in vitro and in vivo HAT activity (Clarke et al. 1999). Thus, rpd3 suppression of the esa1 growth defect is not allele specific and may be a general property for catalytically compromised Esa1. Furthermore, using the RPD3 catalytically dead allele, rpd3-H150A-H151A (Kadosh and Struhl 1998a) in combination with the esa1 mutant showed results consistent with the rpd3Δ (Figure S1, B). Therefore, the growth rescue observed is due to loss of histone deacetylase activity by Rpd3, and not some other function of Rpd3.
To test whether RPD3 deletion could bypass the nonviable esa1Δ phenotype, two tests were conducted. In the first, a plasmid shuffle was performed with a wild-type ESA1 plasmid in the esa1Δ rpd3Δ double mutant. This strain was unable to grow under conditions that select for loss of the wild-type ESA1 plasmid (Figure S1, C). In the second test, an esa1Δ/ESA1 rpd3Δ/rpd3Δ diploid was sporulated, dissected, and examined for viability. All genotypically esa1Δ rpd3Δ double mutants were inviable. Some double mutants germinated and were able to undergo a small number of divisions, but none continued dividing to form colonies (data not shown), similar to esa1Δ itself (Clarke et al. 1999). This analysis confirmed the plasmid shuffle result, demonstrating that rpd3 did not suppress the inviable esa1Δ.
Suppression of esa1's growth defect was mediated exclusively by the Rpd3L complex:
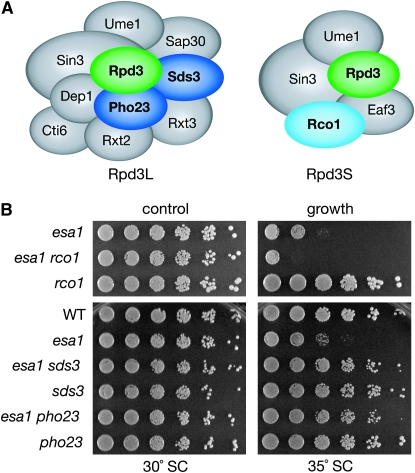
Rpd3S and Rpd3L, the two Rpd3-containing HDAC complexes, each have shared subunits as well as a number of distinct subunits (Carrozza et al. 2005a,b; Keogh et al. 2005) (Figure 2A). Both Rpd3S and Rpd3L also contain proteins that function in additional nuclear complexes. By evaluating subunits that are unique to Rpd3S and Rpd3L, the complexes were dissected genetically to determine whether deletion of both is required, or whether instead the suppression observed in the esa1 rpd3 double mutant is mediated through one specific complex. Double mutants were constructed with esa1 and genes specific to each of the two complexes. These double mutants were then tested for suppression of esa1.
Figure 2.—
Rpd3L is the Rpd3-containing complex responsible for suppression of the growth defect in the esa1 mutant. (A) Cartoon highlighting the unique and shared members of the Rpd3S and Rpd3L complexes. (B) Deletion of RCO1, specific to Rpd3S, did not suppress esa1's growth defect at restrictive temperature. Deletion of PHO23 and SDS3, both specific to Rpd3L, mimicked the suppression seen in esa1 rpd3. Serial dilutions of wild type (LPY5), esa1 (LPY4774), esa1 rco1 (LPY12652), rco1 (LPY12645), esa1 sds3 (LPY12956), sds3 (LPY12958), esa1 pho23 (LPY12729), and pho23 (LPY12732), were plated on SC at permissive (30°) and restrictive temperatures (35°). Cartoon of complexes is modified from Roguev and Krogan (2007).
Rpd3S, the smaller of the two complexes, has only two subunits (Eaf3, Rco1) that distinguish it from Rpd3L (Carrozza et al. 2005b; Keogh et al. 2005). However, Eaf3 is not unique to Rpd3S since it is also a component of NuA4 (Eisen et al. 2001), an Esa1-containing complex. Loss of EAF3 disrupts both NuA4 and Rpd3S, thus RCO1 was chosen instead to disrupt Rpd3S. The Rco1 protein contains a PHD finger and is required for the complex integrity of Rpd3S (Carrozza et al. 2005b). As shown in Figure 2B (top panel), deletion of RCO1 in an esa1 mutant did not suppress the esa1 growth defect. In fact, the esa1 rco1 double mutant displayed a slightly exacerbated growth defect compared to that of esa1. Therefore, the suppression of esa1's growth defect is not mediated through Rpd3S.
Rpd3L contains several subunits distinct from those in Rpd3S. Some are involved in the function of other transcriptional complexes, such as Cti6, which recruits the SAGA HAT complex to chromatin for transcriptional activation (Papamichos-Chronakis et al. 2002). In contrast, Sds3 is a subunit unique to Rpd3L. Sds3 is essential for the integrity of the Rpd3L complex, and Rpd3L dissociates in sds3 mutants, thereby resulting in a loss of all Rpd3L histone deacetylase activity (Lechner et al. 2000; Carrozza et al. 2005a). Deletion of SDS3 in an esa1 mutant mimicked the suppression seen in esa1 rpd3 (Figure 2B, bottom), providing evidence that suppression of esa1 is mediated through Rpd3L.
Pho23 is another Rpd3L-specific protein with a PHD finger and is one of three yeast proteins that belong to the ING tumor suppressor family (Loewith et al. 2001). In contrast to the sds3Δ mutant, the Rpd3L complex is structurally intact in pho23Δ cells and has normal levels of in vitro histone deacetylase activity (Carrozza et al. 2005a). Deletion of PHO23 in the esa1 mutant mimicked the suppression seen in both the esa1 rpd3 and esa1 sds3 double mutants (Figure 2B, bottom). This minor disruption in the Rpd3L complex is able to suppress esa1's growth defect and supports the idea that Pho23 may have a key targeting function that works in opposition to other PHD finger proteins that exist in NuA4 and piccolo.
Comparing the growth at elevated temperatures of esa1 rco1 mutants to the esa1 sds3 and esa1 pho23 strains thus demonstrates that the rescue of esa1's growth defect by deletion of RPD3 is mediated by the Rpd3L complex and not by Rpd3S. This specificity of suppression further establishes functional and not merely structural distinctions between the two Rpd3 complexes. To determine whether the specificity of suppression extended to the diverse biological roles of Esa1, a broader analysis of defective esa1 functions was evaluated.
Disruption of Rpd3L suppressed silencing phenotypes of the esa1 mutant:
Mutants of ESA1 have a wide range of phenotypes, including defects in cell cycle control, transcriptional silencing, and the DNA damage response (Clarke et al. 1999; Bird et al. 2002; Clarke et al. 2006). To determine the involvement of Rpd3L in contributing to these phenotypes, the esa1 rpd3 double mutants along with the complex-specific double mutants were examined for the integrity of these functions using in vivo assays.
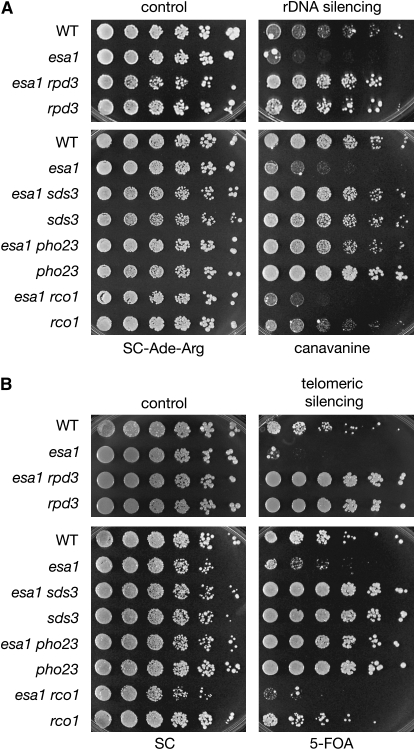
First, rDNA silencing was assayed in the esa1 rpd3 double mutant. Rpd3 has a previously reported increase in rDNA silencing (Sun and Hampsey 1999), confirmed here with the observation that rpd3 increased repression of a CAN1 reporter integrated at a single 25S rDNA locus (Figure 3A). In contrast, esa1 mutants are defective in silencing at the rDNA (Clarke et al. 2006) (Figure 3A). Deletion of RPD3 in combination with esa1 suppressed this rDNA silencing defect. Deletion of RPD3 in the esa1 mutant not only rescued the rDNA silencing defect, but increased silencing in the double mutant to that seen in an rpd3 single mutant (Figure 3A, top). This same trend was observed when the catalytic residues of RPD3 were mutated in combination with esa1 (Figure S2, A). The complex-specific double mutants were next tested for rDNA silencing. The previous observations that Rpd3L-specific mutants display increased rDNA silencing (Sun and Hampsey 1999; Loewith et al. 2001; Keogh et al. 2005) were confirmed. Consistent with the suppression of esa1's growth defect, rDNA silencing was suppressed only when Rpd3L was disrupted in esa1 and not when Rpd3S was disrupted (Figure 3A, compare sds3 and pho23 to rco1 mutants).
Figure 3.—
Disruption of Rpd3L suppresses esa1 silencing phenotypes. (A) Disruption of Rpd3L suppressed the rDNA silencing defect of an esa1 mutant. Wild type (LPY4909), esa1 (LPY4911), esa1 rpd3 (LPY12147), rpd3 (LPY12145), esa1 sds3 (LPY13517), sds3 (LPY13513), esa1 pho23 (LPY13859), pho23 (LPY13854), esa1 rco1 (LPY13505), and rco1 (LPY13501) all have the rDNA∷ADE2-CAN1 reporter to test for rDNA silencing on plates containing canavanine. Decreased growth on canavanine indicates a defect in rDNA silencing. (B) Disruption of Rpd3L suppressed the telomeric silencing defect of an esa1 mutant. Wild type (LPY4917), esa1 (LPY4919), esa1 rpd3 (LPY12211), rpd3 (LPY12093), esa1 sds3 (LPY13540), sds3 (LPY13536), esa1 pho23 (LPY13769), pho23 (LPY13765), esa1 rco1 (LPY13528), and rco1 (LPY13524) all have the TELVR∷URA3 silencing marker to test for telomeric silencing on plates containing 5-FOA. Decreased growth on 5-FOA indicates a defect in telomeric silencing.
Telomeric silencing was next assayed and revealed suppression patterns parallel to those for rDNA silencing. The esa1 mutant is defective in silencing a URA3 reporter gene integrated at telomere VR (Clarke et al. 2006) (Figure 3B). The rpd3 and Rpd3L-specific mutants displayed increased silencing (Figure 3B), confirming previous reports (Vannier et al. 1996; Zhang et al. 1998; Loewith et al. 2001; Carrozza et al. 2005a; Keogh et al. 2005). When genes encoding Rpd3L subunits were deleted in combination with esa1, they all restored telomeric silencing to esa1 mutants, whereas deletion of the Rpd3S-specific RCO1 had no effect on esa1's reduced telomeric silencing (Figure 3B and Figure S2, B). Therefore, Esa1 and Rpd3L share a critical opposing role in silencing at both the rDNA and telomeres.
Rpd3L disruption did not suppress the DNA damage phenotype of the esa1 mutant:
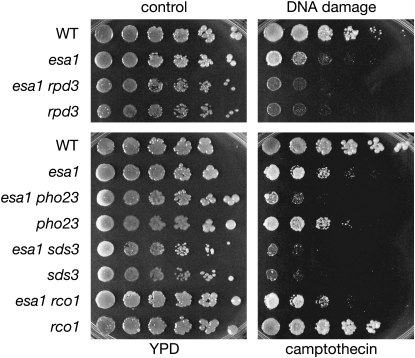
Whereas the growth and silencing phenotypes of esa1 are consistent with Esa1's transcriptional functions, Esa1 also has a role in DNA double-strand break repair. This is readily observed in that esa1 mutants are sensitive to camptothecin (Bird et al. 2002), a phenotype associated with defects in DNA repair and genome integrity. Camptothecin causes double-strand breaks by inhibiting topoisomerase I (Hsiang et al. 1985). Rpd3 also contributes to double-strand break repair, and rpd3 mutants are sensitive to phleomycin, another DNA damaging agent (Jazayeri et al. 2004).
When tested in plate assays, an rpd3 single mutant displayed increased sensitivity to camptothecin, and rpd3 did not suppress esa1's sensitivity to camptothecin (Figure 4). In fact, the esa1 rpd3 double mutant had increased sensitivity to camptothecin compared to esa1 alone. Deletion of Rpd3L- and Rpd3S-specific subunits either exacerbated or had a minimal effect on camptothecin sensitivity in the esa1 mutant (Figure 4).
Figure 4.—
Rpd3L disruption does not suppress esa1's camptothecin sensitivity. The same strains tested in Figure 1 and Figure 2B were plated on a control YPD plate containing DMSO and a plate containing 20 μg/ml of camptothecin in DMSO.
Esa1 and Rpd3 are among several chromatin modifiers that are recruited to the repair of double-strand breaks resulting from DNA damage (Downs et al. 2004; Tamburini and Tyler 2005; Lin et al. 2008). Camptothecin sensitivity in esa1 cells is thought to result from a failure of Esa1 and NuA4 recruitment to double-strand breaks. Therefore, rpd3 as a suppressor of esa1 is unlikely to involve Esa1's role in acetylation at sites of DNA damage.
In addition to the silencing and DNA damage phenotypes, rpd3 mutants have reduced mating efficiency and are cycloheximide sensitive (Vidal and Gaber 1991). To examine whether mutation of ESA1 could reciprocally suppress rpd3 phenotypes, the esa1 rpd3 and the complex-specific double mutants were examined for changes in mating efficiency and cycloheximide sensivitity. Mutation of ESA1 in rpd3 had no effect on the reduced mating efficiency of rpd3, and the same was seen for the Rpd3L-specific mutants pho23 and sds3 (Figure S3, A). The mating defect of rpd3 mutants appeared specific to Rpd3L, shown by the reduced mating of pho23 and sds3 compared to wild-type mating in rco1 (Figure S3, A). Reduced mating efficiency has been observed previously for sap30, another Rpd3L-specific mutant (Zhang et al. 1998).
To determine whether mutation of ESA1 could suppress rpd3's cycloheximide sensitivity, growth of the esa1 rpd3 double mutant was examined on cycloheximide-containing plates. No suppression was observed; in fact the esa1 mutant displayed a previously unreported modest cycloheximide resistance (Figure S3, B). Together, rpd3 in the context of Rpd3L can suppress many but not all esa1 mutant phenotypes. The nature of this functional interaction is not however reciprocal because esa1 mutants do not suppress the rpd3 defects tested.
Deletion of RPD3 restored global histone acetylation levels of shared target residues in the esa1 mutant:
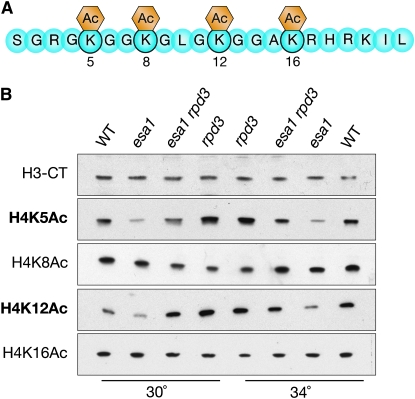
To test whether the genetic relationship discovered between esa1 and rpd3 was observed at the molecular level of histone modification, global histone acetylation was evaluated using isoform-specific antisera for lysines targeted by Esa1 and Rpd3. The enzymatic activities of Esa1 and Rpd3 both target specific lysines on the N-terminal tail of histone H4 (Figure 5A). Esa1 has global effects in vivo on H4K5 acetylation (Clarke et al. 1999) and also acetylates multiple lysines on H4 at sites within the rDNA (Clarke et al. 2006) and at specific gene promoters (Suka et al. 2001). Rpd3 globally deacetylates H4K5 and H4K12 (Rundlett et al. 1996) and is responsible for deacetylation of most histone tail lysine residues at specific gene promoters (Suka et al. 2001).
Figure 5.—
Deletion of RPD3 restores global acetylation levels of specific histone H4 residues in esa1 mutants. (A) Diagram of the histone H4 N-terminal tail highlighting sites of acetylation modifications. (B) Deletion of RPD3 restored global acetylation of H4K5 and H4K12, but not H4K8 and H4K16. Whole cell protein extracts from wild-type (LPY5), esa1 (LPY4774), esa1 rpd3 (LPY12156), and rpd3 (LPY12154) cells at both permissive (30°) and restrictive (34°) temperatures were immunoblotted with an antiserum specific to the C terminus of H3 to control for histone levels, and with H4 antisera to detect the amount of bulk histone acetylation at each lysine residue.
Lysates were collected from cells grown at permissive and slightly elevated temperatures that maintained viability. Immunoblots were performed on these lysates and probed with antisera specific for each acetylated lysine. As a control, histone levels were assayed and found comparable among all strains, as shown by probing for the C terminus of H3 (Figure 5B, top panel). As expected, esa1 mutants displayed decreased bulk histone acetylation, most dramatic for H4K5 (Clarke et al. 1999) and H4K12, and rpd3 mutants had slightly increased acetylation of H4K5 and H4K12 compared to wild type, consistent with earlier reports (Rundlett et al. 1996) (Figure 5B). In the noncatalytic mutants (rco1, sds3, and pho23), global histone acetylation changes were not observed (Figure S4). This result might be expected since an Rpd3 complex is still present in each of these mutants. Thus, it was not surprising that there were only very subtle changes in acetylation in these mutants when combined with esa1 (Figure S4). Additionally, at two H4 lysines that are not shared targets of Esa1 and Rpd3, H4K8 and H4K16, acetylation was not changed in the esa1 rpd3 double mutant.
Finally, in the esa1 rpd3 double mutant, there was almost a complete restoration of the esa1 global acetylation defect at both permissive and elevated temperatures that was strongest for H4K12 (Figure 5B). There was also an intermediate effect on acetylation at H4K5. Together, these results provide a molecular basis for the growth defect and silencing suppression observed in the esa1 rpd3 double mutant (Figures 1 and 3).
Suppression of esa1's growth defect by deletion of RPD3 is mediated through H4K12 acetylation:
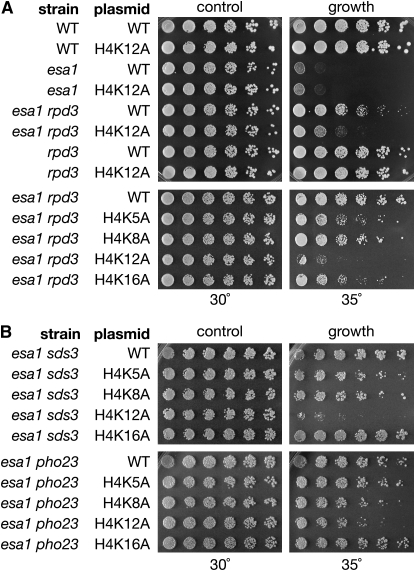
Because the most dramatic change in global histone acetylation in esa1 rpd3 was at H4K12 (Figure 5B), it seemed likely that this particular residue was most critical for the functional interaction between the two enzymes. To evaluate the possibility, mutants were constructed in which each target lysine was changed to alanine, an amino acid residue that cannot be acetylated. The ability of rpd3 to suppress esa1's growth defect was then tested with each histone lysine mutant. In wild-type cells, the H4K12A mutant by itself did not display any growth defects, nor did it affect growth in the esa1 or rpd3 single mutant. However, H4K12A in combination with the esa1 rpd3 double mutant displayed a dramatic reduction in growth at elevated temperature compared to the esa1 rpd3 double mutant (Figure 6A, top). The other H4 lysine mutants (H4K5A, H4K8A, and H4K16A) had minimal effects on the growth of the esa1 rpd3 double mutant (Figure 6A, bottom). The dependence on H4K12 was also observed in the esa1 rpd3-H150A-H151A catalytic mutant (Figure S5). Therefore, the suppression observed in the esa1 rpd3 double mutant is specifically dependent on H4K12 and not H4K5, K8, or K16.
Figure 6.—
Suppression of esa1's growth defect by rpd3 is dependent on H4K12. Strains are deleted for all copies of H3 and H4 and carry a TRP1 plasmid with either wild-type H4 or H4 with one mutated lysine residue. Plasmid retention was required for survival. (A) Serial dilutions of the following strains were plated at permissive (30°) and restrictive temperatures (35°) on SC. Top panel: growth of H4K12A mutants in combination with esa1 rpd3. Strains are wild type (LPY12383), H4K12A (LPY12394), esa1 (LPY12384), esa1 H4K12A (LPY12071), esa1 rpd3 (LPY12707), esa1 rpd3 H4K12A (LPY12714), rpd3 (LPY12695), and rpd3 H4K12A (LPY12702). Bottom panel: growth of esa1 rpd3 mutants in combination with each lysine individually mutated to alanine. Strains are esa1 rpd3 (LPY12707), esa1 rpd3 H4K5A (LPY12708), esa1 rpd3 H4K8A (LPY12711), esa1 rpd3 H4K12A (LPY12714), and esa1 rpd3 H4K16A (LPY12717). (B) Suppression of esa1's growth defect through deletion of noncatalytic Rpd3L subunits was also dependent on H4K12. Top panel: twofold dilutions, starting at an A600 of 0.1, were plated on SC−Trp for assaying growth of esa1 sds3 in combination with each lysine individually mutated to alanine. Strains are esa1 sds3 (LPY14175), esa1 sds3 H4K5A (LPY14176), esa1 sds3 H4K8A (LPY14177), esa1 sds3 H4K12A (LPY14178), and esa1 sds3 H4K16A (LPY14179). Bottom panel: as above except in esa1 pho23 mutant. Strains are esa1 pho23 (LPY14165), esa1 pho23 H4K5A (LPY14166), esa1 pho23 H4K8A (LPY14167), esa1 pho23 H4K12A (LPY14168), and esa1 pho23 H4K16A (LPY14169).
H4K12 was also found to be the key acetylated lysine in suppression of esa1 by disruption of the Rpd3L complex. As shown in Figure 6B, when each lysine was individually mutated in the esa1 sds3 and esa1 pho23 double mutants, only the H4K12A substitution resulted in a loss of suppression, albeit to a more modest degree than in esa1 rpd3. Although the dependence on H4K12 appears subtle in the esa1 pho23 double mutant, this slight effect was observed reproducibly. Notably, in the protein immunoblots H4K5 showed a moderate acetylation increase in the esa1 rpd3 double mutant compared to esa1 (Figure 5B), yet the H4K5A mutant had little impact on the growth of esa1 rpd3, esa1 sds3, or esa1 pho23 (Figure 6).
Thus H4K5 and H4K12 are common targets of global acetylation and deacetylation by both Esa1 and Rpd3. However, the distinction observed here between the growth of esa1 rpd3 in H4K5A vs. H4K12A mutants points to H4K12 as the critical shared target of Esa1 and the Rpd3L complex for regulating growth and viability (Figure 7A).
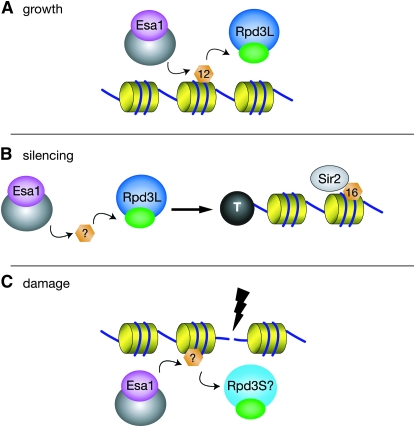
Figure 7.—
A model depicting a critical role for Esa1 and Rpd3L in coordinating the dynamic acetylation of H4K12. (A) Esa1 and Rpd3L control H4K12Ac for general transcriptional targets contributing to cell viability and growth. (B) Esa1 and Rpd3L contribute to rDNA and telomeric silencing. This relationship is not mediated specifically through H4K12 acetylation, but likely through a number of other targets. Sir2 deacetylation of H4K16 appears downstream of the role for Esa1 and Rpd3L. (C) Esa1 and Rpd3S, but not Rpd3L, may control acetylation at sites of DNA damage.
DISCUSSION
The findings presented here tightly link Esa1's acetyltransferase activity and Rpd3's deacetylase activity in critical cellular processes. Loss of Rpd3L specifically alleviated many of the cell's needs for fully functional Esa1 activity, a property not shared by the Rpd3S complex, nor by other class I–III deacetylases (summarized in Table S4). This exclusive relationship between Esa1 and Rpd3L centers on their shared histone target H4K12. In addition, Esa1 works specifically with the Rpd3L complex in maintaining silencing at the rDNA and telomeres, but not in repairing camptothecin-induced double-strand breaks (Figure 7, Table S4).
Esa1 and Rpd3 have both previously been shown to be required for rDNA and telomeric silencing. Esa1 is enriched at the rDNA by chromatin immunopreceiptation and Esa1-dependent changes in H4 acetylation are seen at the rDNA (Clarke et al. 2006). Unlike its role in growth, the rpd3-mediated suppression of esa1's rDNA and telomeric silencing defects was not dependent on H4K12 acetylation (Figure S6). Hence suppression at these loci is mechanistically distinct. Rpd3L's role in rDNA and telomeric silencing involves boundary formation (Zhou et al. 2009) and is dependent on the histone deacetylase Sir2 that targets H4K16 (Sun and Hampsey 1999; Raisner and Madhani 2008). Therefore the observed dependence on H4K16 acetylation (Figure S6) was not surprising. This dependence on Sir2 and H4K16 deacetylation has led to the idea that Rpd3 has an indirect effect in silencing, possibly through altering Sir2 activity (Sun and Hampsey 1999) or the expression of other genes involved in silencing. Because esa1's rDNA and telomeric silencing defects were suppressed by disruption of Rpd3L (Figure 3), and were dependent on H4K16 acetylation (Figure S6), it is likely that Esa1 and Rpd3L's role in silencing is upstream of Sir2 (Figure 7B).
It is becoming evident that histone modifying enzymes also target many nonhistone substrates (reviewed in Sterner and Berger 2000). Indeed, recent data indicate that such nonhistone substrates exist for NuA4 (Lin et al. 2009), including Yng2, which is also a substrate of Rpd3 (Lin et al. 2008). Further studies should provide additional insight into the range and roles of nonhistone substrates in Esa1 and Rpd3 functions, perhaps revealing a more direct link for their influence on rDNA and telomeric silencing. In this case however, we have shown that H4K12 is a key shared target for the contributions of Esa1 and Rpd3 to cell growth and viability.
A critical role for dynamic acetylation and deacetylation of H4K12 by Esa1 and Rpd3L:
The cell contains numerous HATs and HDACs that together acetylate many lysine residues on histones. The intricacies of histone acetylation and deacetylation result from several features: each HAT and HDAC often targets multiple lysine residues, different HATs and HDACs have overlapping acetylation targets, and other post-translational modifications may influence activity or substrate recognition. For example, a simple mutation of H4K12 did not suppress esa1 defects (Figure 6A), even though it is a key target of Esa1. This is because Esa1 has many other histone targets, including other lysines on H4, H2A, and H2A.Z, and lack of acetylation of these also contributes to esa1's growth defect.
Defining roles for specific histone acetylation sites is further complicated by the genomewide data that acetylation of H4K5, H4K8, and H4K12 are redundant in transcription (Dion et al. 2005). One point in support of this idea is that the H4K12A single mutant displayed no obvious growth defects (Figure 6A). A previous study defined the H3 and H4 N termini as the functional targets of Rpd3 in regulation of transcription (Sabet et al. 2004). The connections between Esa1, Rpd3L, and H4K12 presented here strengthen this functional importance through further identification of a key specific lysine (H4K12) in the H4 N terminus, and the acetyltransferase responsible (Esa1). Since in the absence of Esa1 and Rpd3L, H4K12 acetylation became particularly important for cell viability (Figure 6), these specific links define a model whereby control of H4K12 acetylation is essential for transcriptional regulation of a subset of genes by Esa1 and Rpd3L for cell viability (Figure 7A).
Among the several genomewide ChIP data sets that define Esa1 and Rpd3 binding (Reid et al. 2000; Kurdistani et al. 2002; Robert et al. 2004), little overlap has been observed between regions strongly enriched for Esa1 and those enriched for Rpd3. This may be due to the fact that Esa1 and Rpd3 both exist in multiple complexes in the cell, creating noise in the data sets. Esa1-bound loci would include both NuA4 and piccolo, thereby conflating their occupancy sites. Likewise, genomewide ChIP that has been performed does not allow discrimination between sites of Rpd3L vs. Rpd3S occupancy. When analyzed at specific loci, Rpd3S functions at downstream regions (Carrozza et al. 2005b; Keogh et al. 2005), thus it is likely that the genomewide binding of Rpd3 found at downstream regions can be attributed to Rpd3S and binding at promoters can be attributed to Rpd3L. However, because the genomic binding maps were generated with nontiling arrays, resolving the differences in Rpd3L and Rpd3S binding with available data sets is not possible. The differences in function between Rpd3L and Rpd3S in relation to Esa1 provide a new tool for refining understanding of the two complexes.
Distinguishing complexes and their functional interactions:
Esa1 and Rpd3 each act as the catalytic subunit of two multiprotein histone modifying complexes. The two Rpd3 complexes, Rpd3S and Rpd3L, are composed of distinct subunits that allow them to be genetically dissected. Several recent articles have examined different roles for Rpd3S and Rpd3L (Carrozza et al. 2005a,b; Keogh et al. 2005; Biswas et al. 2008; Knott et al. 2009). We have established that disruption of Rpd3L function is specifically responsible for the genetic suppression of esa1 mutants for cell viability and silencing phenotypes (Figures 2 and 3).
Two different roles for histone modifying enzymes in the DNA damage response have been uncovered. One role is to participate in the transcriptional response through the activation of DNA repair genes. For example, Esa1 and Rpd3 are both required for transcriptional activation of the damage-inducible genes HUG1 and RNR3 (Sharma et al. 2007). The identification of these as shared targets for activation raises a useful distinction because rpd3 cannot suppress esa1's sensitivity to DNA damage. It seems likely therefore that Esa1 and Rpd3 target genes relevant to changes in H4K12 acetylation are those that Esa1 activates and Rpd3 represses. Identification of these genes should prove of great interest.
The other function for histone modifying enzymes in DNA repair is more direct: chromatin modification targeted to the site of DNA damage. Along with several other HATs and HDACs, Esa1, some members of NuA4, and Rpd3 itself all bind at double-strand breaks, followed by changes in acetylation of nearby chromatin (Bird et al. 2002; Downs et al. 2004; Tamburini and Tyler 2005; Lin et al. 2008). Our observations showing that deletion of Rpd3L-specific subunits does not suppress repair defects of esa1 mutants make it unlikely that Rpd3L functions together with Esa1 at sites of DNA damage. However, since Rpd3 is present at double-strand breaks and is required for nonhomologous end joining (Jazayeri et al. 2004; Tamburini and Tyler 2005), perhaps Rpd3S and Esa1 coordinate acetylation at sites of DNA damage (Figure 7C).
By constructing specific double deletion mutants, it was possible to refine understanding of Rpd3S and Rpd3L functions beyond earlier reports. In contrast, Esa1 exists in the NuA4 and piccolo complexes, yet because piccolo is a subcomplex of NuA4, it has not yet been possible to disrupt piccolo without also disrupting NuA4 function. Therefore, it remains to be determined whether rpd3 suppression of esa1 is mediated through NuA4 or piccolo.
Future studies should provide additional insight into distinctions between NuA4 and piccolo that may allow this question to be answered. One idea comes from studies examining the chromatin modifying complexes SLIK/SALSA and SAGA. These complexes share most subunits, including the histone acetyltransferase Gcn5. It was found that the shared subunit Spt7 exists in a C-terminally truncated form in the smaller SLIK/SALSA complex, allowing for construction of specific SPT7 alleles that favor a specific complex (Sterner et al. 2002; Wu and Winston 2002). Analogous to this shift between SLIK/SALSA and SAGA, the discovery of specific alleles of piccolo components that favor activity of one complex over another may allow for future dissection of Esa1's interactions with Rpd3.
One possibility is that NuA4 and Rpd3S, which share the chomodomain protein Eaf3, work together, whereas piccolo and Rpd3L are also a functional pair. Some data supporting this idea can be extrapolated from genomewide studies. For example, double mutants of RPD3 and genes encoding NuA4-specific subunits show reduced fitness, whereas a double deletion mutant of RPD3 and EPL1, which is in both piccolo and NuA4, shows synthetic rescue (Lin et al. 2008). However, because deletion of EPL1 disrupts both piccolo and NuA4, it is difficult to make a clear distinction between the two.
The composition of NuA4, piccolo, Rpd3L, and Rpd3S is evolutionarily conserved (reviewed in Doyon and Côté 2004; Yang and Seto 2008). One particular class of proteins in both is the PHD finger-containing ING family of tumor suppressors. Yng2 is a yeast ING protein that is a subunit of both piccolo and NuA4 (Loewith et al. 2000), whereas Pho23 is another yeast ING protein that is a subunit of Rpd3L (Loewith et al. 2001; Carrozza et al. 2005a). The precise functions of Yng2 and Pho23 in their complexes are unknown, but analogous to the opposing activities between Esa1 and Rpd3, Yng2 and Pho23 have opposite effects on p53-dependent transcriptional activation, shown in an experiment where human p53 was expressed in yeast to drive transcription (Nourani et al. 2003). This opposing effect on activity of a human protein in transcription emphasizes the conserved nature underlying the partnership between the Esa1 and Rpd3 complexes reported here. In addition, the identification of H4K12 as a critical shared acetylation target uncovers the importance of the dynamic acetylation and deacetylation of a particular histone residue in the context of Esa1 and Rpd3L function.
Dynamic and reciprocal histone modifications are increasingly recognized as key regulatory switches. This principle was highlighted in a recent study investigating histone ubiquitination in metazoan development. Coordinate control of H2B ubiquitination in Drosophila by the ubiquitin ligase dBRE1 and the ubiquitin protease Scrawny was found to be essential for regulating gene silencing to promote cellular differentiation (Buszczak et al. 2009). Our studies identify links between Esa1 and Rpd3L specifically in the acetylation and deacetylation of H4K12. Further, they reveal a critical distinct characteristic of the Rpd3L complex in relation to Esa1, and identify roles for specific histone residues in promoting cell viability. Future functional dissection of Rpd3 and Esa1 multiprotein complexes will deepen understanding of how such chromatin modifiers control important and diverse cellular processes.
Acknowledgments
We thank E. M. Scott, B. Wood, S. Torigoe, M. Ruault, D. Stillman, M. Smith, and M. Grunstein for providing strains and reagents. We also thank R. M. Garza, S. J. Jacobson, M. R. Koch, M. Ruault, and E. M. Scott for critical reading of the manuscript, and members of the Pillus lab for advice throughout the course of this study. This work was supported by the National Institutes of Health and the University of California Cancer Research Coordinating Committee.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103846/DC1.
References
- Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith et al., 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. [DOI] [PubMed] [Google Scholar]
- Alejandro-Osorio, A. L., D. J. Huebert, D. T. Porcaro, M. E. Sonntag, S. Nillasithanukroh et al., 2009. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 10: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant et al., 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, J. G., C. J. Viggiani, D. G. Gibson and O. M. Aparicio, 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov, N., and J. Côté, 2007. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26: 5395–5407. [DOI] [PubMed] [Google Scholar]
- Babiarz, J. E., J. E. Halley and J. Rine, 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., J. K. Tong and S. L. Schreiber, 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97: 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Biswas, D., S. Takahata and D. J. Stillman, 2008. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S). Mol. Cell. Biol. 28: 4445–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley et al., 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17: 1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak, M., S. Paterno and A. C. Spradling, 2009. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 323: 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza, M. J., L. Florens, S. K. Swanson, W. J. Shia, S. Anderson et al., 2005. a Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731: 77–87, discussion 75–76. [DOI] [PubMed] [Google Scholar]
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson et al., 2005. b Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Choy, J. S., and S. J. Kron, 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22: 8215–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. S., J. E. Lowell, S. J. Jacobson and L. Pillus, 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. S., E. Samal and L. Pillus, 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 17: 1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- Dang, W., K. K. Steffen, R. Perry, J. A. Dorsey, F. B. Johnson et al., 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadal, E., M. Zapater, P. M. Alepuz, L. Sumoy, G. Mas et al., 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374. [DOI] [PubMed] [Google Scholar]
- De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter et al., 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384: 589–591. [DOI] [PubMed] [Google Scholar]
- Decker, P. V., D. Y. Yu, M. Iizuka, Q. Qiu and M. M. Smith, 2008. Catalytic-site mutations in the MYST family histone acetyltransferase Esa1. Genetics 178: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M. F., S. J. Altschuler, L. F. Wu and O. J. Rando, 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102: 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger et al., 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16: 979–990. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., and J. Côté, 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt et al., 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276: 3484–3491. [DOI] [PubMed] [Google Scholar]
- Fritze, C. E., K. Verschueren, R. Strich and R. Easton Esposito, 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang, Y. H., R. Hertzberg, S. Hecht and L. F. Liu, 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260: 14873–14878. [PubMed] [Google Scholar]
- Jazayeri, A., A. D. McAinsh and S. P. Jackson, 2004. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101: 1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1998. a Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1998. b Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18: 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten, M. M., S. Dorland and D. J. Stillman, 1997. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol. 17: 4852–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny et al., 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan et al., 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20: 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377. [DOI] [PubMed] [Google Scholar]
- Knott, S. R., C. J. Viggiani, S. Tavare and O. M. Aparicio, 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev. 23: 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani, S. K., D. Robyr, S. Tavazoie and M. Grunstein, 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31: 248–254. [DOI] [PubMed] [Google Scholar]
- Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter et al., 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 275: 40961–40966. [DOI] [PubMed] [Google Scholar]
- Lin, Y. Y., J. Y. Lu, J. Zhang, W. Walter, W. Dang et al., 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136: 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. Y., Y. Qi, J. Y. Lu, X. Pan, D. S. Yuan et al., 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22: 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol and D. Young, 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20: 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman et al., 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276: 24068–24074. [DOI] [PubMed] [Google Scholar]
- Megee, P. C., B. A. Morgan and M. M. Smith, 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Millar, C. B., F. Xu, K. Zhang and M. Grunstein, 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, L., J. P. Lambert, M. Gerdes, A. S. Al-Madhoun, I. S. Skerjanc et al., 2008. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 28: 2244–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss, J., and J. C. Wang, 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA 85: 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani, A., L. Howe, M. G. Pray-Grant, J. L. Workman, P. A. Grant et al., 2003. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J. Biol. Chem. 278: 19171–19175. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou and D. Tzamarias, 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9: 1297–1305. [DOI] [PubMed] [Google Scholar]
- Raisner, R. M., and H. D. Madhani, 2008. Genomewide screen for negative regulators of sirtuin activity in Saccharomyces cerevisiae reveals 40 loci and links to metabolism. Genetics 179: 1933–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, J. L., V. R. Iyer, P. O. Brown and K. Struhl, 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani et al., 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy et al., 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev, A., and N. J. Krogan, 2007. SIN-fully silent: HDAC complexes in fission yeast. Nat. Struct. Mol. Biol. 14: 358–359. [DOI] [PubMed] [Google Scholar]
- Ruault, M., and L. Pillus, 2006. Chromatin-modifiying enzymes are essential when the Saccharomyces cerevisiae morphogenesis checkpoint is constitutively activated. Genetics 174: 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner et al., 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner and M. Grunstein, 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835. [DOI] [PubMed] [Google Scholar]
- Sabet, N., S. Volo, C. Yu, J. P. Madigan and R. H. Morse, 2004. Genome-wide analysis of the relationship between transcriptional regulation by Rpd3p and the histone H3 and H4 amino termini in budding yeast. Mol. Cell. Biol. 24: 8823–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertil, O., A. Vemula, S. L. Salmon, R. H. Morse and C. V. Lowry, 2007. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol. Cell. Biol. 27: 2037–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V. M., R. S. Tomar, A. E. Dempsey and J. C. Reese, 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 27: 3199–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti et al., 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95: 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., R. Belotserkovskaya and S. L. Berger, 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99: 11622–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., and S. L. Berger, 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64: 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383. [DOI] [PubMed] [Google Scholar]
- Suka, N., Y. Suka, A. A. Carmen, J. Wu and M. Grunstein, 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8: 473–479. [DOI] [PubMed] [Google Scholar]
- Sun, Z. W., and M. Hampsey, 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152: 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini, B. A., and J. K. Tyler, 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25: 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, F., and D. E. Gottschling, 2002. Assays for gene silencing in yeast. Methods Enzymol. 350: 165–186. [DOI] [PubMed] [Google Scholar]
- Vannier, D., D. Balderes and D. Shore, 1996. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 144: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, M., and R. F. Gaber, 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer and M. Grunstein, 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10: 1223–1233. [DOI] [PubMed] [Google Scholar]
- Voth, W. P., Y. W. Jiang and D. J. Stillman, 2003. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20: 985–993. [DOI] [PubMed] [Google Scholar]
- Wu, P. Y., and F. Winston, 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22: 5367–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. J., and E. Seto, 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9: 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst et al., 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1: 1021–1031. [DOI] [PubMed] [Google Scholar]
- Zhou, J., B. O. Zhou, B. A. Lenzmeier and J. Q. Zhou, 2009. Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation. Nucleic Acids Res. 37: 3699–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]