Abstract
It is well established that higher eukaryotes use alternative splicing to increase proteome complexity. In contrast, Saccharomyces cerevisiae, a single-cell eukaryote, conducts predominantly regulated splicing through retention of nonfunctional introns. In this article we describe our discovery of a functional intron in the PTC7 (YHR076W) gene that can be alternatively spliced to create two mRNAs that code for distinct proteins. These two proteins localize to different cellular compartments and have distinct cellular roles. The protein translated from the spliced mRNA localizes to the mitochondria and its expression is carbon-source dependent. In comparison, the protein translated from the unspliced mRNA contains a transmembrane domain, localizes to the nuclear envelope, and mediates the toxic effects of Latrunculin A exposure. In conclusion, we identified a definitive example of functional alternative splicing in S. cerevisiae that confers a measurable fitness benefit.
IN higher eukaryotes alternative splicing is pervasive; in humans the majority of genes are alternatively spliced to form multiple proteins (Modrek et al. 2001; Johnson et al. 2003). In contrast, only 5% of the genes in Saccharomyces cerevisiae contain introns and >95% of those intron-containing genes possess only a single intron (Nash et al. 2007). The simple architecture of the yeast genome constrains its ability to utilize alternative splicing and has encouraged the view that alternative splicing is absent in S. cerevisiae. Currently no conclusive examples of functional alternative splicing exist; most confirmed instances of alternative splicing in yeast downregulate gene expression, a process that is often referred to as “regulated splicing.” In this process, nonfunctional introns are not spliced out of the transcript and premature stop codons are included in the fully processed mRNA. The stop codons activate the nonsense-mediated decay (NMD) pathway and the mRNA is degraded before it can be translated (Gonzalez et al. 2001).
The transition from vegetative growth to meiosis illustrates how regulated splicing improves yeast fitness. DNA breakage and recombination could be toxic during vegetative growth, and therefore entrance into meiosis is tightly controlled. Consequently, all 13 meiosis-specific intron-containing genes are regulated post-transcriptionally with splicing repressed during vegetative growth and induced during sporulation (Engebrecht et al. 1991; Nakagawa and Ogawa 1999; Juneau et al. 2007). Other examples of regulated splicing include YRA1 and MTR2 (RNA export) (Preker et al. 2002; Parenteau et al. 2008) and RPL30 (ribosomal) (Li et al. 1996).
In contrast to regulated splicing, there is only one example in S. cerevisiae where splicing results in the expression of multiple proteins, SRC1 (Grund et al. 2008). SRC1 contains a single intron located at the 3′ end of the pre-mRNA. The intron has two alternative 5′-splice sites (Rodriguez-Navarro et al. 2002). Splicing at the most highly conserved 5′-splice site (Bon et al. 2003) results in the translation of a full-length protein that spans the nuclear membrane twice, while splicing at the less conserved 5′-splice site results in a truncated protein that spans the nuclear membrane only once and has significantly reduced activity (Grund et al. 2008). It has been proposed that truncated SRC1 may provide a unique cellular function, but current data do not conclusively support this hypothesis. However, we now have convincing evidence that the serine/threonine phosphatase PTC7 is alternatively spliced to produce two different proteins with distinct cellular functions.
Previously, we developed a high-resolution array-based genomic assay to find novel introns in yeast (Juneau et al. 2007). One of the introns we discovered is in PTC7, a conserved type 2C protein phosphatase known to localize to the mitochondria and predicted to contain a transmembrane domain (Ramos et al. 2000; Jiang et al. 2002). The intron is notable because it is divisible by three and lacks stop codons (Juneau et al. 2007; Zhang et al. 2007); consequently, neither splicing nor retention of the intron would be predicted to disrupt translation of PTC7.
We now show that PTC7 is alternatively spliced into two mRNA isoforms, which express two functionally distinct proteins that localize to different compartments within S. cerevisiae. One isoform of PTC7 is created by the removal of the intron while the other isoform results from intron retention. Comparative sequence analysis of PTC7 across several hemiascomycetous yeast species showed that splicing and translation of the intron is possible in all 12 intron-containing PTC7 orthologs. The protein coded by the spliced mRNA (Ptc7s) localizes to the mitochondria, while the protein that is translated from unspliced mRNA (Ptc7u) localizes to the nuclear envelope. We also found that Ptc7s expression and modification are regulated by carbon metabolism whereas Ptc7u effectively mediates the deleterious effects of Latrunculin A, a compound that disrupts actin filaments. Together these data provide compelling evidence that S. cerevisiae performs functional alternative splicing.
MATERIALS AND METHODS
Media and growth conditions:
Yeast extract/peptone/dextrose (YPD) media and 30° growth conditions we used for most experiments are described in Guthrie and Fink (1991). We used YP–raffinose and YP–galactose for microscopy experiments requiring GAL1 promoter induction as described in Guthrie and Fink (1991). We also used YP–glycerol (YPG), YP–lactose (YPL), YP–ethanol (YPE), and YP without additional carbon (YPØ). The concentration of the various carbon sources, when present, was 2% (w/v) except in the case of YPE, which had 1.5% ethanol (v/v). Latrunculin A was obtained from Sigma–Aldrich (St. Louis) (catalog no. L5163). Concentrated stocks were dissolved in DMSO and stored at −20°.
Strains and plasmids:
For construction of PTC7 epitope-tagged and untagged splicing mutants we used the isogeneic S288c strains BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0), and BY4743 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0; 4741/4742) (Open Biosystems, Huntsville, AL).
We used the kanMX4-URA3 module of the pCORE plasmid (Storici et al. 2001) (provided by M. Resnick, National Institutes of Health) to create the intron-plus and -minus ptc7 strains. We used the pFA6a-3HA-kanMX6 plasmid (Longtine et al. 1998) to add three tandem hemagglutinin (3HA) (YPYDVPDYA) epitopes to the carboxyl termini of the Ptc7 protein constructs used for Western analysis. We used plasmids pFA6a-His3MX6-PGAL1, pFA6a-GFP(S65T)-kanMX6, pBS34, and pBS7 to create GAL-driven fluorescent Ptc7 constructs labeled with green fluorescent protein (GFP) (Heim and Tsien 1996; Longtine et al. 1998), mCherry (Shaner et al. 2004), or Venus (Nagai et al. 2002) on the carboxyl terminus for microscopy and Western analysis of promoter-specific splicing. The plasmids pBS34 and pBS7 were obtained from the Yeast Resource Center at the University of Washington with permission from Roger Y. Tsien at the University of California, San Diego.
Strain construction:
All ptc7 strains altered to contain additional tags (HA, GFP, mCherry, or Venus) or galactose-driven promoters were constructed as described in Longtine et al. (1998).
Marker-free intron-minus and -plus strains were constructed as described in Juneau et al. (2006), using the plasmids and methodology described in Storici et al. (2001). The intron-minus ptc7 strain contained a complete deletion of the intron. The intron-plus strain was constructed such that the wild-type intron was replaced with an intronic sequence that maintained the protein-coding sequence, but was incapable of splicing because the splicing signals were deleteriously altered. The sequence of the intron within the intron-plus ptc7 strain is as follows (mutations are in lowercase, and 5′-, 3′-, and all potential branch-point splice signals are underlined): GaATGTTTAT TGTTTTATTC ATTGGTGTct TgATTGCTAA ATTTGCGGGA CAAATGtTgA TCGACTCAGA aACcAAtTTT TCTCATATCA TtG.
RNA and cDNA sample preparation:
RNA was extracted with hot phenol from log-phase yeast growing in YPD as previously described (Schmitt et al. 1990). Total RNA was treated for 10 min at 37° with RNase-free DNase I (GE Healthcare Life Sciences, catalog no. 27-0514-01). RNA was simultaneously repurified and DNased a second time, using QIAGEN's RNase-Free DNase Set (QIAGEN, Valencia, CA; catalog no. 79254) and RNeasy Mini Kit (QIAGEN, catalog no. 74104). Single-stranded cDNA was synthesized as previously described (Juneau et al. 2007).
Western blot analysis:
Whole-cell lysate was isolated as previously described (Neklesa and Davis 2008). Lysates were normalized to 5 mg/ml protein, determined using a Bradford Assay (Bio-Rad, Hercules, CA; catalog no. 500-0006). Lysate was mixed 3:1 with SB Loading buffer (Invitrogen, Carlsbad, CA; catalog no. NP0007) and incubated for 10 min at 70° before running on an XCell SureLock Mini-Cell gel box (Invitrogen, catalog no. EI0002), at 200 V, using SDS 10% acrylamide gels (Invitrogen catalog no. NP0301) and MOPs running buffer (Invitrogen, catalog no. NP0001). Size-separated protein was transferred to nitrocellulose (Invitrogen, catalog no. LC2001) at 24 V for 1.5 hr. Protein normalization was confirmed by Ponceau staining of the nitrocellulose membrane. Briefly, membranes were stained for 5 min with 0.1% (w/v) Ponceau S, 2% (v/v) glacial acetic acid in water before being destained in water (3–10 min). Western blots were blocked for >1 hr in 5% (w/v) milk solubilized in 150 mm NaCl, 2.5 mm Tris Base (pH 7.4), 0.05% Tween (TBS-T) buffer. The blots were stained either with 2.5 μg of rabbit anti-HA antibody (Bethyl, catalog no. A190-208A) or 20 μg rabbit polyclonal antibody to GFP (Abcam, catalog no. ab290) for 1 hr in 20 ml TBS-T; blots were then washed three times for 5 min in TBS-T and stained with 5 μl of HRP-conjugated anti-rabbit antibody (Cell Signaling Technology, catalog no. 7074) in 15 ml TBS-T and washed three more times for 10 min before ECL visualization (GE Healthcare, catalog no. RPN2109).
Phenotypic growth analysis:
Phenotypic analysis was carried out in Tecan (Zurich, Switzerland) GENios microplate readers as previously described (Juneau et al. 2006). Yeast were grown in YPD with or without 1.3 μm Latrunculin A (Sigma–Aldrich, catalog no. L5163). Growth rates were determined by comparing average doubling times as previously described (Lee et al. 2005).
Fluorescent microscopy protocols:
Yeast strains containing fluorescently tagged Ptc7 under the control of the GAL1 promoter were grown overnight in YP–Raffinose. Cells were diluted to a concentration of 0.15 OD in YP–galactose and grown to a final concentration of 0.8–1.0 OD. Cells were fixed in 1.23% formaldehyde before staining with DAPI (Sigma, catalog no. D9542-1MG) and phalloidin (Invitrogen, Alexa Fluor 488 phalloidin catalog no. A12379). Cells were not fixed when stained with MitoTracker Red CMXRos (Invitrogen, catalog no. M7512).
DAPI staining:
Cells were washed three times with TBS-T before being stained with DAPI at a concentration of 100 ng/ml in TBS-T for 1 hr at room temperature in a roller. After staining with DAPI, cells were washed once in TBS-T, vortexed for 1 hr in TBS-T, and washed again in TBS-T.
Phalloidin staining:
Cells were washed three times with TBS-T and resuspended in 100 μl TBS-T, and 5 μl of full-strength Alexa Fluor 488 phalloidin were added. Cells were vortexed for 5 min before incubating in a roller in the dark overnight. Cells were washed three times with TBS-T and visualized.
MitoTracker staining:
Cells were grown to 0.3 OD in YPG and MitoTracker Red CMXRos was added to a final concentration of 3 nm. Cells were grown for another hour and then washed twice in YPG and once in PBS, plus 2% dextrose before visualizing.
Sequence comparisons:
ClustalW2 sequence alignments were completed using T-Coffee (Notredame et al. 2000) and color-coded diagrams were prepared using ESPript (Gouet et al. 1999) (Figure 2). We quantified the percentages for protein identity, protein similarity, and DNA identity using MatGAT (Campanella et al. 2003); we report the percentages for sequence similarity and identity for each species compared to S. cerevisiae in Table 1.
Figure 2.—
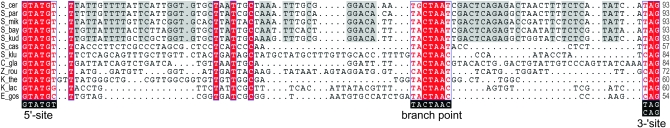
The intronic splice sites are conserved across several hemiascomycetous yeast species. PTC7 intron sequences from Saccharomyces cerevisiae (S_cer), S. paradoxus (S_par), S. mikatae (S_mik), S. bayanus (S_bay), S. kudriavzevii (S_kud), S. castellii (S_cas), S. kluyveri (S_klu), Candida glabrata (C_gla), Zygosaccharomyces rouxii (Z_rou), Kluyveromyces thermotolerans (K_the), K. lactis (K_lac), and Eremothecium gossypii (E_gos) were collectively aligned using T-Coffee (Notredame et al. 2000). Identical nucleotides are shown as white letters highlighted in red. Sequences conserved within intron splice signals are denoted by red letters. The most commonly used splice signal sequences in S. cerevisiae (Bon et al. 2003) are displayed as white letters in black boxes and are aligned below the intronic sequences. The conservation between the five most closely related species (S_cer, S_par, S_mik, S_kud, and S_bay) is highlighted in gray.
TABLE 1.
PTC7 protein sequence analysis
| Species | Ptc7u identitya | Ptc7u similaritya | Intron identityb | Intron similarityb | Transmembrane start/endc | Transmembrane intronic/totald | Ptc7s MTS probe | Ptc7u MTS probe |
|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | 100 | 100 | 100 | 100 | 17/39 | 21/23 | 0.939 | 0.484 |
| S. paradoxus | 92.2 | 96.3 | 86.7 | 93.3 | 17/39 | 21/23 | 0.955 | 0.500 |
| S. mikatae | 88.8 | 94.7 | 76.7 | 93.3 | 17/39 | 21/23 | 0.943 | 0.672 |
| S. bayanus | 84.8 | 93.9 | 63.3 | 86.7 | 17/39 | 21/23 | 0.981 | 0.208 |
| S. kudriavzevii | 76.8f | 91.3f | 66.7 | 83.3 | 17/39 | 21/23 | 0.675f | 0.554f |
| S. castellii | 60.3 | 77.0 | 26.7 | 43.3 | 16/38 | 18/23 | 0.954 | 0.760 |
| S. kluyveri | 56.8 | 72.5 | 16.7 | 46.7 | 17/39 | 23/23 | 0.993 | 0.905 |
| C. glabrata | 55.0 | 74.9 | 30.0 | 50.0 | 12/34 | 17/23 | 0.870 | 0.450 |
| Z. rouxii | 56.5 | 71.4 | 29.0 | 43.3 | 31/53 | 23/23 | 0.991 | 0.755 |
| K. thermotolerans | 57.4 | 69.5 | 16.7 | 36.7 | 21/40 | 17/19 | 0.997 | 0.850 |
| K. lactis | 51.6 | 71.4 | 26.7 | 36.7 | 13/35 | 19/23 | 0.962 | 0.669 |
| E. gossypii | 48.5 | 64.4 | 16.7 | 36.7 | 13/32 | 17/20 | 0.985 | 0.712 |
| D. hansenii | 44.4 | 65.2 | — | — | 13/30 | — | — | 0.481 |
| Y. lipolytica | 31.8 | 52.9 | — | — | — | — | — | 0.977 |
Percentages of protein sequence identity and similarity are reported for each species with respect to S. cerevisiae for the complete Ptc7u protein sequence.
Percentages of protein sequence identity and similarity are reported for each species with respect to S. cerevisiae for the protein sequence coded for by the intron.
The amino acid position for the beginning (start) and end of each predicted transmembrane domain.
The number of transmembrane amino acids that are contained within the intron (intronic) compared to the total amino acid length of the predicted transmembrane domain (total).
The probability that Ptc7s or Ptc7u contains an MTS that would target the protein to the mitochondria.
Only 209 nucleotides of the PTC7 sequence were available from the draft sequence of S. kudriavzevii; the sequence contains the complete first exon and intron, but only 61 nucleotides of the second exon. Because only 39 amino acids are available for analysis of Ptc7s and 69 amino acids for Ptc7u, the calculated MTS probabilities are likely inaccurate.
RESULTS
RNA and protein are expressed from both spliced and unspliced PTC7 isoforms:
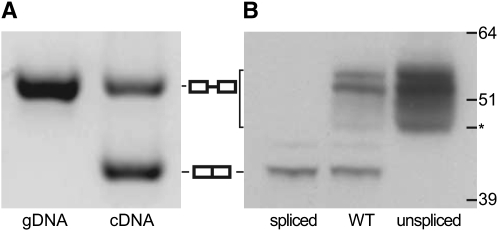
To ascertain if both spliced and unspliced mRNA isoforms are expressed in yeast, we reverse transcribed RNA, which was treated twice with DNase and PCR amplified across the exon–exon splice junction. We observed that wild-type yeast grown in YPD produced two mRNA products (Figure 1A). The PCR product from the unspliced mRNA comigrated with the PCR product prepared from genomic DNA (gDNA), while the PCR product from the spliced mRNA migrated as expected for a double-stranded DNA that is 93 bp smaller than the unspliced DNA. We concluded that both the spliced and the unspliced mRNAs are expressed in yeast during vegetative growth.
Figure 1.—
Both RNA isoforms of PTC7 are translated. (A) PCR products produced by primers surrounding the intronic sequence of PTC7 were separated using gel electrophoresis; the gel was stained with SYBR Green I. The image is inverted for clarity; bands of DNA appear dark on a light background. The left lane shows the PCR product resulting from amplification of genomic DNA (gDNA). The right lane shows the PCR product resulting from the amplification of cDNA, reverse transcribed from unspliced (top band) and spliced (bottom band) mRNA. Graphical representations of the spliced (two adjacent boxes) and unspliced (two boxes connected with a line) products of PTC7 are shown in the middle. (B) An immunoblot analysis of HA-tagged Ptc7 from three yeast strains: wild type (WT), spliced (the PTC7 intron was deleted from the genome), and unspliced (the intron was rendered unable to splice while the amino acid sequence was maintained). The molecular weights of a protein standard are specified in kilodaltons on the right side of the gel. An asterisk (*) marks the fastest mobility band for Ptc7u.
Because it was possible that one of the two PTC7 mRNA isoforms was not translated, we investigated the translational profile via immunoblot analysis. We tagged three splicing variants of PTC7 (wild type, spliced, and unspliced) with the HA epitope on the carboxyl termini (C termini) of each protein (Longtine et al. 1998). The spliced variant protein (Ptc7s) was created by deleting the PTC7 intron from the yeast genome; the unspliced variant protein (Ptc7u) was made by mutating the splicing signals within the intron while maintaining its amino acid coding sequence (see materials and methods for details). The wild-type Ptc7 protein migrates as a series of bands on an SDS–PAGE gel (Figure 1B, middle lane). The fastest (lowest) band is consistent with the predicted molecular weight of Ptc7s (41.1 kDa, including the 3.3-kDa HA epitope tag). The fastest migrating band was found exclusively in wild-type and intron-minus PTC7 strains, but not in the strain that can express only Ptc7u (Figure 1B); thus, we designated it as Ptc7s. In comparison, the slowest (topmost) migrating wild-type Ptc7 bands were observed only in yeast that could express Ptc7 from unspliced mRNA (Figure 1B, middle and right lanes); accordingly, these bands were labeled Ptc7u. The fastest band within the subset of slow Ptc7u bands migrates at a molecular weight predicted for unmodified Ptc7u (44.5 kDa; see asterisk, Figure 1B). Our immunoblot analysis provides strong evidence that alternative splicing of PTC7 results in the expression of two discrete proteins. Several Ptc7u bands display a slower mobility than expected, which could result from post-translational modification; we have not characterized any putative protein modifications beyond showing they are not sensitive to phosphatase treatment (data not shown) and therefore are not due to phosphorylation.
Intronic splice-site sequences, transmembrane domains, and mitochondrial targeting sequences are conserved between PTC7 orthologs:
We compared orthologs of 14 PTC7 genes found in hemiascomycetous yeast (Karolchik et al. 2008; Sherman et al. 2009). Introns were located in 12 of the 14 PTC7 sequences (Figure 2), transmembrane domains were predicted within all 12 intron-containing Ptc7u orthologs (Figure 3 and Table 1), and mitochondrial targeting signals (MTSs) were predicted within all 12 Ptc7s orthologs (Table 1). These data demonstrated that the intron/exon architecture of PTC7 is highly conserved and that splicing changes the functional composition of the protein.
Figure 3.—
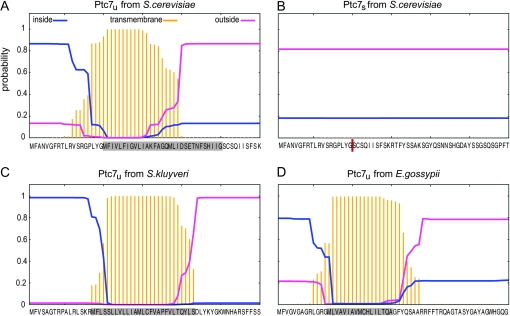
The transmembrane domain within Ptc7 is conserved across Saccharomyces and eliminated upon intron removal. We used TMHMM 2.0 (Krogh et al. 2001) to predict the probability and position of putative transmembrane domains within PTC7. The dark yellow bars plot the probability that an amino acid is part of a transmembrane domain. Blue lines show the probability of an amino acid being inside the cytoplasm and pink lines show the probability for being outside the cytoplasm. The first 60 amino acids of each protein are specified on the bottom; the gray boxes mark the location of the introns; the red bar indicates the position of the exon–exon junction after intron removal. Although we analyzed both Ptc7s and Ptc7u from all 14 yeast species listed in Table 1, only four proteins are presented in this detailed graphical format: (A) Ptc7u from S. cerevisiae, (B) Ptc7s from S. cerevisiae, (C) Ptc7u from S. kluyveri, and (D) Ptc7u from E. gossypii.
We searched for splice signals in the 13 identified orthologs of PTC7 from S. cerevisiae and found introns in 11 of the sequences. All 12 PTC7 introns were divisible by three, lacked stop codons, and contained the required splice signals (Figure 2). No splice signals were found in either Debaryomyces hansenii or Yarrowia lipolytica, the two most distantly related orthologs (Table 1). We next carried out a multiple sequence alignment on all 12 PTC7 introns (Notredame et al. 2000). There was a high degree of similarity across the entire intron among the five most closely related Saccharomyces yeast species (S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus) (Figure 2, gray highlight); these yeast species belong to the sensu stricto complex (Edwards-Ingram et al. 2004) and a large degree of sequence conservation was expected, which likely reflects functional conservation. Outside of the sensu stricto complex, sequence conservation was concentrated at the three major splice signals (5′-splice site, branch point, and 3′-splice site) (Figure 2). Two of the identified splice signals, the 5′-splice site in Kluyveromyces lactis (GTATGG) and the branch point in Zygosaccharomyces rouxii (CACTAAT), have not been previously identified in S. cerevisiae (Bon et al. 2003), but they are quite similar to known signals, so splicing is plausible. Notably, the distance between the branch point and the 3′-splice site in PTC7 was somewhat variable (5–32 nucleotides); this was to be expected since this distance is species dependent in hemiascomycetous yeast where the mean distance is known to range between 1.8 and 45.8 (Bon et al. 2003).
Two previous studies on PTC7 from S. cerevisiae identified a predicted transmembrane domain near the amino terminus (N terminus) of the protein (Ramos et al. 2000; Jiang et al. 2002). We used TMHMM 2.0, a membrane protein topology prediction method (Krogh et al. 2001), to look for predicted transmembrane domains within all 14 orthologs of PTC7. Transmembrane domains were predicted in all 12 of the intron-containing orthologs, as well as in D. hansenii (Table 1). The only PTC7 ortholog that lacks both the intron and the transmembrane domain is also the ortholog that shares the least homology with PTC7 from S. cerevisiae (Table 1). Interestingly, in every case the predicted transmembrane domain overlapped the amino acid sequences coded by the intron, suggesting that intron splicing (removal) would disrupt the transmembrane domain. Indeed, no transmembrane domain is predicted to form in any one of the Ptc7s orthologs (Figure 3B, data shown for S. cerevisiae only).
Because Ptc7 was previously found to localize to the mitochondria, we calculated the probability of finding an MTS at the N terminus of each spliced and unspliced ortholog (Claros and Vincens 1996); the data demonstrated that Ptc7s was much more likely to contain an MTS than Ptc7u (0.961 vs. 0.633 on average) (Table 1). When we looked at the two PTC7 orthologs that lack introns, we found that D. hansenii, which contains a transmembrane domain, had a low probability of having an MTS (0.481) while Y. lipolytica, which lacks the transmembrane domain, had a much higher probability of containing an MTS (0.977).
These data strongly suggest that the transmembrane domain and the MTS are mutually exclusive and that alternative splicing may alter Ptc7 localization within yeast.
Ptc7u localizes to the nuclear envelope while Ptc7s localizes to the mitochondria:
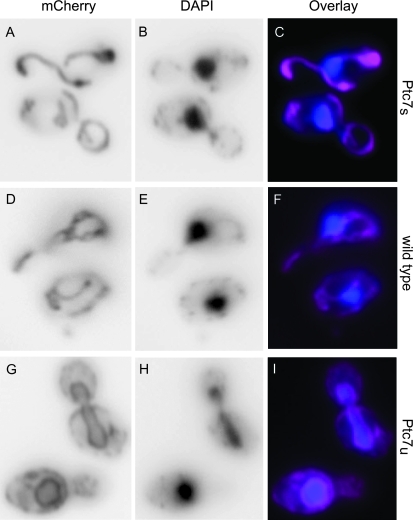
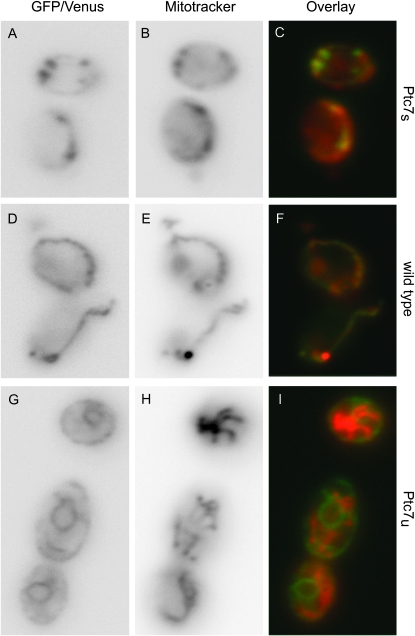
The differential localization of proteins translated from spliced and unspliced PTC7 mRNA (Ptc7u and Ptc7s, respectively) was striking; Ptc7u clearly localized to the nuclear envelope (Figure 4), while Ptc7s colocalized with mitochondria (Figure 5). We visualized the isoforms of Ptc7 by tagging all three splicing variants of PTC7 (wild type, Ptc7s, and Ptc7u) on their C termini with fluorescent proteins: mCherry (Shaner et al. 2004), Venus (Nagai et al. 2002), or GFP (Heim and Tsien 1996; Longtine et al. 1998). To increase protein concentration within the cell to levels that were readily visualized, we found it necessary to replace the native promoter with a galactose-inducible promoter (Guthrie and Fink 1991; Longtine et al. 1998). Localization to the nuclear envelope was validated by costaining cells with DAPI. Localization to the mitochondria was verified by staining cells with MitoTracker Red CMXRos (Invitrogen).
Figure 4.—
Ptc7u localizes to the nuclear envelope. We used fluorescent microscopy to establish the cellular localization of Ptc7u in S. cerevisiae. We created three GAL1-inducible variants of Ptc7 (Ptc7s, wild type, and Ptc7u) tagged on the C-terminal end with mCherry (Shaner et al. 2004). C, F, and I show the overlaid data for Ptc7 and DAPI; DAPI is a blue-emitting fluorescent stain activated upon DNA binding, which identifies the nucleus. Ptc7 is colored pink and DAPI is blue. (A–C) Fluorescent data for yeast expressing only Ptc7s: (A) mCherry, (B) DAPI, and (C) mCherry and DAPI data overlaid. (D–F) Fluorescent data for yeast expressing wild-type Ptc7: (D) mCherry, (E) DAPI, and (F) mCherry and DAPI data overlaid. (G–I) Fluorescent data for yeast expressing only Ptc7u: (G) mCherry, (H) DAPI, and (I) mCherry and DAPI data overlaid.
Figure 5.—
Ptc7s localizes to the mitochondria. In an experiment similar to the one described in Figure 4, we used fluorescent microscopy to establish the cellular localization of Ptc7s in S. cerevisiae. Three galactose-induced fluorescently labeled variants of Ptc7 were visualized in cells costained with MitoTracker red CMXRos, a fluorescent stain activated upon mitochondrial uptake (Invitrogen). In C, F, and I, the data for Ptc7 and MitoTracker are overlaid, Ptc7 is green, and MitoTracker is red. (A–C) Fluorescent data for yeast expressing only Ptc7s: (A) GFP, (B) MitoTracker, and (C) GFP and MitoTracker data overlaid. (D–F) Fluorescent data for yeast expressing wild-type Ptc7: (D) GFP, (E) MitoTracker, and (F) GFP and MitoTracker data overlaid. (G–I) Fluorescent data for yeast expressing only Ptc7u: (G) Venus, (H) MitoTracker, and (I) Venus and MitoTracker data overlaid.
Protein expressed from unspliced mRNA clearly localized to the nuclear envelope (Figure 4, G–I) and membrane localization is maintained throughout cell division (Figure 4G). Although we cannot rule out mislocalization due to overexpressed Ptc7u, we did not observe similar nuclear envelope localization with overexpressed Ptc7s, nor did we observe any obvious fitness defect upon galactose induction (data not shown). Instead, we attribute the prominent nuclear envelope localization of Ptc7u to the transmembrane domain, which is present in the unspliced isoform, but not in the spliced isoform.
In contrast to the strong nuclear envelope localization of Ptc7u, Ptc7s localized to the mitochondria (Figure 5, A–C). This result was not unexpected because previous reports showed that wild-type Ptc7 localizes to the mitochondria (Ramos et al. 2000). Predictably, we also observed mitochondrial localization of wild-type Ptc7 (Figure 5, D–F). We expected to see both mitochondrial and nuclear envelope localization for wild-type Ptc7 because wild-type PTC7 expressed both Ptc7s and Ptc7u proteins (Figure 1B). However, an immunoblot analysis of galactose-induced PTC7 substantiated the microscopy data and showed that the vast majority of protein expressed from GAL1-induced wild-type PTC7 was Ptc7s (supporting information, Figure S1, A and B, middle lanes). Indeed, it was only upon extensive overexposure of the blot that small amounts of Ptc7u were observed (Figure S1, B, middle lane). Notably, we did not see the same skewed Ptc7s/Ptc7u ratio when wild-type yeast were grown in galactose (data not shown), which suggests that splicing may be dependent upon the promoter or regulated by gene expression levels.
We conclude that alternative splicing of PTC7 directs protein localization and that Ptc7s, which lacks a putative transmembrane domain, localizes to the mitochondria while Ptc7u, which contains a putative transmembrane domain, localizes to the nuclear envelope.
PTC7 splicing and modification are sensitive to carbon source availability:
Because the spliced form of PTC7 localizes to the mitochondria, we asked how PTC7 expression and splicing were affected in fermentable and nonfermentable carbon sources and found clear evidence that protein expression and post-translational modification of Ptc7s were carbon source dependent.
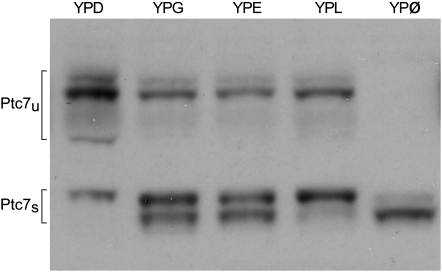
Yeast containing HA-tagged Ptc7 were grown in different media (YPD, YPG, YPE, YPL, or YPØ). When yeast were grown in fermentable media (YPD), Ptc7u was the prominent protein isoform (Figure 6; far left lane). In contrast, more Ptc7s was expressed than Ptc7u in nonfermentable media (YPG, -E, and -L) (Figure 6). Interestingly, we did not observe any carbon-specific splicing differences (data not shown), suggesting that protein expression is not regulated by mRNA levels. This observation was not entirely unexpected since the overall correlation between transcript and protein levels in yeast is weak and for the majority of genes it is effectively zero, sometimes negative (Gygi et al. 1999; Foss et al. 2007).
Figure 6.—
Expression of Ptc7s is regulated by carbon source availability. A strain containing wild-type PTC7, tagged with HA on its C terminus, was grown in various yeast-peptone-based (YP-) media. Proteins were assayed by Western analysis. The YP-based media contained one of the following: 2% dextrose (YPD), 2% glycerol (YPG), 1.5% ethanol (YPE), 2% lactose (YPL), or no additional carbon than what is naturally found in YP (YPØ). The protein products (Ptc7u or Ptc7s) are specified on the left.
We did observe carbon source-specific protein modification; Ptc7s ran as a doublet on SDS–PAGE in all nonfermentable media. The slower Ptc7s band was the only band observed in YPD, while a faster-migrating band appeared in all nonfermentable growth conditions and was the predominant band in YPØ (Figure 6). The difference in mobility of the two Ptc7s bands corresponds to an estimated size difference of ≤1 kDa. We have not further characterized the modification(s) that caused the mobility shift in Ptc7s beyond showing they are not sensitive to phosphatase treatment (data not shown). Nevertheless, we propose that the observed carbon source-dependent protein expression and modification, combined with the mitochondrial localization, provide compelling evidence that Ptc7s is functionally relevant and is likely involved in carbon metabolism.
Yeast lacking Ptc7u are more sensitive to Latrunculin A, a compound that affects actin polymerization:
To better understand the functionality of both PTC7 isoforms, we used a chemical probe to intensify phenotypic differences between strains expressing alternative isoforms. Specifically, we carried out chemical sensitivity growth assays and observed that yeast missing the Ptc7u protein isoform were much more sensitive to Latrunculin A (Lat-A). Lat-A is a drug that binds actin and disrupts filament formation; this suggests that Ptc7u may be involved indirectly in actin filament function.
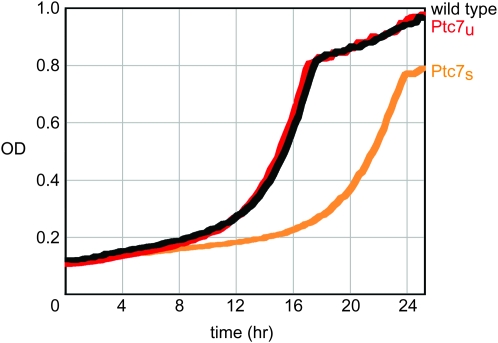
To select the best chemical probe, we used the yeast chemogenomic data from Hillenmeyer et al. (2008) (http://fitdb.stanford.edu) to identify candidate drugs that might induce a growth defect in strains containing altered PTC7 genes. We tested the three highest-scoring compounds for PTC7: lithium chloride, alverine citrate, and Lat-A. Lat-A was the only compound, of the three, that produced a growth defect specific to changes in PTC7 copy number; both the homozygous and the heterozygous deletion strains of PTC7 showed slower growth than wild-type yeast in the presence of Lat-A (data not shown). Interestingly, when we tested yeast strains that express either Ptc7u or Ptc7s exclusively, the strain that expressed Ptc7s grew much slower than wild-type yeast; doubling times were 4.21 and 3.03 hr, respectively (Figure 7, light orange vs. black lines). In contrast, the strain expressing Ptc7u grew at a nearly identical rate to wild type in the presence of Lat-A.
Figure 7.—
Yeast lacking Ptc7u are sensitized to Latrunculin A exposure. The graph shows the growth curves for three homozygous diploid variants of PTC7: wild type in black, PTC7s/PTC7s in light orange, and PTC7u/PTC7u in red. Time in hours is plotted along the x-axis and the optical density (OD), measured at 595 nm, is plotted along the y-axis. Cells were grown in YPD containing 1.3 μm Lat-A. OD readings were recorded every 15 min. The doubling time for each strain in Lat-A is as follows: wild type, 3.03 hr; PTC7u/PTC7u, 2.94 hr; and PTC7s/PTC7s, 4.21 hr.
The Lat-A growth data provided preliminary evidence that Ptc7u might be involved in actin filament function. We further explored the relationship between actin and Ptc7u by using microscopy to look for colocalization of Ptc7 and actin (see materials and methods for details). We saw no obvious colocalization of Ptc7u and actin patches or cables (data not shown). We did, however, uncover genetic evidence for an interaction between Ptc7 and actin. Specifically, a synthetic genetic array experiment (Tong et al. 2001) revealed a significant positive/alleviating interaction between a ptc7-null mutant and a temperature-sensitive actin allele (act1-120) (M. Costanzo, personal communication). These genetic data, combined with our observation that Lat-A affects cells lacking Ptc7u to a much greater extent than cells lacking Ptc7s, provide strong evidence that Ptc7u is functionally distinct from Ptc7s.
DISCUSSION
Here we identify the first unambiguous example of functional alternative splicing in S. cerevisiae. PTC7, a protein phosphatase, expresses two different mRNA isoforms (Figure 1A), each of which are translated into two distinct proteins (Figure 1B) with disparate functions (Figures 6 and 7) and unique localization patterns (Figures 4 and 5). Intronic conservation of splice sites, transmembrane domain positioning, and MTS occurrence (Figures 2 and 3 and Table 1) suggest that alternative splicing extends the functionality of PTC7 and improves fitness in hemiascomycetous yeast.
Introns and the spliceosomal machinery required for intron processing would likely be lost during evolution if splicing did not confer a fitness advantage to S. cerevisiae. Indeed, introns have been shown to increase the fitness of yeast in two ways: (1) regulated splicing post-transcriptionally regulates gene expression (Engebrecht et al. 1991; Li et al. 1996; Nakagawa and Ogawa 1999; Preker et al. 2002; Juneau et al. 2007; Parenteau et al. 2008) and (2) introns can function to enhance the transcriptional and translational output of the genes they occupy (Juneau et al. 2006). Here we provide compelling evidence that introns also improve the fitness of yeast by increasing protein diversity through alternative splicing.
The question arises, Will there be other examples of alternative splicing found in S. cerevisiae? PTC7 is the only gene known to contain an in-frame intron lacking stop codons (Nash et al. 2007). In-frame introns have been predicted, but could not be validated in yeast grown under standard conditions in YPD (Davis et al. 2000). It could be that splicing is condition specific, as was found for introns in meiotic genes (Engebrecht et al. 1991; Nakagawa and Ogawa 1999; Juneau et al. 2007); this suggests that mapping introns under a variety of growth conditions could be worthwhile. Additionally, introns do not need to be in frame to participate in functional alternative splicing; start or stop sites could be altered via splicing. For example, alternative splicing of an intron at the 3′ end of SRC1 results in the expression of two proteins, a full-length protein and a truncated protein (Grund et al. 2008). The truncated protein is less active and it is not known whether shortened Src1 performs a unique function or is simply a less functional form of the protein resulting from anomalous splicing. Further experimentation should confirm whether SRC1 is indeed another example of functional alternative splicing.
We provide data showing that PTC7 is alternatively spliced, but the exact function of each protein isoform and how they are regulated remain to be determined. On the basis of their predicted sequences, both PTC7 protein isoforms are capable of acting as phosphatases. Ptc7s and Ptc7u both contain all 11 conserved type 2C protein phosphatase (PP2C) motifs (Jiang et al. 2002), suggesting that both proteins operate as serine/threonine phosphatases, but with distinct cellular roles. Expression of the two protein isoforms may be promoter dependent: overexpression with either the MET25 (Ramos et al. 2000) or the GAL1 inducible promoters produces significantly more mitochondrial Ptc7s than Ptc7u (Figure 5 and Figure S1). Promoter-specific alternative splicing has been identified in Schizosaccharomyces pombe (Moldon et al. 2008); S. cerevisiae likely shares some of the same regulatory mechanisms.
We conclude from these studies that S. cerevisiae does indeed utilize alternative splicing to increase the complexity of its proteome. Further research on yeast will reveal and/or confirm additional examples beyond PTC7, where intron retention results in multiple, functionally distinct proteins.
Acknowledgments
We thank Taavi Neklesa and Shawn Hoon for experimental advice; Sarah Pierce and Michelle Nguyen for technical assistance; Michael Costanzo for sharing unpublished data; Marilyn Fukushima for comprehensive editing of the manuscript; and Richard Hyman, Michael Mindrinos, and Sujatha Krishnakumar for helpful comments on the manuscript. The authors declare no conflict of interest. This work was supported by National Institutes of Health grant HG000205 to R.W.D.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.105155/DC1.
References
- Bon, E., S. Casaregola, G. Blandin, B. Llorente, C. Neuveglise et al., 2003. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 31: 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella, J. J., L. Bitincka and J. Smalley, 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M. G., and P. Vincens, 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779–786. [DOI] [PubMed] [Google Scholar]
- Davis, C. A., L. Grate, M. Spingola and M. Ares, Jr., 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28: 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards-Ingram, L. C., M. E. Gent, D. C. Hoyle, A. Hayes, L. I. Stateva et al., 2004. Comparative genomic hybridization provides new insights into the molecular taxonomy of the Saccharomyces sensu stricto complex. Genome Res. 14: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht, J. A., K. Voelkel-Meiman and G. S. Roeder, 1991. Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Foss, E. J., D. Radulovic, S. A. Shaffer, D. M. Ruderfer, A. Bedalov et al., 2007. Genetic basis of proteome variation in yeast. Nat. Genet. 39: 1369–1375. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C. I., A. Bhattacharya, W. Wang and S. W. Peltz, 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25. [DOI] [PubMed] [Google Scholar]
- Gouet, P., E. Courcelle, D. I. Stuart and F. Metoz, 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Grund, S. E., T. Fischer, G. G. Cabal, O. Antunez, J. E. Perez-Ortin et al., 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J. Cell Biol. 182: 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, New York/London/San Diego.
- Gygi, S. P., Y. Rochon, B. R. Franza and R. Aebersold, 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, R., and R. Y. Tsien, 1996. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6: 178–182. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer, M. E., E. Fung, J. Wildenhain, S. E. Pierce, S. Hoon et al., 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., M. Whiteway, C. Ramos, J. R. Rodriguez-Medina and S. H. Shen, 2002. The YHR076w gene encodes a type 2C protein phosphatase and represents the seventh PP2C gene in budding yeast. FEBS Lett. 527: 323–325. [DOI] [PubMed] [Google Scholar]
- Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch et al., 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144. [DOI] [PubMed] [Google Scholar]
- Juneau, K., M. Miranda, M. E. Hillenmeyer, C. Nislow and R. W. Davis, 2006. Introns regulate RNA and protein abundance in yeast. Genetics 174: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K., C. Palm, M. Miranda and R. W. Davis, 2007. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc. Natl. Acad. Sci. USA 104: 1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., R. M. Kuhn, R. Baertsch, G. P. Barber, H. Clawson et al., 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 36: D773–D779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., B. Larsson, G. von Heijne and E. L. Sonnhammer, 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Lee, W., R. P. St. Onge, M. Proctor, P. Flaherty, M. I. Jordan et al., 2005. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., J. Vilardell and J. R. Warner, 1996. An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc. Natl. Acad. Sci. USA 93: 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Modrek, B., A. Resch, C. Grasso and C. Lee, 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldon, A., J. Malapeira, N. Gabrielli, M. Gogol, B. Gomez-Escoda et al., 2008. Promoter-driven splicing regulation in fission yeast. Nature 455: 997–1000. [DOI] [PubMed] [Google Scholar]
- Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba et al., 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and H. Ogawa, 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, R., S. Weng, B. Hitz, R. Balakrishnan, K. R. Christie et al., 2007. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 35: D468–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa, T. K., and R. W. Davis, 2008. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proc. Natl. Acad. Sci. USA 105: 15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., D. G. Higgins and J. Heringa, 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Parenteau, J., M. Durand, S. Veronneau, A. A. Lacombe, G. Morin et al., 2008. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol. Biol. Cell 19: 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P. J., K. S. Kim and C. Guthrie, 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, C. W., U. Guldener, S. Klein, J. H. Hegemann, S. Gonzalez et al., 2000. Molecular analysis of the Saccharomyces cerevisiae YHR076w gene. IUBMB Life 50: 371–377. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., J. C. Igual and J. E. Perez-Ortin, 2002. SRC1: an intron-containing yeast gene involved in sister chromatid segregation. Yeast 19: 43–54. [DOI] [PubMed] [Google Scholar]
- Schmitt, M. E., T. A. Brown and B. L. Trumpower, 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18: 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer et al., 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sherman, D. J., T. Martin, M. Nikolski, C. Cayla, J. L. Souciet et al., 2009. Genolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 37: D550–D554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici, F., L. K. Lewis and M. A. Resnick, 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., J. R. Hesselberth and S. Fields, 2007. Genome-wide identification of spliced introns using a tiling microarray. Genome Res. 17: 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]