Abstract
Rice plant architecture is an important agronomic trait and a major determinant in high productivity. Panicle erectness is the preferred plant architecture in japonica rice, but the molecular mechanism underlying domestication of the erect panicle remains elusive. Here we report the map-based cloning of a major quantitative trait locus, qPE9-1, which plays an integral role in regulation of rice plant architecture including panicle erectness. The R6547 qPE9-1 gene encodes a 426-amino-acid protein, homologous to the keratin-associated protein 5-4 family. The gene is composed of three Von Willebrand factor type C domains, one transmembrane domain, and one 4-disulfide-core domain. Phenotypic comparisons of a set of near-isogenic lines and transgenic lines reveal that the functional allele (qPE9-1) results in drooping panicles, and the loss-of-function mutation (qpe9-1) leads to more erect panicles. In addition, the qPE9-1 locus regulates panicle and grain length, grain weight, and consequently grain yield. We propose that the panicle erectness trait resulted from a natural random loss-of-function mutation for the qPE9-1 gene and has subsequently been the target of artificial selection during japonica rice breeding.
THE worldwide explosion of the human population necessitates an increase in grain yield, which poses a substantial challenge (Rosegrant and Cline 2003). Improvement of plant architecture is considered as a viable approach to increase grain yield, because crop plants with desirable architecture are able to produce much higher yields (Wang and Li 2008). The most striking example arose in the late 1950s, when selection for the semi-dwarf stature in rice and wheat greatly improved plant architecture and yield potential (Peng et al. 1999; Monna et al. 2002; Sasaki et al. 2002; Spielmeyer et al. 2002). Tiller, panicle, and leaf morphology also play important roles in shaping high-yield crop architecture. Most plant architecture traits are controlled by quantitative trait loci (QTL) derived from naturally occurring allelic variation. Rice (Oryza sativa L.) is the most important food crop in the world (White 1994). It is the staple of diet for heavily populated Asian countries as well as many African countries. Numerous QTL or major genes controlling plant architecture traits have been identified and several have recently been cloned (Li et al. 2004; Ashikari et al. 2005; Fan et al. 2006; Xie et al. 2006; Song et al. 2007; Yu et al. 2007; Jin et al. 2008; Shomura et al. 2008; Tan et al. 2008; Xing et al. 2008; Xue et al. 2008). Cloning and functional characterization of these genes not only addresses fundamental questions in plant development, but also facilitates bridging the gap between gene identification and breeding application by improving the precision and efficiency of selection.
Rice panicle architecture not only contributes to grain yield, but also to the ecological conditions of cultivated populations and the physicochemical properties of different varieties (Xu et al. 1996; Yuan 1997; Chen et al. 2001). Presently, most japonica rice varieties cultivated in China exhibit the panicle erectness (PE) type of inflorescence (Zhang et al. 2002b). PE varieties typically bear short, erect panicles and leaves, which benefit ventilation and light penetration. As a result, populations of PE varieties show higher photosynthetic rates and material production capacity (Liu et al. 2001; Zhang et al. 2002a; Chen et al. 2007). Additionally, PE rice varieties show increased lodging and fertilizer resistance due to decreased plant height (Xu et al. 1995). Therefore, PE is the preferred plant architecture for high-yield japonica rice.
Since development of the first rice PE variety, Guihuahuang, in the early 1960s, a large number of PE japonica varieties have been released in China, including the most well known, Liaojing 5. PE varieties have increased yield potential compared to panicle drooping varieties and therefore are the preferred. PE serves as the most suitable morphological index, and has subsequently been brought into super high-yield breeding. The development and cultivation of PE varieties is considered the third landmark trait after dwarf and hybrid rice in the history of Chinese rice breeding (Zhang et al. 2002b).
The genetic mechanisms controlling PE have received some attention. Initially, PE was reported to be governed by a recessive gene (Zhu and Gu 1979), while other studies suggested a major gene with dominant or additive effects, and polygenic modifications serving to regulate PE (Xu et al. 1995; Wang et al. 1997). Chen et al. (2006) proposed that panicle angle was controlled by two major genes with additive-dominance-epistatic effects and also polygenes with additive-dominance-epistatic influences, using major gene–polygene mixed inheritance models and a joint analysis method. Pedigree analysis of PE varieties indicated that two-thirds of the varieties possessed genes from the Italian Balilla variety and shared a close relationship with Liaojing 5 (Zhang et al. 2002b). The dominant EP gene was first reported from chromosome 9, between the two SSR markers RM5833-11 and RM5686-23, at a genetic distance of 1.5 and 0.9 cM, respectively (Kong et al. 2007). In a previous study, we identified and characterized qPE9-1, a major QTL on chromosome 9 responsible for the erect panicle trait using a double-haploid (DH) population derived from a cross between Wuyunjing 8 and Nongken 57 varieties (Yan et al. 2007).
However, despite some progresses of the molecular mechanisms governing rice PE, the complexity of the trait results in substantial gaps in our understanding of its regulation. Here we report on a major QTL, qPE9-1, which encodes a keratin-associated protein 5-4, regulates rice PE, and plays pleiotropic roles in an array of plant architecture and yield traits.
MATERIALS AND METHODS
Plant materials:
The PE variety Wuyunjing 8 was crossed and backcrossed three times with a panicle drooping indica variety R6547 to produce an advanced backcross population. The two parents differ significantly in various agronomic traits, particularly in panicle architecture. R6547 exhibits long, drooping panicles and spindled grains, whereas Wuyunjing 8 bears short, erect panicles and round grains. A pair of near-isogenic lines (NILs) for the qPE9-1 locus, designated R6547 (qPE9-1) and R6547 (qpe9-1), was selected from the BC3F6 generation to analyze genetic effects. The flanking markers c15 and H58 (Yan et al. 2007) were used to tag the chromosome segment containing the qPE9-1 locus in every backcross generation. Applying similar methods, NILs with the japonica background were developed from the BC3F4 generation for comparative analysis using Wuyujing 3 and Wuyunjing 8 as the recurrent parents and R6547 as the donor. Thirteen indica varieties, 27 japonica varieties, and seven accessions of wild rice species (supporting information, Table S1) were collected for coding sequence analysis. An additional 50 varieties widely grown in China were used for distribution detection by H90 marker analysis (data not shown). These materials were grown and examined under normal field conditions at the experimental field of Yangzhou University, Yangzhou, China.
Phenotype data collection:
All panicle traits were measured during the mature stage. The panicle curvature was presented by the angle included between the lines connecting panicle pedestal with panicle tip and the elongation line of stem. For all above traits, more than 10 representative plants of each line and variety in the middle of each plot were sampled, and the main stem panicle of each plant was chosen for trait measurement. Paddy grains were dried naturally after harvesting and stored at room temperature for at least 1 month before testing. Fully filled grains were used for measuring grain length, width, thickness, and weight. Ten randomly selected grains from each plant were lined up lengthwise along a vernier caliper to measure grain length and then arranged by breadth to measure grain width. Grain thickness was determined for each grain individually using a vernier caliper. All the values were averaged and used as the measurements for each plant. Grain weight was calculated on the basis of 100 grains and converted to 1000-grain weight.
Fine-mapping of qPE9-1:
The BC3F2 segregation population (R6547 background) was used to fine map qPE9-1. Several BC3F1 plants carrying the heterozygous region flanking the qPE9-1 locus were selected using MAS, and their self-pollinated progeny were used for fine mapping qPE9-1. Four hundred twenty-two plants with extreme drooping panicles from the BC3F2 segregating population were chosen to screen recombinants. The candidate genes from Wuyunjing 8 and R6547 were sequenced and analyzed. The newly developed molecular markers covering the qPE9-1 locus were based on indica–japonica differences. Primer sequences are provided in Table S2.
RNA extraction and gene expression analysis:
Total RNA was extracted from different tissues during the heading stage. RNA extraction followed the Trizol reagent protocol provided by the manufacturer (Invitrogen) with subsequent DNaseI (TaKaRa) treatment. Approximately 1 μg of total RNA from each sample was used for first-strand cDNA synthesis. RT–PCR and quantitative real-time PCR was conducted to amplify qPE9-1 transcript using first-strand cDNA. OsActin was also amplified as a control. Quantitative real-time PCR was carried out on an ABI 7500 real-time system (Applied Biosystems) with the SYBR Premix Ex Taq system (TaKaRa). Each set of experiments was repeated three times. The relative amount of the qPE9-1 transcript is presented as 2−ΔCT according to the ΔCT method described in the real-time PCR Applications Guide. The ΔCT value was obtained by subtracting the CT (threshold cycle) number of the OsActin gene from that of the qPE9-1 gene (ΔCT = CTqPE9-1-CTOsActin). The ΔCT value was converted to the linear form in terms of 2−ΔCT for statistical analysis. Primer sequences are provided in Table S3.
Transgenic analysis:
A 2794-bp DNA fragment containing the qPE9-1 promoter region (1513 bp before ATG) and the entire coding region (1281 bp) from R6547 was cloned into pCAMBIA1301 to generate a p-GPE construct for complementary tests. A p-gpe construct containing the qpe9-1 promoter region (1513 bp before ATG) and the entire coding region (656 bp) from Wuyunjing 8 was also generated.
To generate the RNAi construct p-RNAi, a gene fragment of qPE9-1 was amplified from R6547 cDNA. A hairpin structure with two inverted repeat fragments was subsequently constructed and transferred into the plant binary vector p1301UbiNOS, expressing under the control of the maize ubiquitin promoter (Shi et al. 2007).
The full coding region of qPE9-1 was amplified from R6547 cDNA and was inserted into the p1301UbiNOS vector to generate an overexpression construct p-PEOX. The coding region of qpe9-1 was also amplified from Wuyunjing 8 cDNA and inserted into the p1301UbiNOS vector to generate p-peox.
For mutation sites analysis, a clone from the PAC library of Nipponbare genomic DNA named AP005419 was digested with restriction endonucleases EcoRI and BamHI. Then a 14.3-kb genomic DNA fragment containing the entire qPE9-1 coding region and upstream and downstream sequence was purified and inserted into the plant binary vector pCAMBIA1301 to generate p-FL. Concurrently, a 9.8-kb genomic DNA fragment containing only the qPE9-1 partial coding region and downstream sequence was digested with HindIII and inserted into pCAMBIA1301 to generate p-CK for comparison analysis.
The qPE9-1 1513-bp promoter region was amplified for qPE9-1 expression pattern analysis. The amplification product was subcloned into pCAMBIA1301-GUS to generate the qPE9-1 promoter–GUS fusion construct.
All constructs were transformed by Agrobacterium tumefaciens-mediated transformation (Hiei et al. 1994). All transgenic lines were assayed in the second (T1) or third (T2) generations. All primer sequences are provided in Table S3.
Subcellular localization:
To determine its exact subcellular location, qPE9-1 cDNA was fused in-frame with GFP into the p163-GFP vector to generate CaMV35S:qPE9-1∷GFP. CaMV35S∷GFP was used as a control. The expression constructs were transfected into rice Nipponbare protoplasts. The transformed protoplasts were examined using a confocal microscope (Leica TCS SP5 confocal system). Primer sequences are provided in Table S3.
RESULTS
Development of NILs and phenotypic analysis:
An advanced R6547 background population was generated to isolate the qPE9-1 gene for PE. The phenotypic distribution of panicle architecture in the BC3F2 population is shown in Figure S1. Our results demonstrated that panicle curvature, panicle length, grain length, and 1000-grain weight were not independently inherited, i.e., shorter panicles with small grains were always associated with erect panicles. A bimodal distribution of panicle curvature in the BC3F2 population suggested this trait is controlled by a semidominant QTL (Figure S1, A), which is consistent with the previous studies (Zhang et al. 2002a; Jin et al. 2003; Chen et al. 2008). Panicle and grain length also showed a semidominant distribution model in the BC3F2 population (Figure S1, B andC). Because grain length is always associated with plant yield, we also analyzed the 1000-grain weight distribution in the BC3F2 population, which exhibited a similar distribution model (Figure S1, D).
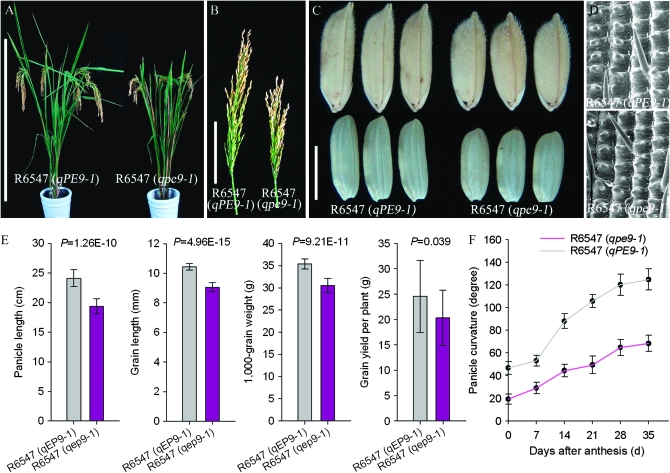
A pair of NILs, R6547 (qPE9-1) and R6547 (qpe9-1), was developed from a BC3F6 generation. The NILs possessed nearly all the genetic background of R6547, with the exception of the introgressed fragment. An array of plant architecture and yield traits was compared between the pair of NILs. During the heading stage, panicle curvature in R6547 (qpe9-1) was less than R6547 (qPE9-1) (Figure 1, A and F). Panicle curvature differences between NILs were more obvious with grain filling. Finally, panicle curvature in R6547 (qpe9-1) was 54.8% of that detected in R6547 (qPE9-1) (Figure 1F). We observed a substantial decrease in panicle length (−19.7%) and grain length (−13.2%) in R6547 (qpe9-1) compared with R6547 (qPE9-1) (Figure 1, A–C, and E). We also observed a significant decrease in grain weight (−13.7%) and grain yield per plant (−17.2%) in R6547 (qpe9-1) (Figure 1E). Scanning electron microscopy (SEM) showed that the outer glume epidermal cells of R6547 (qpe9-1) were shorter than those of R6547 (qPE9-1) (Figure 1D). This result suggested that qPE9-1 may regulate rice cell size.
Figure 1.—
Performance of lines R6547 (qPE9-1) and R6547 (qpe9-1). (A) Plant phenotype of lines R6547 (qPE9-1) and R6547 (qpe9-1). Bar, 100 cm. (B) Main panicle of lines R6547 (qPE9-1) and R6547 (qpe9-1). Bar, 10 cm. (C) Grains and brown rice of lines R6547 (qPE9-1) and R6547 (qpe9-1). Bar, 5 mm. (D) Scanning electron microscopy (SEM) of rice glume epidermis from lines R6547 (qPE9-1) and R6547 (qpe9-1). Bar, 200 μm. (E) Comparison of panicle length, grain length, 1000-grain weight, and grain yield per plant between lines R6547 (qPE9-1) and R6547 (qpe9-1). (F) Dynamic change of panicle curvature after anthesis. Data are means ± SD (n = 10–15). A Student's t-test was applied to generate P-values.
The additional agricultural traits measured are listed in Table S4. We observed an obvious shorter leaf, uppermost internode and plant height in R6547 (qpe9-1) compared with those in R6547 (qPE9-1). However, no obvious differences were detected in grain width, grain thickness, the number of spikelets on the main panicle, primary branch and secondary branch. Two pairs of NILs were developed using MAS and two PE japonica varieties (Wuyujing 3 and Wuyunjing 8) as the recurrent parents, and similar results were generated (Figure S2 and Table S5). These results demonstrated that qPE9-1 regulated panicle curvature and an array of other plant architecture and yield traits, and the effect-increasing allele was derived from R6547.
Map-based cloning of qPE9-1:
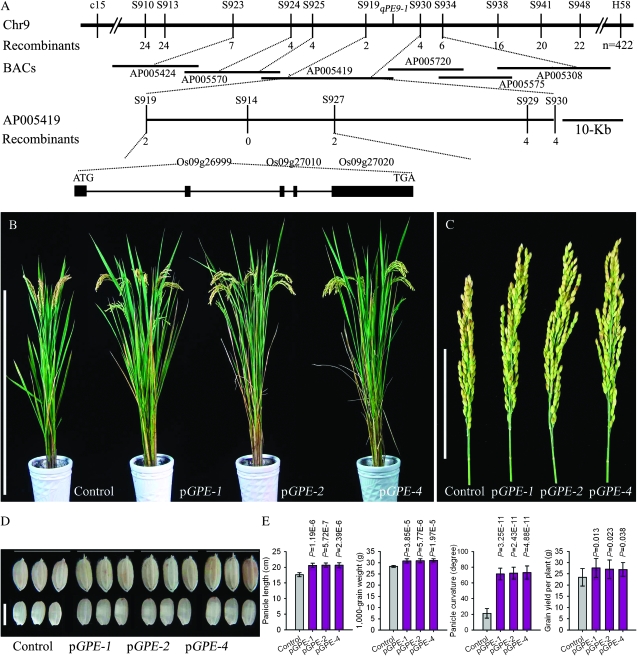
Using a total of 422 individuals with extreme drooping panicles from the 2552 BC3F2 plants, we finally delimited qPE9-1 within a ∼32-kb window between the S919 and S927 markers (Figure 2A). The 32-kb DNA fragment located on a single PAC clone (AP005419) containing three genes, Os09g26999, Os09g27010, and Os09g27020, in the Nipponbare genome according to the TIGR Rice Genome Annotation Database (Figure 2A). Os09g27010 encodes a protein kinase APK1B; Os09g27020 an unclassified retrotransposon protein; and Os09g26999 encodes a protein consisting of threeVon Willebrand factor type C [VWFC] domains, one transmembrane domain, and one 4-disulfide-core domain. The VWFC domains are also present in the OVATE protein and GS3 protein, which have been associated with tomato fruit shape and rice grain length regulation, respectively (Liu et al. 2002; Fan et al. 2006). Os09g26999 demonstrated obvious genetic effects on panicle and grain length, therefore the gene was considered a strong candidate for qPE9-1.
Figure 2.—
Map-based cloning of qPE9-1. (A) The qPE9-1 gene was finally delimited to a 32-kb genomic DNA region between S919 and S927, and cosegregated with S914. Numbers represent recombination events. Three candidate genes were located within this region in the Nipponbare genome according to the TIGR Rice Genome Annotation Database, one of which was qPE9-1. The gene structure of qPE9-1 is indicated. (B) Phenotypic characters of pGPE1 lines and control. Bar, 100 cm. (C) Main panicle phenotypes in pGPE1 lines and control. Bar, 10 cm. (D) Grains and brown rice in pGPE1 lines and control. Bar, 5 mm. (E) Panicle length, 1000-grain weight, panicle curvature, and grain yield comparisons per plant between pGPE1 lines and control. Panicle curvature was detected 28 days after anthesis. Data are means ± SD (n = 10–20). A Student's t-test was applied to generate P-values.
To validate Os9g26999 as a candidate gene, we carried out a functional complementary test. The p-GPE construct containing the promoter and entire coding region of qPE9-1 allele from R6547 was transformed into recipient Wuyunjing 8. All plantlets regenerated from the p-GPE transformed calli were confirmed positive, and the transgenic lines were further identified. All lines showed a significant increase in panicle curvature compared with the control and exhibited a drooping panicle (Figure 2, A and E). We also observed a significant increase in panicle length and 1000-grain weight (Figure 2, B–E). Furthermore, compared with the control, grain yield per plant increased in transgenic lines (Figure 2E). The p-gpe construct containing the promoter and entire coding region of qpe9-1 allele from Wuyunjing 8 was simultaneously transformed into Zhonghua 11 (an easy regenerated japonica variety with a drooping panicle). But we observed no change in the transgenic lines (data not shown). These results demonstrated that in R6547, Os9g26999 is a key gene (qPE9-1) for PE and has pleiotropic effects controlling plant architecture and yield traits and the qpe9-1 in Wuyunjing 8 is a loss-of-function allele.
Sequence analysis and natural variation of qPE9-1:
Alignment of qPE9-1 cDNA with its genomic DNA revealed that qPE9-1 contained five exons and four introns, encoding a protein of 426 amino acid residues (Figures 2A and 3). FASTA analysis indicated that the qPE9-1 protein is homologous to the keratin-associated protein (KAP) 5-4 family in human. Results further established that qPE9-1 contains three VWFC domains (residues 99–153, 276–316, and 339–385), one transmembrane domain (residues 88–106), and one 4-disulfide-core domain (residues 153– 166) (http://www.ebi.ac.uk/InterProScan/) (Figure 3). An additional gene controlling grain size (GS3) has been identified in rice and also carries VWFC and transmembrane domains (Fan et al. 2006), demonstrating the importance of these structures. The fact that the two QTL exhibiting similar protein domains and genetic effects suggested a similar molecular mechanism controls grain size. Thirteen single nucleotide polymorphisms (SNP1–SNP13) and four insertion–deletion polymorphisms (InDel1–InDel4) were detected on the qPE9-1 locus between R6547 and Wuyunjing 8 (Figure S3). All sequence polymorphisms were delimited in noncoding regions, with the exception of SNP13 and InDel4. SNP13 results in a cystine-to-tyrosine substitution at site 105 (C105Y); and the Wuyunjing 8 allele, due to its InDel4 (637-bp deletion and 12-bp insertion) in exon 5, encodes a presumably truncated protein that lacks 231 C-terminal residues (Figure 3 and Figure S3). The missing C-terminal amino acids cover the two rear VWFC domains (Figure 3), rendering the truncated protein nonfunctional.
Figure 3.—
Predicted sequences and structure of the qPE9-1/qpe9-1 protein in R6547 and Wuyunjing 8. Predicted sequence analysis of the qPE9-1 protein revealed several known regions and domains (http://www.ebi.ac.uk/InterProScan/). The amino acids marked with black lines indicate the VWFC domains, the amino acids marked blue showed the 4-disulfide-core domain, and the predicted transmembrane domain was marked red.
Mutation sites analysis:
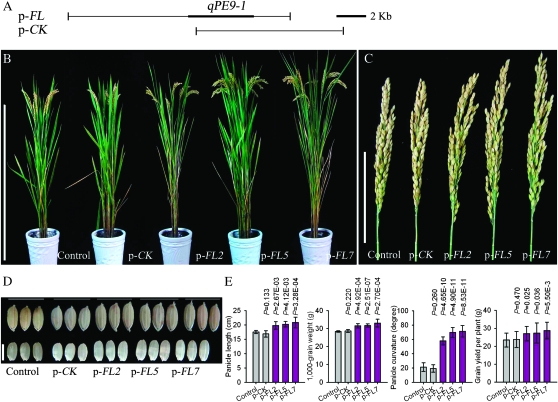
Two mutations are present in rice PE varieties and we were interested which mutation is associated with rice PE. Therefore, we obtained the qPE9-1 allele DNA sequence from Nipponbare, a famous panicle drooping japonica variety. Sequence analysis indicated that Nipponbare and Wuyunjing 8 only differed at InDel4 in the qPE9-1 coding region (see below). We generated a p-FL construct, which covered the entire coding region of the upstream and downstream Nipponbare allele sequence, and transformed it into Wuyunjing 8 (Figure 4A). The p-CK construct containing a partial coding region and three-flanking region of the qPE9-1 gene was used for comparative analysis (Figure 4A). The complementary test showed image results of all 18 p-FL independent transgenic lines consistent with that of pGPE lines (Figure 4, B–E). In contrast, transgenic plants carrying a p-CK construct showed no noticeable change in panicle and plant architecture (Figure 4). On the basis of these results, we concluded that the Nipponbare and R6547 qPE9-1 alleles are functional and the premature stop codon resulting from InDel4 is responsible for PE.
Figure 4.—
Mutation sites analysis. (A) Genomic fragments containing the entire or partial qPE9-1 allele from the Nipponbare genomic PAC were cloned into a pCAMBIA1301 vector to generate p-FL and p-CK vectors. The p-CK was used for comparison and contained only a partial coding and three-flanking region of the qPE9-1 allele. (B) Phenotypes of transgenic plants and control. Bar, 100 cm. (C) Panicle phenotype of transgenic plants and control. Bar, 10 cm. (D) Grain and brown rice of transgenic plants and control. Bar, 5 mm. (E) Comparison of panicle length, 1000-grain weight, panicle curvature, and grain yield per plant of transgenic plants and control. Panicle curvature was detected 28 days after anthesis. Data are means ± SD (n = 10–20). A Student's t-test was applied to generate P-values.
RNAi and overexpression experiments:
Transgenic plants expressing different qPE9-1 levels were also generated in our study. All 18 transgenic Zhonghua 11 lines carrying p-RNAi showed reduced expression levels and exhibited typical erect and short panicles, and a significant decrease in 1000-grain weight (Figure S4 and Figure S5). All 16 transgenic Zhonghua 11 lines carrying p-PEOX showed increased expression levels and displayed the opposite phenotype (Figure S4 and Figure S5). In contrast, both transgenic Zhonghua 11 plants expressing p-peox and Wuyunjing 8 plants carrying p-RNAi showed no obvious change in plant and panicle architecture (data not shown). Taken together, we conclude that qPE9-1 acts as a functional allele and its loss-of-function mutation leads to PE. These results also implied that PE is a comprehensive trait resulting from short panicles, small grain length, and reduced weight.
Expression pattern and subcellular localization of qPE9-1:
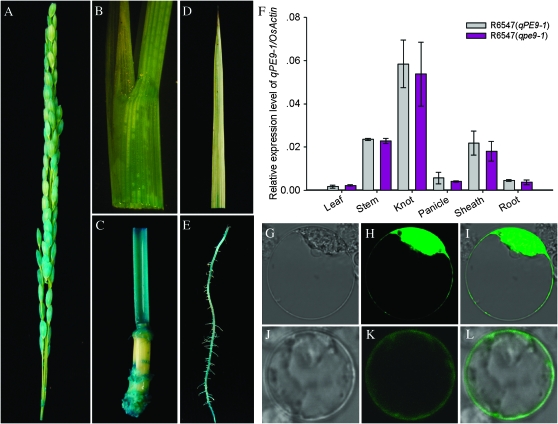
qPE9-1 expression pattern was analyzed using transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of the qPE9-1 gene promoter region (Figure 5). GUS activity was detected mainly in elongating and dividing tissues, including the shoot apical meristem, and the divisional and elongating zones of stems and knots (Figure 5C). GUS activity was also detected in panicle, sheath, leaf, and root tissues (Figure 5, A, B, D, and E). Quantitative real-time PCR analysis was consistent with GUS staining (Figure 5F). Differences in qPE9-1 expression levels between R6547 (qPE9-1) and R6547 (qpe9-1) lines were not observed, suggesting that the genomic sequence changes did not affect expression. Subcellular localization of the qPE9-1 protein was identified using the chimerical fusion protein CaMV35S:qPE9-1∷GFP, and CaMV35S∷GFP alone was used as a control. Confocal microscopy showed that transient expression of the qPE9-1∷GFP fusion protein in rice protoplast was located in the membrane (Figure 5, J–L). However, expression in the control was distributed throughout the entire cell (Figure 5, G–I). These results suggested that the qPE9-1 protein is a membrane protein.
Figure 5.—
Expression pattern and subcellular localization of qPE9-1. (A) GUS activity in young panicle. (B) GUS activity in sheath. (C) GUS activity in stem and knot. (D) GUS activity in leaf. (E) GUS activity in root. (F) Transcript levels of qPE9-1 relative to OsActin in various tissues detected by quantitative real-time PCR. (G–L) CaMV35S∷GFP (G–I) and CaMV35S:qPE9-1∷GFP (J–L) in rice protoplast. The (G and J) photographs were taken in an optic field to examine cell morphology (light), (H and K) were taken in a dark field to localize green fluorescence (GFP), and (I and L) were taken in combination (merge).
The deletion associated with the PE trait defines a domestication-related gene:
The predicted qPE9-1 coding region was sequenced for 13 indica and 27 japonica rice varieties and seven accessions of wild rice (Table S1). Comparison of the predicted coding sequences showed that exclusive of the two mutation sites (SNP13 and InDel4) discussed above, two new mutation sites, SNP14 and SNP15, located in the InDel4 region were detected in the fifth exon. SNP14 (A to T) resulted in an amino acid change (histidine to leucine) and SNP15 (A to T) replaced serine by cystine. Sequence analysis revealed that only PE varieties, including Guihuahuang and its donor parent Balilla, carried the common qpe9-1 allele of Wuyunjing 8. Most indica rice varieties possessed the qPE9-1 allele in common with R6547, and wild rice accessions shared the other alleles. Four japonica varieties with drooping panicles (including Nipponbare and Zhonghua11), shared an allele similar to qPE9-1 in R6547 (Table S1). Furthermore, a gene-tagged marker H90 anchoring the InDel4 region (Yan et al. 2007) was used to examine the distribution of qPE9-1/qpe9-1 in 18 panicle drooping varieties and 32 PE varieties. All drooping panicle varieties carried qPE9-1, while all PE varieties carried qpe9-1 (data not shown). The single allele in PE varieties supported strong selection at the qpe9-1 locus during rice domestication. The qpe9-1 locus may have arisen from a naturally occurring mutation and was conserved due to its preferred phenotype, similar to sd1 in indica rice.
DISCUSSION
qpe9-1 is a key gene involved in rice PE formation:
Crop morphological traits are closely associated with yield potential. Idealized plant architecture with a specific combination of morphological traits deemed favorable for photosynthesis, growth, and grain yield was defined by Donald (1968). In Japan, all cultivated japonica varieties bear a drooping panicle, including the most famous variety Nipponbare. However in China, japonica PE varieties have become predominant. In cultivated populations of PE varieties, individual competition is reduced to a minimum. PE is considered as high-yielding plant architecture in japonica rice due to panicle and plant architecture that significantly optimizes canopy structure (Liu et al. 2001; Zhang et al. 2002a; Chen et al. 2007). In the present study, we identified and characterized a major panicle and plant architecture QTL designated qPE9-1. Our results provided strong evidence that a deletion in qPE9-1 leads to PE in rice and has pleiotropic effects on an array of rice traits, including shortened panicle, reduced grain length, and weight. SEM analysis indicated that qPE9-1 may regulate rice cell size.
Complex traits such as PE are based on naturally occurring variations governed by several genes at quantitative trait loci and their interactions with other genomewide loci. Therefore, for accurate QTL analysis, phenotypic differences in nontarget traits should be minimized in mapping populations. Previous studies identified and characterized the PE trait using various mapping populations and several different genetic modes were proposed. However, the majority of these studies evaluated PE in primary mapping populations, which included F2:3, doubled haploid lines (DHs), and BC1F1. These populations are not suitable for fine mapping or cloning QTL because of excessive genetic background noise, although they are easy to develop. As a result, a lack of congruence in the results of former studies regarding the PE trait is widespread in the literature (Zhu and Gu 1979; Chen et al. 2006; Kong et al. 2007; Yan et al. 2007). To overcome these inconsistencies, we generated three pairs of NILs with varied genetic backgrounds to assure more reliable results. These data together with the transgenic experiments clearly demonstrated that a semidominant gene controls the PE trait and the R6547 and Nipponbare allele is functional.
Gene structure of qPE9-1:
Our results revealed that the qPE9-1 encodes a putative homologous gene of keratin-associated protein 5-4 in human. The KAPs form a matrix where intermediate filaments (IFs) are embedded. The complex forms the bulk of keratin fiber, the main structural protein of certain tissues such as hair in humans and wool in animals (Gillespie and Marshall 1980). KAPs fall into three general families; high sulfur proteins, ultrahigh sulfur proteins, and high-glycine-tyrosine proteins in humans. The keratin-associated protein 5-4 is an ultrahigh sulfur protein (Crewther et al. 1965; Parry et al. 2006). To date, the function of the homologous KAP genes in plants has not been characterized and qPE9-1 cloning will provide an opportunity to investigate the function of these genes. The qPE9-1 protein contains three VWFC domains and one transmembrane domain. The VWFC domain has been found in a rice grain size gene GS3 and the OVATE gene in tomato (Liu et al. 2001; Fan et al. 2006). GS3 is a negative regulator that prevents increases in grain size and a nonsense mutation in the second exon of the gene results in large grains. OVATE determines the conversion of fruit from round to pear shape and is a recessive trait. These results confirmed the wide range of roles for the VWFC domain in fruit/grain shape regulation and also indicated the conserved molecular nature of the domain across species.
The qpe9-1 was the target of artificial selection during domestication:
Balilla, an Italian PE variety, was introduced to China in 1958 and named Beijing 5 (Zhang et al. 2002b). Taihu Institute of Agricultural Sciences in Jiangsu Province successfully developed another PE variety, Suzhou 63-2, from the progeny of a natural hybrid of Balilla. Guihuahuang was subsequently developed from Suzhou 63-2 progeny and released into cultivation. In 1974, the first PE variety from Liaoning Province, Qianchonglang, was developed by crossing Balilla with other indica and japonica varieties (Yan et al. 2007). The famous PE variety Liaojing 5 was developed from Balilla progeny in 1976 and then widely introduced into cultivation. Liaojing 5 displayed high yield potential and was novel for many traits, including panicle and leaf architecture (Zhang et al. 2002b). Since then, an increasing number of PE varieties have been developed and released. To date, japonica varieties displaying the PE phenotype have been widely cultivated throughout most of the japonica growing regions, from Zhejiang to Liaoning Province in China (Yan et al. 2007). Zhang et al. (2002b) found that two-thirds of the PE varieties in cultivation have genes from Balilla and share a close relationship with Liaojing 5. In this study, we sequenced the qPE9-1 allele from 13 indica and 27 japonica varieties, and seven accessions representing different species of wild rice. Our results found that all PE varieties, including Balilla, Guihuahuang, Liaojing 5, and Wuyunjing 8 shared the qpe9-1 allele, while the panicle drooping varieties possessed the wild-type qPE9-1 allele. The distribution of the qPE9-1/qpe9-1 alleles in 18 panicle drooping varieties and 32 PE varieties was examined using a gene-tagged marker, yielding the same results. The single qpe9-1 allele in all PE varieties confirmed this result and demonstrated that strong selection for the qpe9-1 locus has occurred during rice domestication.
The breeding value of qpe9-1:
Our study indicated that all PE varieties analyzed carried the qpe9-1 allele. The gene confers desirable plant architecture, and was therefore conserved during selection. Furthermore, the gene was found to be a key regulator of plant architecture in the high-yielding japonica varieties and serves an important role in PE formation and shaping plant architecture. The qpe9-1 allele itself is a paradox, conducive to development of the rice architecture, but exhibiting negative effects on individual plant yield. Previous studies also observed that the grain yield per plant of erect panicle type was significantly lower than that of drooping panicle type (Zhou et al. 2006; Chen et al. 2008). Yield is a complex polygenic trait and difficult to be selected directly, while plant architecture traits are easily observed and readily selected during rice improvement. Although leading to a decrease in grain yield per plant, qpe9-1 improves plant architecture and population quality in PE japonica rice. These qualities provide new insights into the complex relationship between plant architecture and yield. Currently, all japonica rice in cultivation is PE varieties. However, PE indica varieties are not yet available. Here, we also noted differences between panicle drooping NILs: panicle curvature in the indica genetic background NILs was less than that in the japonica background NILs. These observations suggested that more than one different gene is responsible for PE in rice subspecies. The qPE9-1 cloning strategy employed in this study remains a viable approach to isolate other genes determining the PE trait in indica rice.
Acknowledgments
We thank Bin Han (National Center for Gene Research, China) for providing the PAC clone. This study was financially supported by grants from the State Key Program of Basic Research of People's Republic of China (nos. 2005CB120807 and 2006CB101703); the National Natural Science Foundation of People's Republic China (no. 30530118), and the Key Program of the Bureau of Education, Jiangsu Province, People's Republic of China (no. 05KAJ2012).
Sequence data from this article have been deposited with the GenBank Data Libraries: FJ501956, qPE9-1 allele of R6547 (initially named PAY1); FJ554569, qpe9-1 allele of Wuyunjing 8 (initially named pay1).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102681/DC1.
References
- Ashikari, M., H. Sakakibara, S. Lin, T. Yamamoto, T. Takashi et al., 2005. Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Chen, W. F., Z. J. Xu, L. B. Zhang, W. Z. Zhang and D. R. Ma, 2007. Theories and practices of breeding japonica rice for super high yield. Scientia Agricultura Sinica 40: 869–874. [Google Scholar]
- Chen, W. F., Z. J. Xu, W. Z. Zhang, Z. L. Bu and S. R. Yang, 2001. Creation of new plant type and breeding rice for super high yield. Acta Agronomica Sinica 27: 665–672. [Google Scholar]
- Chen, X. G., J. B. Liu and D. L. Hong, 2006. Genetic analysis on panicle angle and number of spikelets per panicle by using six generations of three crosses derived from erect×curve panicles in japonica rice (Oryza sativa L.). Acta Agronomica Sinica 32: 1143–1150. [Google Scholar]
- Chen, Y. Z., S. L. Wei, C. Liu, W. Y. Zhou, Z. F. Ma et al., 2008. Exploitation of molecular markers for erect panicle trait and study on breeding Indica erect panicle varieties in rice. Southwest China J. Agric. Sci. 21: 6–11. [Google Scholar]
- Crewther, W. G., R. D. B. Fraser, F. G. Lennox and H. Lindley, 1965. The chemistry of keratins. Adv. Protein Chem. 20: 191–346. [DOI] [PubMed] [Google Scholar]
- Donald, C. M. 1968. The breeding of crop ideotypes. Euphytica 17: 385–403. [Google Scholar]
- Fan, C., Y. Xing, H. Mao, T. Lu, B. Han et al., 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. M., and R. C. Marshall, 1980. Variability in the proteins of wool and hair, pp. 67–77 in Proceedings 6 International Wool Textile Research Conference II. Pretoria, South Africa.
- Hiei, Y., S. Ohta, T. Komari and T. Kumashiro, 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium tumeficience and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Huang, X., Q. Qian, Z. Liu, H. Sun, S. He et al., 2009. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Jin, J., W. Huang, J. P. Gao, J. Yang, M. Shi et al., 2008. Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Jin, X. H., J. Y. Wang, Z. J. Xu, W. Z. Zhang, W. F. Chen et al., 2003. Genetic multiple effects of erect panicle type rice. J. Shenyang Agric. Univ. 34: 332–335. [Google Scholar]
- Kong, F., J. Y. Wang, J. C. Zou, L. X. Shi, D. M. Jin et al., 2007. Molecular tagging and mapping of the erect panicle gene in rice. Mol. Breed. 19: 297–304. [Google Scholar]
- Li, J., M. Thomson and S. R. McCouch, 2004. Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., J. Van Eck, B. Cong and S. D. Tanksley, 2002. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 99: 13302–13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Z. J. Xu, W. F. Chen, J. Q. Li, L. X. Li et al., 2001. Effect of different N-level on canopy photosynthetic characteristics of rice varieties with erect panicle type. J. Shenyang Agric. Univ. 32: 8–12. [Google Scholar]
- Monna, L., N. Kitazawa, R. Yoshino, J. Suzuki, H. Masuda et al., 2002. Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9: 11–17. [DOI] [PubMed] [Google Scholar]
- Parry, D. A., T. A. Smith, M. A. Rogers and J. Schweizer, 2006. Human hair keratin-associated proteins: sequence regularities and structural implications. J. Struct. Biol. 155: 361–369. [DOI] [PubMed] [Google Scholar]
- Peng, J., D. E. Richards, N. M. Hartley, G. P. Murphy, K. M. Devos et al., 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Rosegrant, M. W., and S. A. Cline, 2003. Global food security: challenges and policies. Science 302: 1917–1919. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., M. Ashikari, M. Ueguchi-Tanaka, H. Itoh, A. Nishimura et al., 2002. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702. [DOI] [PubMed] [Google Scholar]
- Shi, Z., J. Wang, X. Wan, G. Shen, X. Wang et al., 2007. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226: 99–108. [DOI] [PubMed] [Google Scholar]
- Shomura, A., T. Izawa, K. Ebana, T. Ebitani, H. Kanegae et al., 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Song, X. J., W. Huang, M. Shi, M. Z. Zhu and H. X. Lin, 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Spielmeyer, W., M. H. Ellis and P. M. Chandler, 2002. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., X. Li, F. Liu, X. Sun, C. Li et al., 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Wang, B. L., Y. H. Dong and S. Wang, 1997. Studies on genetic activities of semidwar sm and erect-panicle in rice. J. Shenyang Agric. Univ. 28: 83–87. [Google Scholar]
- Wang, Y., and J. Li, 2008. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- White, P. T., 1994. Rice: the essential harvest. Natl. Geogr. 185: 48–79. [Google Scholar]
- Xie, X., M. H. Song, F. Jin, S. N. Ahn, J. P. Suh et al., 2006. Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor. Appl. Genet. 113: 885–894. [DOI] [PubMed] [Google Scholar]
- Xing, Y. Z., W. J. Tang, W. Y. Xue, C. G. Xu and Q. Zhang, 2008. Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice. Theor. Appl. Genet. 116: 789–796. [DOI] [PubMed] [Google Scholar]
- Xu, Z. J., W. F. Chen, L. B. Zhang and S. R. Yang, 1995. Advance in estimation and utilization of rice erect panicle. J. Shenyang Agric. Univ. 26: 335–341. [Google Scholar]
- Xu, Z. J., W. F. Chen, H. F. Zhou, B. L. Zhang and S. R. Yang, 1996. The physiological and ecological characters and application prospects of erect panicle rice population. Chinese Sci. Bull. 41: 1122–1126. [Google Scholar]
- Xue, W., Y. Xing, X. Weng, Y. Zhao, W. Tang et al., 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yan, C. J., J. H. Zhou, S. Yan, F. Chen, M. Yeboah et al., 2007. Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor. Appl. Genet. 115: 1093–1100. [DOI] [PubMed] [Google Scholar]
- Yu, B., Z. Lin, H. Li, X. Li, J. Li et al., 2007. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52: 891–898. [DOI] [PubMed] [Google Scholar]
- Yuan, L. P., 1997. Hybrid rice breeding for super high yield. Hybrid Rice 12: 1–6. [Google Scholar]
- Zhang, W. Z., Z. J. Xu, W. F. Chen, L. B. Zhang, X. H. Jin et al., 2002. a The research progress on erect panicle type of rice. J. Shenyang Agric. Univ. 33: 471–475. [Google Scholar]
- Zhang, W. Z., Z. J. Xu, L. B. Zhang, W. F. Chen, F. L. Qiu et al., 2002. b Analysis on evolution for the erect panicle type varieties of rice. J. Shenyang Agric. Univ. 33: 161–166. [Google Scholar]
- Zhou, W. Y., D. L. Bai, X. Q. Yang, X. Y. Li, Y. F. Liu et al., 2006. Inheritance of erect panicle type and its effect on the agronomic characteristics of indica rice. Guangxi Agric. Sci. 37: 490–493. [Google Scholar]
- Zhu, L. H., and M. H. Gu, 1979. The inheritance of rice grain shattering. Hereditas 1: 17–19. [Google Scholar]