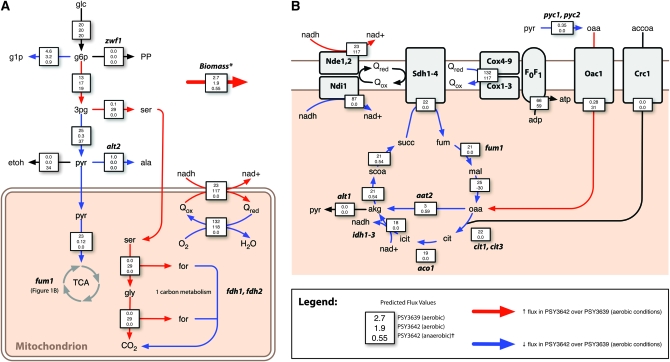
Figure 1.—
Constraint-based modeling predicts mutations that redirect flux through serine/glycine biosynthesis and C1 metabolism, leading to increased aerobic formic acid secretion. Arrows denote key cytoplasmic (A) and mitochondrial (B) reactions for which predicted flux is higher (red) or lower (blue) in PSY3642 compared to PSY3639 (see text for details). Boxes superimposed over arrows contain flux values for three simulated conditions: (i) PSY3639 (aerobic), (ii) PSY3642 (aerobic), and (iii) PSY3642 (anaerobic)(†, anaerobic flux values, given in A only). Flux values are relative to a constant glucose uptake rate of 20 mmol gDW−1 hr−1. See Table S2 for a complete list of predicted fluxes and metabolites required for growth (*, the biomass equation, given in Table S2). Abbreviations (not provided in the text): glc, glucose; g1p, glucose-1-phosphate; g6p, glucose-6-phosphate; ser, serine; etoh, ethanol; pyr, pyruvate; ala, alanine; gly, glycine; for, formate; nad, nicotinamide adenine dinucleotide; Q, quinone; accoa, acetyl-CoA; cit, citrate; isocit, isocitrate; succ, succinate; fum, fumarate; and mal, malate.