Abstract
The B-type cyclin Clb5 is involved primarily in control of DNA replication in Saccharomyces cerevisiae. We conducted a synthetic genetic array (SGA) analysis, testing for synthetic lethality between the clb5 deletion and a selected 87 deletions related to diverse aspects of cell cycle control based on GO annotations. Deletion of the spindle checkpoint genes BUB1 and BUB3 caused synthetic lethality with clb5. The spindle checkpoint monitors the attachment of spindles to the kinetochore or spindle tension during early mitosis. However, another spindle checkpoint gene, MAD2, could be deleted without ill effects in the absence of CLB5, suggesting that the bub1/3 clb5 synthetic lethality reflected some function other than the spindle checkpoint of Bub1 and Bub3. To characterize the lethality of bub3 clb5 cells, we constructed a temperature-sensitive clb5 allele. At nonpermissive temperature, bub3 clb5-ts cells showed defects in spindle elongation and cytokinesis. High-copy plasmid suppression of bub3 clb5 lethality identified the C-terminal fragment of BIR1, the yeast homolog of survivin; cytologically, the BIR1 fragment rescued the growth and cytokinesis defects. Bir1 interacts with IplI (Aurora B homolog), and the addition of bub3 clb5-ts significantly enhanced the lethality of the temperature-sensitive ipl1-321. Overall, we conclude that the synthetic lethality between clb5 and bub1 or bub3 is likely related to functions of Bub1/3 unrelated to their spindle checkpoint function. We tested requirements for other B-type cyclins in the absence of spindle checkpoint components. In the absence of the related CLB3 and CLB4 cyclins, the spindle integrity checkpoint becomes essential, since bub3 or mad2 deletion is lethal in a clb3 clb4 background. clb3 clb4 mad2 cells accumulated with unseparated spindle pole bodies. Thus, different B-type cyclins are required for distinct aspects of spindle morphogenesis and function, as revealed by differential genetic interactions with spindle checkpoint components.
CELL cycle progression is achieved by series of activations of cyclins/cyclin-dependent kinase (CDK) complexes (Morgan 2003). CDK becomes active only when it is associated with cyclins. The process has to proceed sequentially and in a timely fashion. In Saccharomyces cerevisiae, there are six B-type cyclins, Clb1–6 (Nasmyth 1993). Clb1–4 are mitotic cyclins (Surana et al. 1991), and Clb5,6 are S-phase cyclins (Epstein and Cross 1992; Schwob and Nasmyth 1993). While different cyclins/CDK complexes promote distinct cell cycle events, these B-type cyclins also share overlapping functions. The primary role of Clb5,6 is to trigger DNA replication (Epstein and Cross 1992; Schwob and Nasmyth 1993). Mitotic cyclins Clb1–4 trigger entering into mitosis (Fitch et al. 1992; Richardson et al. 1992), and they also have functions in spindle pole body (SPB) separation (Fitch et al. 1992) and spindle elongation (Rahal and Amon 2008). Clb2 inhibits mitotic exit; therefore, degradation of Clb2 is required for mitotic exit (Wasch and Cross 2002).
CLB5 is a nonessential gene, although Clb5,6 are the primary drivers of DNA replication in wild-type cells (Schwob and Nasmyth 1993). Clb-Cdk1 activity also inhibits rereplication within a single cell cycle by phosphorylation of the prereplicative complex (Labib et al. 1999; Drury et al. 2000; Nguyen et al. 2000, 2001; Liku et al. 2005). Binding of Clb5 to Orc6 also contributes to preventing DNA rereplication (Wilmes et al. 2004). The Clb5 hydrophobic patch mutant, Clb5-hpm, cannot bind to Orc6 (Wilmes et al. 2004).
There are several known mitotic functions for Clb5. When clb5 was combined with cdc28-4 (CDC28 is the only CDK in S. cerevisiae), cells exhibited defects in nuclear positioning (Segal et al. 1998) and spindle polarity (Segal et al. 2000). Phosphorylation of Fin1 by Clb5-Cdk1 inhibits Fin1 association with the spindle, which affects spindle integrity (Woodbury and Morgan 2007). Consistently, Clb5 is present long after completion of replication and is degraded at the metaphase–anaphase transition by Cdc20/APC (anaphase promoting complex) (Shirayama et al. 1999).
Synthetic genetic array analysis (SGA; Tong et al. 2001) can identify novel functions or pathways controlled by a nonessential protein. This analysis carried out with clb5 led to a study of the interaction of different B-type cyclins with components of the spindle assembly checkpoint. The spindle assembly checkpoint ensures the proper attachment between mitotic spindles and kinetochores. The checkpoint thus inhibits anaphase entry when spindles do not attach to the kinetochores properly. Components of the spindle checkpoint are mitotic-arrest-defective genes (MAD1, MAD2, MAD3) and the budding uninhibited by benzimidazole genes (BUB1 and BUB3) (Amon 1999). There is a functional difference between BUB and MAD genes. Deletion of BUB1 or BUB3 causes chromosome mis-segregation compared to the deletion of MAD genes (Warren et al. 2002). Bub1p and Bub3p are recruited to the kinetochore in early mitosis independently from spindle–kinetochore attachment status, whereas Mad1p and Mad2p are bound to kinetochores in response to the unattached kinetochores (Gillett et al. 2004). Thus, unlike Mad1p and Mad2p, Bub1p and Bub3p have functions that are independent and distinct from their checkpoint function in chromosome segregation. In this study, we discuss the genetic interactions between CLB5 and spindle checkpoint genes, emphasizing the difference between Mad and Bub proteins.
MATERIALS AND METHODS
Strains and plasmids:
The deletion sets used in this study were obtained from EuroScarf and are derivatives of BY4741 (Winzeler et al. 1999). All other strains used are derivatives of W303 (strain list in Table 1). Standard methods were used for mating and tetrad analysis. The deletion strains mad2 and bub3 were generated by PCR-mediated homologous recombination using genomic DNA obtained from haploid deletion sets to produce the deletion strains in a W303 background. DNA transformation was performed by the lithium acetate method (Gietz et al. 1992). The ORC6-rxl, clb5-hpm, and clb5pCLB2 alleles were described previously (Cross et al. 1999; Cross and Jacobson 2000; Wilmes et al. 2004). A clb5 temperature-sensitive mutant was generated by error-prone mutagenesis by PCR (Leung et al. 1989). The PCR reaction buffer contains 16.6 mm (NH4)2SO4; 67 mm Tris–HCl, pH.8.8; 6.7 μm EDTA, pH.8.0, in 0.7 mg/ml BSA; 10 mm β-mercaptoethanol (BME); 10% DMSO; MnCl2 6 mm; and 1-μm primers of 1 mm dNTP each and 1 μl Taq Polymerase] in a 100-μl reaction. The CLB5 was amplified from the CLB5-HA-TRP1 plasmid (FC408) using the primers 5′-GATGATAATAGTAGTAATACTGGTGG-3′ and 5′-GCTTTAGGTG ATTGAGTCTC TTGAAG-3′. The amplified PCR products with potential mutations and FC408 plasmid digested with BamHI and EcoRI were cotransformed into a bub3 clb5 strain containing a CLB5-URA3 plasmid. The transformants were selected on SC-Trp plates followed by an SC + FOA selection at 23° to select the loss of the CLB5-URA3 plasmid. A transformant that was inviable upon transfer to 37° contained a plasmid-borne temperature-sensitive clb5 gene, which we named clb5-1, containing two substitutions, T246S and F428L, determined by sequencing. This allele was transferred to an integrating TRP1 plasmid and integrated into the genome at trp1 in a clb5 background to generate a clb5-1 strain. To generate a conditional clb3 strain, we placed CLB3 under control of GALL, an attenuated version of the GAL1 promoter (Mumberg et al. 1994, 1995); GAL1-CLB3 was shown previously to be lethal (Lew et al. 1993). To do this, CLB3 was amplified by PCR using genomic DNA to add HindIII sites at both 5′- and 3′-ends. The primers were 5′-CCCAAGCTTATGCATCATAACTCACA-3′ and 5′-CCCAAGCTTTTAGTTAGATCTTTCTA-3′. The PCR product was cloned into the GALLp305 plasmid (Mumberg et al. 1994) cut with HindIII. This GALL-CLB3 plasmid was sequenced to confirm that there were no missense mutations. The GALL-CLB3 plasmid was cut with BstEII to target integration to leu2 and transformed into a clb3∷TRP1 clb4∷HIS3 strain. The clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 strain was then crossed with mad2∷KanMX to generate mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3. Since GALL-CLB3 strains showed significant growth defects upon galactose induction, a hormone-inducible system was employed to regulate CLB3 gene expression (Louvion et al. 1993; Picard 2000). Human estrogen receptor (HER) fused to the GAL4 promoter under control of the ADH1 promoter can regulate gene expression in response to the addition of estradiol (Picard 2000).
TABLE 1.
Strain list
| Strain name | Genetic background | Origin |
|---|---|---|
| RUY051 | MATα clb5∷URA3 mfa∷MFA1pr-HIS3 URA3 ura3-1 leu2,3-112 his3-11 trp1-1 can1-100 lys2-801 | This study |
| RUY112 | MATamad1∷KanMX ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY156 | MATα mad2∷KanMX ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY154 | MATα bub1∷KanMX ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY155 | MATα bub3∷KanMX ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY135 | MATα clb5-hpm ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | Cross and Jacobson (2000) |
| RUY388 | MATα clb5∷CLB2 [pRS416] ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | Cross et al. (1999) |
| RUY292 | MATabub3∷KanMX clb5∷URA3 [clb5-1∷TRP1] | This study |
| RUY351 | MATα bub3∷KanMX clb5∷URA3 TRP1∷clb5-1 ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY360 | MAT? bub3∷KanMX clb5∷URA3 TRP1∷clb5-1 TUB1-GFP∷HIS3 CDC10-GFP∷LEU2 ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY430 | MATa bub3∷KanMX clb5∷URA3 TRP1∷clb5-1 TUB1-GFP∷HIS3 MYO1-GFP∷KanMX ura3-1 leu2,3-112 his3-11 trp1-1 can1-100 | This study |
| RUY443 | MATa bub3∷KanMX clb5∷URA3 TRP1∷clb5-1 ipl1-321 ura3-1 leu2,3-112 his3-11 trp1-1 can1-100 | ipl1-321 is from Sue Biggins |
| RUY590 | MATα clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY605 | MAT? mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 | This study |
| RUY610 | MATα clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 GAL4-HER-URA3 ura3-1 leu2,3-112 his3-11 trp1-1 can1-100 | This study |
| RUY650 | MATabar1 clb3∷TRP1 clb4∷HIS3 PDS1-13xMYC∷LEU2 ura3-1 leu2,3-112 his3-11 trp1-1 can1-100 | This study |
GAL4-HER plasmid marked by URA3 was cut with PakI in the ADH1 promoter and was transformed into the mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3, and the transformants were selected on D-Ura plates, generating mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 GAL4-HER∷URA3.
Modified synthetic genetic array analysis:
Our genetic screen is a method modified from SGA analysis (Tong et al. 2001). Genes that are related to the DNA damage checkpoint response, DNA metabolism, and cell cycle regulation were selected on the basis of the Saccharomyces Genome Database Gene Ontology annotations (the selected 87 deletion strains are listed in the supporting information, Table S1). A clb5 deletion strain was used as a query strain to be crossed with the selected deletions using the large-patch-format screen (some deletions were not tested due to their initial slow growth; see Table S1) (Archambault et al. 2005). The query strain MATαclb5∷URA3 mfa∷MFA1pr-HIS3 trp1 ade2 can1 leu2 his3 lys2 ura3 was spread on yeast extract–peptone–dextrose plates. The selected deletion mutant arrays (genotype MATa TRP1 ADE2 met15 leu2 ura3 his3 geneX∷kanMX) were mated to the query cells, and the cells were incubated for 1 day. The resulting zygotes were selected on SC-Min plus Leu and His to allow the growth of diploid cells, which were then printed to sporulation medium plates and incubated for 5 days at 22°. To select haploid MATa mfa∷MFA1pr-HIS3 clb5∷URA3 spore, progeny spores were selected onto haploid selection medium (Tong et al. 2001). The MATa haploids were then printed to YPD medium containing G418 and grown for 1 day. Finally, double mutants were selected on SC-His-Ura-Arg plus canavanine plus G418 for 2 days, and the proliferation of the haploid clb5∷URA3 geneXΔ∷KanMX cells was scored visually. We confirmed the deletion strains by PCR to test if the gene is correctly disrupted. This large-patch format allowed us to perform the screening with fewer false-positive results; we found previously that the small-dot format using a pin tool gave a very high false-positive rate in our hands (Archambault et al. 2005). In this study, we did not have any false-positive hits using the large-patch method.
Serial dilutions:
Cells were incubated overnight in 3 ml liquid media, and cell concentration was normalized on the basis of OD measurement. Five 10-fold serial dilutions were plated using a multiple pipetter. The plates were incubated at the indicated temperature for 2 days.
Microscopy:
Fluorescent images were collected at room temperature using Axiovert 200 (Carl Zeiss, Thornwood, NY) fitted with a cooled charge-coupled device camera (Orca ER; Hamamatsu) with a ×60 objective lens (NA 1.4). The images were analyzed with Openlab software (Improvision, Coventry, UK). For Figure 4B, the images were processed by custom software written in Matlab, which provides nonlinear contrast enhancement to bring out spindle morphology and draws cell outlines on the basis of background staining in the original images. These images are presented in Figure 4B solely for illustration because the long spindles were difficult to reproduce photographically from the original images; the quantitative data were obtained by scoring the original images, which were very clear upon direct inspection.
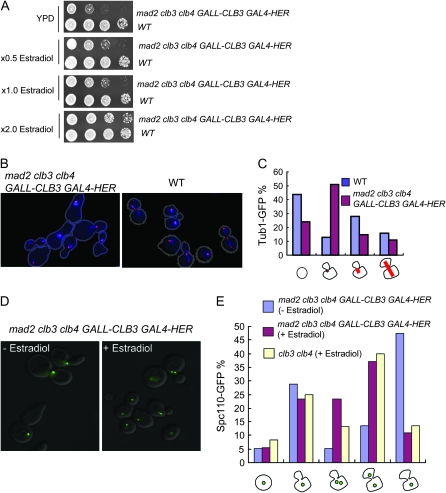
Figure 4.—
The mad2 clb3 clb4 strain has defects in SPB separation and spindle assembly. (A) A conditional mutant of the mad2 clb3 clb4 strain was generated. The mad2 clb3 clb4 GALL-CLB3 GAL4-HER or wild-type strain was plated on YPD, YPD plus 0.5× estradiol, YPD plus 1× estradiol, and YPD plus 2× estradiol. The plates were grown at 30° for 2 days to test viability. (B) The mad2 clb3 clb4 GALL-CLB3 GAL4-HER TUB1-CFP or TUB1-CFP strain was incubated in YPD with estradiol first, and the estradiol was removed for 6 hr to turn off CLB3. The cells were then briefly fixed with formaldehyde and stained with DAPI. They were observed under a fluorescent microscope with a cyan fluorescent protein (red image) and DAPI (blue image) channel. The images were merged and contrast enhanced using MatLab software to visualize the fluorescent signals for reproduction. These manipulations improved image reproducibility when printed but had no effect on the qualitative interpretation (original images available upon request). (C) The cells were categorized into four groups in which cells have no spindles, spindles as a dot, short spindles, or elongated spindles. At least 100 cells were counted for each sample, and the percentage of each category was calculated. Note that this scoring was performed with the unenhanced images. (D) The mad2 clb3 clb4 GALL-CLB3 GAL4-HER SPC110-GFP or CLB3 CLB4 SPC110-GFP strain was grown in YPD with estradiol, and then estradiol was removed for 6 hr. The cells were observed under a fluorescent microscope to visualize Spc110-GFP. The fluorescent image and DIC image were merged. (E) Cell images such as those in D were counted and categorized into four groups (from left to right below the x-axis): single cells with one SPB; small-budded cells with one SPB; small-budded cells with two SPB; large-budded cells with two separated SPB in the mother and daughter cells; and large-budded cells with one SPB.
Time-lapse microscopy:
Cells were prepared and imaged as described (Bean et al. 2006; Di Talia et al. 2007). Briefly, the fluorescent image of the microcolonies was collected with fluorescence time-lapse microscopy at 37°. The images were acquired every 3 min for 8 hr with a Hamamatsu Orca-ER camera. Visual Basic software integrated with ImagePro Plus was used to automate image acquisition and microscope control.
Multicopy suppressor screening:
bub3 clb5-1 cells were transformed with a YEp13 genomic DNA library from ATCC (Broach 1979). The transformants were first obtained at permissive temperature on SC-LEU plates and then replica plated to SC-LEU plates at 37°. The colonies that grew at 37° were selected, and the transforming genomic plasmids were rescued and end sequenced to identify the genomic segment. Plasmid rescue was confirmed by retransformation.
DAPI staining:
Cells were fixed with freshly made 4% paraformaldehyde solution for 10 min at room temperature. The fixed cells were washed with 30% ethanol for 1 min. The cells were then washed with sorbitol–phosphate buffer (1.2 m sorbitol/100 mm KPO4/100 μm MgCl2) twice and finally stained with 0.5 μg/ml DAPI solution (Niepel et al. 2005).
α-Factor block release and Western blotting:
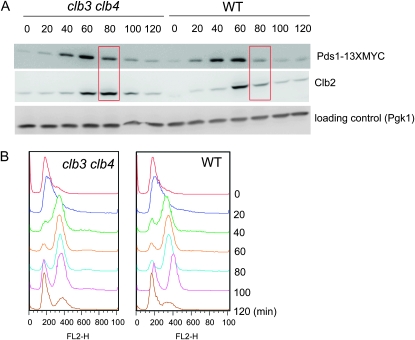
Log-phase YPD cultures were arrested by adding α-factor at a final concentration of 50 nm for 2.5 hr at 30°. At least 100 cells were counted and >99% of the cells were unbudded. α-Factor was removed by centrifugation, the cells were resuspended in YPD, and samples were collected every 20 min for 120 min. α-Factor was added back 40 min after release from the block to provide a single-cycle experiment. Pds1-13XMYC was visualized using anti-MYC antibody (9E10) (Santa Cruz, CA) by Western blotting analysis. In the same sample, Clb2 expression was also monitored using anti-Clb2 antibody (laboratory stock).
RESULTS
clb5 deletion results in a requirement for the spindle integrity checkpoint proteins BUB1 or BUB3:
We conducted a modified SGA screening using selected haploid deletion sets to identify genes required for cell growth in the clb5 deletion strain. Sixty-nine deletions in genes with functions related to the DNA damage checkpoint and DNA metabolism, and 18 deletions in genes with functions related to cell cycle regulation, were selected based on Saccharomyces Genome Database Gene Ontology annotations (Table S1). These 87 deletion mutants were used for the SGA analysis (materials and methods). Only bub1, bub3, and mck1 caused synthetic lethality in combination with clb5. These results from SGA analysis were confirmed by tetrad analysis (Figure 1A and data not shown). The genetic interaction between bub1 and bub3 with clb5 was reported previously by SGA analysis using bub1 or bub3 as query strains (Daniel et al. 2006). However, there is no study to show the molecular mechanism of the lethality between spindle assembly checkpoint genes and B-type cyclins.
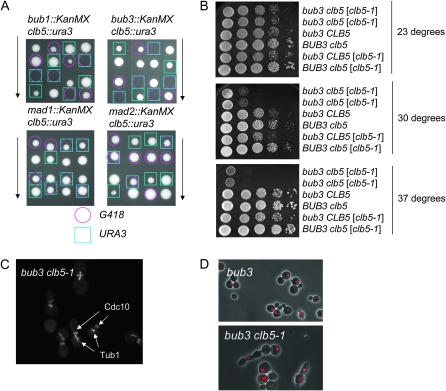
Figure 1.—
Cytokinesis and spindle elongation defects in bub3 clb5-1 cells. (A) Tetrad analysis was performed using bub1, bub3, mad1, or mad2 deletion strains crossed to a clb5 deletion strain. The colonies with a pink circle are bub1, bub3, mad1, or mad2 deletions marked by G418 resistance. The colonies with a blue square are clb5 deletion marked by URA3. (B) bub3 clb5 (clb5-1) (i.e., with the genomic copy of CLB5 disrupted, but carrying an episomal plasmid with the clb5-1 temperature-sensitive allele; see text) were inviable at 37°. Strains with indicated genotypes were plated on YPD plates at 10-fold serial dilution in horizontal lanes. The plates were incubated at 23°, 30°, or 37° for 2 days. (C) bub3 clb5-1 cells are defective in spindle elongation. The bub3 clb5-1 CDC10-GFP TUB1-GFP strain was incubated at 23° first, and then the temperature was shifted to 37° for 4 hr. Cells were observed under a fluorescent microscope to visualize Cdc10-GFP and Tub1-GFP. (D) bub3 clb5-1 cells showed cytokinesis defects at 37°. The bub3 clb5-1 strain was incubated at 23° first, and then the temperature was shifted to 37° for 6 hr. The cells were subject to ethanol fixation followed by RNase treatment. Propidum iodide was added to stain DNA. Fluorescent images and phase-contrast images were merged.
Bub1 and Bub3 are spindle checkpoint proteins. Bub3 binds to Mad2 to inhibit Cdc20 activity and arrest the cell cycle until the bipolar spindle is formed (Hwang et al. 1998; Kim et al. 1998). The spindle checkpoint is activated and arrests the cell cycle at the metaphase–anaphase transition (Amon 1999; Pinsky and Biggins 2005). Thus, cells can faithfully segregate their sister chromatids during mitosis. Bub1 also recruits Sgo1 to the kinetochore. Bub1 kinase and Sgo1 have roles in maintaining efficient sister-chromatid biorientation during mitosis (Fernius and Hardwick 2007). Perhaps related to this common role, SGO1, like BUB1, is essential in the absence of Clb5.
We tested if clb5 cells require other spindle checkpoint components such as Mad1 or Mad2 for viability. Mad1 binds to Mad2 and recruits Mad2 to the kinetochore (Chen et al. 1998; Ikui et al. 2002). In contrast to the results above, the clb5 mad1 or clb5 mad2 double-mutant cells were viable (Figure 1A). We next examined if the hydrophobic patch region in Clb5, a substrate-binding region in cyclins (Schulman et al. 1998; Cross and Jacobson 2000), is required for viability in the bub3 cells. The clb5-hpm bub3 cells either died or formed small colonies in tetrad analysis (Table 2, boldface). In addition, the sgo1 clb5-hpm double mutant is inviable; thus the hydrophobic patch-binding motif in Clb5 is required for viability in the absence of sgo1.
TABLE 2.
Summary of genetic interactions
| Δbub3 | Δmad2 | Δsgo1 | |
|---|---|---|---|
| ORC6-rxl | Viable | Viable | Viable |
| Δclb5, Δclb6 | Δclb5 lethal | Viable | Δclb5 lethal |
| Δclb2, Δclb4 | Viable | Viable | Viable |
| Δclb1, Δclb3, Δclb4 | Δclb3 Δclb4 lethal | Δclb3 Δclb4 lethal | Viable |
| clb5-hpm | Sick | Viable | Sick |
| clb5pCLB2 | Lethal | Viable | Viable |
Boldface indicates the genetic interaction between clb5 mutants and bub3 or sgo1. Underlining indicates the genetic interaction between the clb3 clb4 mutant and bub3 or mad2.
Clb5 interacts with Orc6 through the hydrophobic patch region in Clb5 (Wilmes et al. 2004). We considered the possibility that the requirement for the Clb5 hydrophobic patch in the absence of BUB3 might be specifically related to lack of Clb5-Orc6 interaction. However, ORC6-rxl, encoding a mutant Orc6 that does not bind Clb5, did not cause synthetic lethality in the bub3 strain (Table 2). Therefore, synthetic lethality between bub3 and clb5 likely required protein binding between Clb5 and other protein(s). Numerous proteins have been shown to interact with Clb5 in a hydrophobic-patch-dependent manner (Loog and Morgan 2005).
Next we tested cyclin specificity using the clb5∷CLB2 allele, in which the mitotic cyclin CLB2 is under the control of the CLB5 promoter (Cross et al. 1999), providing a control for the decrease in total B-type cyclin expression due to deletion of CLB5. Interestingly, the bub3 clb5 clb5∷CLB2 double mutant was inviable (Table 2). We conclude that Clb2 cannot substitute for Clb5 to provide the function required in the absence of BUB2, even when expressed early in the cell cycle.
clb5 bub1 and clb5 bub3 strains were defective in spindle elongation and cytokinesis:
A conditional mutant was generated to further study the lethality in the bub3 clb5 double mutant. We first attempted to make the bub3 clb5 GAL-CLB5 strain so that we could remove Clb5 by transfer from galactose to glucose. However, the bub3 clb5 GAL-CLB5 strain was not fully viable on galactose, suggesting that the bub3 deletion causes sensitivity to the Clb5 expression level. We therefore generated a temperature-sensitive mutant of CLB5 by PCR mutagenesis. The bub3 clb5 mutant containing the clb5-1 temperature-sensitive allele on a low-copy-number plasmid [bub3 clb5 (clb5-1)] was viable at the permissive temperature of 23°, but inviable at 37° (Figure 1B). The clb5-1 mutation was then integrated at the TRP1 locus in a bub3 background. The cell morphology of the resulting bub3 clb5-1 cells was examined at 37°. To visualize the spindle and septin ring in the cells, we employed Tub1 tagged with green fluorescent protein (TUB1-GFP) and Cdc10 tagged with GFP (CDC10-GFP). Although Tub1-GFP and Cdc10-GFP proteins are in the same color, they can be distinguished on the basis of their localization. Cdc10 is present in a ring at the bud neck throughout the budded period, and tubulin is assembled into a spindle that elongates during anaphase in wild-type, bub3, or clb5 cells. bub3 clb5-1 cells formed short spindles near the bud neck (Figure 1C); most of these cells contained a septin ring at the bud neck. To visualize spindle movement, we employed time-lapse microscopy with 3-min resolution. The short spindles moved back and forth between mother and daughter cells around the bud neck area (File S1). Thus, the cells failed to elongate spindles. When the bub3 clb5-1 cells were incubated for a longer period at the nonpermissive temperature, the cells exhibited defects in cytokinesis or cell separation (Figure 1D). bub3 clb5-1 cells underwent several cell divisions at 37° but were not capable of continued proliferation, as expected.
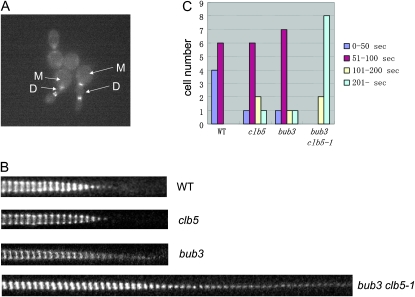
To distinguish between cytokinesis and cell separation defects, bub3 clb5-1 cells were incubated at 37° for 8 hr and treated with zymolase after the fixation. Before the zymolase treatment, 19.4% of the cells exhibited bibudded cells. After the zymolase treatment, bibudded cells did not separate, and 19.1% of the cells still remained attached (data not shown). Therefore, the bub3-clb5-1 combination caused a cytokinesis defect rather than a cell separation defect. Cytokinesis is a multi-step process. The only type II myosin in S. cerevisiae, Myo1, is recruited to the bud neck before bud emergence and stays at the bud neck until anaphase. Myo1 contraction is a key step for cytokinesis progression. Myo1-GFP was visualized using time-lapse microscopy to determine which cytokinesis step is defective in bub3 clb5-1 cells at the nonpermissive temperature (Figure 2A). Mutant cells frequently rebud before they contract the Myo1 ring on the bud neck between mother and daughter cells. This suggests that cells had defects in Myo1 contraction. This result was confirmed by karyograph experiments. Images were taken every 30 sec and the Myo1-GFP at the bud neck area was analyzed. In the wild type, Myo1 contracted within 10 min. In contrast, the bub3 clb5 mutants did not contract the Myo1 ring even after 20 min (Figure 2B). A single mutant, bub3 or clb5, contracted Myo1 normally similar to wild-type cells. The results were quantified on the basis of how long Myo1-GFP stayed on the bud neck. Ten cells for each genotype were monitored by time-lapse microscope. Myo1-GFP disappeared from the bud neck within 100 sec of initiation of contraction in all wild-type cells, whereas none of the bub3 clb5-1 cells completed contraction of the Myo1-GFP within 100 sec. Most of them required >200 sec for ring disappearance (Figure 2C).
Figure 2.—
Myo1 ring contraction is defective in the bub3 clb5-1 mutant. (A) Cells were incubated in YPD liquid media at 23° and put on SC-complete media containing agarose at low density. A movie was taken at 37° as described in materials and methods. The image is from the picture after 8.75 hr at 37°. M indicates mother cells, and D indicates daughter cells. (B) Myo1-GFP images at the bud neck were taken every 30 sec with time-lapse fluorescent microscopy. The images were trimmed to focus on the bud neck area, and at least 40 images were put next to each other to follow the myosin ring contraction. (C) The time length of Myo1-GFP expression on the bud neck was analyzed in the time-lapse images. Ten cells for each genotype were monitored to monitor how many seconds Myo1-GFP stayed on the bud neck.
Overexpression of the C-terminal fragment of Bir1 suppressed bub3 clb5 lethality:
To understand the lethality in bub3 clb5 cells, we conducted a multi-copy suppressor screen. A Yep13 yeast genomic DNA library was transformed into the bub3 clb5-1 cells, and we looked for plasmids that complemented or suppressed the temperature sensitivity of bub3 clb5-1. We identified three different plasmids (Figure 3A). Two of them had overlapping regions that contained the BUB3 sequence, an expected positive. Another plasmid contained the C-terminal region of BIR1 (a yeast homolog of survivin) and the C-terminal region of GRR1 (an F-box protein component of the SCF ubiquitin–ligase complex). Both the BIR1 and GRR1 fragments in this plasmid lack the promoter regions as well as the N-terminal coding sequence. The C-terminal fragment of Bir1 was fused to the GPD promoter. The GPD promoter allows cells to express protein at a high level constitutively (Mumberg et al. 1995). The resulting plasmid, C′-BIR1/GPDpRS423, rescued lethality in bub3 clb5-1 at the nonpermissive temperature (Figure 3B). Interestingly, the C′-BIR1/GPDpRS423 also rescued lethality in sgo1 clb5-1 at 37° (data not shown). This indicates that lethality in bub3 clb5 and sgo1 clb5 might be due to the same mechanism. Presumably, the C-terminal BIR1 fragment in the Yep13 plasmid, lacking its own promoter, was expressed from cryptic promoters in the vector.
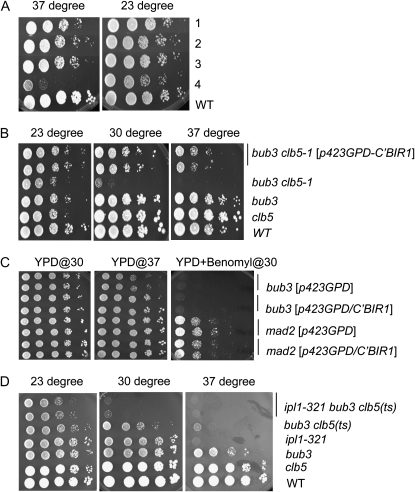
Figure 3.—
The C-terminal fragment of BIR1 suppressed lethality of bub3 clb5-1. (A) Three types of dosage-suppressor plasmids were identified for the clb5-1 bub3 strain. Row 1, part of BIR1 and part of GRR1 (3×); row 2, CIN5, STI1, BUB3, and fragment chromosome from XIV (3×); row 3, AHC1, HST3, and BUB3 (2×); row 4, control (bub3 clb5-1). The numbers in parentheses indicate how many times the plasmid was obtained by the screening. The plasmids obtained from the screening were transformed into the bub3 clb5-1 strain, and cell viability was tested by serial dilution at either 23° or 37°. (B) The C-terminal fragment of BIR1 under the GPD promoter rescued lethality in the bub3 clb5-1 mutant at 37°. The pRS423-GPD-C′BIR1 plasmid was transformed into the bub3 clb5-1 strain. The transformant was grown, and cell viability was tested by serial dilution methods. The indicated strains were plated on YPD and incubated at 23°, 30°, or 37° for 2 days. (C) The C-terminal fragment of BIR1 under the GPD promoter rescued the temperature-sensitive phenotype in the bub3 deletion at 37°. The experiment was performed similarly to B. (D) ipl1-321 enhanced lethality in bub3 clb5-1. The indicated strains were serially diluted and plated on YPD. They were incubated at 23°, 30°, or 37° for 2 days.
bub3 cells grew slowly at 37°. The slow growth phenotype at 37° was rescued when the C-terminal fragment of BIR1 was overexpressed (Figure 3C). However, tests of benomyl sensitivity (benomyl partially depolymerizes microtubules, creating a requirement for the spindle integrity checkpoint) showed that the C-terminal fragment of BIR1 does not rescue the spindle checkpoint defect in bub3 cells.
Bir1 binds to Sli15 (INCENP) and Ipl1 (Aurora B) to constitute the chromosomal passenger complex (CPC). The CPC plays a role in chromosome segregation and cytokinesis, and it is conserved from yeast to higher eukaryotes (Norden et al. 2006; Ruchaud et al. 2007). Ipl1 is a kinase and Sli15 and Bir1 are nonenzymatic subunits. Ipl1 is a key component in the CPC, and Bir1 regulates Ipl1 function (Kelly and Funabiki 2009). We tested if reduction of Ipl1 activity affects bub3 clb5-1 lethality. Cell viability in bub3 clb5-1 at 37°, although already low, was further reduced when it was combined with ipl1-321 (Figure 3D). Thus, loss of Ipl1 did not bypass, and could enhance, lethality in bub3 clb5.
Overall, we conclude that the requirement for Bub1, Bub3, and Sgo1 but not for Mad2 in a clb5 or clb5-hpm background, and the genetic interactions with chromosome passenger complex proteins, is due to a function of Clb5 related to kinetochore function and/or cytokinesis, rather than to a requirement for Clb5 in the absence of the spindle integrity checkpoint.
B-type cyclin requirements in spindle checkpoint mutants:
Our SGA analysis was carried out only with clb5. Therefore, we asked whether other B-type cyclins are required in cells mutant for various spindle checkpoint components. The ORC6-rxl, clb5 clb6, clb2 clb4, clb1 clb3 clb4, clb5-hpm, or clb5pCLB2 strain was crossed with bub3, mad2, or sgo1. The results are summarized in Table 2. In addition to the genetic interaction between bub3 and clb5 described above, we found that bub3 was essential in the absence of CLB3 and CLB4 (Table 2, underlining). The clb3 clb4 strain also required Mad2 for viability (Table 2, underlining). This indicates that lethality in clb3 clb4 bub3 or clb3 clb4 mad2 could be specifically dependent on the spindle integrity checkpoint, since clb3 clb4 cells, unlike clb5 cells, are dependent on Mad2 as well as on Bub3.
clb3 clb4 mad2 cells accumulate with unseparated spindle pole bodies:
A conditional mutant was generated to further study the lethal phenotype of mad2 clb3 clb4 cells. A hormone-inducible system was used to regulate CLB3 gene expression (Louvion et al. 1993; Picard 2000). The GAL4-HER promoter under control of the ADH1 promoter can regulate gene expression in response to the addition of estradiol (Picard 2000). We used this system rather than galactose to induce GALL-CLB3 because full induction of GALL-CLB3 with galactose was toxic (data not shown). The mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 GAL4-HER strain was viable when incubated in YPD supplemented with 200 nm estradiol, but showed growth defects when estradiol was removed from the medium (Figure 4A), and partial growth defects were observed at reduced estradiol levels. Tub1-CFP was observed as a dot in the mad2∷KanMX clb3∷TRP1 clb4∷HIS3 LEU2∷GALL-CLB3 GAL4-HER strain when incubated without estradiol. The phenotype was distinct from that in bub3 clb5-1 (Figure 4, B and C). We tested a similar strain containing Spc110-GFP, a SPB component, to monitor SPB separation. Spc110-GFP was observed in a single dot in most cells, suggesting a failure of SPB duplication or separation in the mad2 clb3 clb4 strain (Figure 4D).
Transient activation of the spindle checkpoint in the absence of Clb3 and Clb4:
Dependence of clb3 clb4 cells on MAD2 and BUB3, which are nonessential genes in a CLB3 CLB4 background, suggests that the spindle integrity checkpoint may be transiently activated in MAD2 clb3 clb4 cells. Spindle checkpoint activation is achieved by Mad2 binding to Cdc20, an activator for the APC. The APC promotes the degradation of Pds1 (securin), which is an inhibitor of Esp1 (separase). Separase cleaves cohesion to promote sister-chromatid separation during anaphase. Therefore, spindle checkpoint activation can be indirectly monitored by the timing of Pds1 degradation. The cell cycle was arrested at the G1 phase by α-factor block, and Pds1 levels were monitored by Western blot after release, using Myc-tagged Pds1. We observed about a 20-min delay in Pds1 degradation in the absence of Clb3 and Clb4, even though DNA replication was completed on schedule (Figure 5). Similarly, the timing of Clb2 degradation was delayed in the clb3 clb4 strain compared to wild-type cells; Cdc20-APC is responsible for initial Clb2 degradation (Yeong et al. 2000) (Figure 5A). This result suggests that the spindle checkpoint was activated transiently in the clb3 clb4 strain. This is likely due to some slowing or defect in the spindle assembly in the absence of Clb3 and Clb4, resulting in a Mad2-dependent delay that is required for viability. clb3,4,5,6 strains were reported to have a severe defect in spindle morphogenesis (Schwob and Nasmyth 1993).
Figure 5.—
Spindle integrity checkpoint activation in the clb3 clb4 double mutant. (A) Western blotting analysis showed that Pds1 degradation was delayed in the clb3 clb4 strain. Cells were synchronized at G1 phase by α-factor, and the arrest was released by removing the α-factor from the media. Samples were taken every 20 min for 120 min, and Pds1-13XMYC and Clb2 were visualized. Anti-3-phosphoglycerate kinase (Pgk) antibodies were used for the loading control. The experiments were repeated twice, and the same results were obtained from both experiments. (B) FACS analysis was performed using the same samples obtained from the α-factor block and release. The samples were fixed with ethanol and stained with propidium iodide.
CLB2 rescued lethality in clb3 clb4 mad2 cells:
To better understand the lethality of clb3 clb4 mad2 cells, suppressor gene screening was performed. We obtained three types of plasmids, MAD2, CLB4, and CLB2. We did not pick up CLB3, probably because overexpression fo Clb3p is toxic (see above). Clb2, the main mitotic cyclin, can substitute for Clb3 and Clb4 to restore viability in the absence of MAD2.
We next asked if the C-terminal BIR1 fragment can rescue the lethality of clb3 clb4 mad2 cells. In contrast to bub3 clb5, clb3 clb4 mad2 lethality was not suppressed after the overexpression of the C-terminal BIR1 fragment. This result further supports the idea that the lethality in bub3 clb5 and in clb3 clb4 mad2 is caused by distinct mechanisms.
DISCUSSION
A role of Bub kinases in maintaining viability in the absence of Clb5:
Bub1 and Bub3 kinases likely have a separate function from spindle checkpoint activation. Deletion of Bub1 or Bub3 in budding yeast causes a severe chromosome mis-segregation rate compared to the deletion of Mad2 (Warren et al. 2002). In higher eukaryotes, Bub proteins are recruited to the kinetochore before Mad2 recruitment, and Bub proteins stay on the kinetochore after spindle attachment to the kinetochore. Bub kinases may control kinetochore–microtubule interactions during prometaphase (Logarinho and Bousbaa 2008). Bub1 is known to recruit Shugosin (Sgo1) at the kinetochore. However, Sgo1 is not a spindle checkpoint protein; rather, it protects cohesion during meiosis, and Bub1 kinase and Sgo1 control sister-chromatid biorientation during mitosis (Fernius and Hardwick 2007). sgo1 clb5 synthetic lethality was observed (Table 2), and the sgo1 clb5-1 mutant showed cytokinesis defects similar to the bub3 clb5 strain (data not shown). This result implies that clb5 deletion results in a stringent requirement for genes that are involved in chromosome segregation. It has been reported that Clb5 has a function that regulates nuclear positioning (Segal et al. 1998) and spindle polarity (Segal et al. 2000).
During cytokinesis, the septin ring at the bud neck serves as a scaffold for localization of the contractile ring, Myo1 (Guertin et al. 2002). In cytokinesis, Myo1 is contracted followed by actin recruitment. The septin ring finally splits, and cell division is triggered by the final steps, furrow ingression and abscission. Our results show that the bub3 clb5 cells exhibited cytokinesis defects due to myosin ring contraction failure. A link between the cortical cycle and the nuclear cycle has been proposed. For example, the mitotic exit network (MEN) promotes mitotic exit and also contracts the actomyosin ring by targeting MEN kinase Dbf2/Mob1 to the cleavage site (Rauter and Barral 2006). It would be of interest to investigate whether defects in the nuclear cycle, such as chromosome mis-segregation, could trigger cytokinesis defects at a specific stage of mitosis.
A role of the CPC in spindle formation and cytokinesis:
It has been shown that the 10-kDa region at the C terminus in Bir1 is essential and is required for its localization as a CPC. The fragment also contains a binding region for Sli15 (Widlund et al. 2006). This small C-terminal fragment is almost exactly the segment that we recovered as a high-copy suppressor of bub3 clb5 inviability. In contrast, full-length BIR1 did not fully rescue bub3 clb5 lethality (data not shown), which raised two possibilities. We considered the possibility that the C-terminal fragment of Bir1 was acting as a dominant-negative mutant. To test this, the temperature-sensitive ipl1-321 allele was introduced into the bub3 clb5-1 background. If Ipl1 activity is high in the bub3 clb5-1 strain, and the dominant-negative mutant suppresses the Ipl1 activity, then introduction of ipl1-321 into bub3 clb5-1 should suppress lethality. However, ipl1-321 strongly enhanced the temperature sensitivity of bub3 clb5-1. Therefore, Ipl1 activity might be low in bub3 clb5 and restored by the C-terminal fragment of Bir1. The C-terminal fragment of Bir1 may be expressed at a higher level than full-length Bir1 (Widlund et al. 2006).
Although Bir1 and Sli15 are reported to be phosphorylated by Cdks, preliminary Western blot analysis suggests no effect on this phosphorylation of clb5 deletion, as judged by band shift (data not shown). We also tested Ipl1 localization in the clb5 cells. Ipl1-GFP was visualized in the synchronized clb5 deletion strain. However, Ipl1p was normally localized to spindles and kinetochore during mitosis in the clb5 strain (data not shown). We conclude that Clb5p does not control Ipl1 localization.
The C-terminal fragment of BIR1 also suppressed sgo1 clb5 lethality. In fission yeast, there are two shugoshin homologs, Sgo1 and Sgo2. Sgo1 primarily functions in meiosis to protect cohesion, whereas Sgo2 has a role in mitosis of maintaining proper chromosome segregation (Kawashima et al. 2007). Sgo2 and Bir1 make a complex that promotes CPC localization to the kinetochore. This allows cells to enable the tension-generating attachment between kinetochore and spindles. Forced localization of Bir1 to centromeres partially suppressed the defects of sgo2 (Kawashima et al. 2007). These interactions are consistent with our results linking Sgo1 and Bir1 via genetic interactions with CLB5 and BUB3.
Requirement for the spindle checkpoint in the absence of CLB3 and CLB4:
It has been reported that the mitotic cyclins Clb1, Clb2, and Clb3 trigger spindle elongation and activate APC/Cdc20 (Rahal and Amon 2008). In their study, they used the conditional mutant of clb1 clb2, clb2 clb3, or clb1 clb2 clb3 to reveal that Pds1 degradation and Scc1 cleavage did not occur in these strains. In our experiments with the clb3 clb4 double mutant, Pds1 degradation and spindle elongation eventually occurred, but the requirement for the spindle integrity checkpoint and the delay in Pds1 degradation suggests that spindle morphogenesis is slow or defective in these cells. The lethal phenotype seen in mad2 clb3 clb4 could be due to entry into mitosis with a defective spindle; we speculate that this then results in failure of SPB separation in subsequent cell cycles on the basis of the fact that it takes 3–6 hr to show the SPB separation defects. Suppressor gene screening using a conditional mad2 clb3 clb4 strain revealed that overexpression of CLB2 rescued lethality of mad2 clb3 clb4 cells. Clb2 may have an overlapping role for a function in spindle morphogenesis normally carried out primarily by Clb3 and Clb4.
Acknowledgments
We thank Sue Biggins for the ipl1-321 strain and Trisha Davis for the BIR1 plasmid. We also thank Ying Lu for the critical reading of the manuscript. A.E.I. was supported by National Institutes of Health (NIH) training grant T32 CA09673. This work was supported by NIH grant R01 GM047238 to F.R.C.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.105148/DC1.
References
- Amon, A., 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9: 69–75. [DOI] [PubMed] [Google Scholar]
- Archambault, V., A. E. Ikui, B. J. Drapkin and F. R. Cross, 2005. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 25: 6707–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, J. M., E. D. Siggia and F. R. Cross, 2006. Coherence and timing of cell cycle start examined at single-cell resolution. Mol. Cell 21: 3–14. [DOI] [PubMed] [Google Scholar]
- Broach, J. R., J. N. Strathern and J. B. Hicks, 1979. Transformation in yeast: development of a hybrid cloning vector and isolation of the can1 gene. Gene 8: 121–133. [DOI] [PubMed] [Google Scholar]
- Chen, R. H., A. Shevchenko, M. Mann and A. W. Murray, 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143: 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., and M. D. Jacobson, 2000. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20: 4782–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., M. Yuste-Rojas, S. Gray and M. D. Jacobson, 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4: 11–19. [DOI] [PubMed] [Google Scholar]
- Daniel, J. A., B. E. Keyes, Y. P. Ng, C. O. Freeman and D. J. Burke, 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 172: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia, S., J. M. Skotheim, J. M. Bean, E. D. Siggia and F. R. Cross, 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448: 947–951. [DOI] [PubMed] [Google Scholar]
- Drury, L. S., G. Perkins and J. F. Diffley, 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10: 231–240. [DOI] [PubMed] [Google Scholar]
- Epstein, C. B., and F. R. Cross, 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6: 1695–1706. [DOI] [PubMed] [Google Scholar]
- Fernius, J., and K. G. Hardwick, 2007. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 3: e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth et al., 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., A. St. Jean, R. A. Woods and R. H. Schiestl, 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett, E. S., C. W. Espelin and P. K. Sorger, 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin, D. A., S. Trautmann and D. McCollum, 2002. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66: 155–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, L. H., L. F. Lau, D. L. Smith, C. A. Mistrot, K. G. Hardwick et al., 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Ikui, A. E., K. Furuya, M. Yanagida and T. Matsumoto, 2002. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115: 1603–1610. [DOI] [PubMed] [Google Scholar]
- Kawashima, S. A., T. Tsukahara, M. Langegger, S. Hauf, T. S. Kitajima et al., 2007. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 21: 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A. E., and H. Funabiki, 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. H., D. P. Lin, S. Matsumoto, A. Kitazono and T. Matsumoto, 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science 279: 1045–1047. [DOI] [PubMed] [Google Scholar]
- Labib, K., J. F. Diffley and S. E. Kearsey, 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1: 415–422. [DOI] [PubMed] [Google Scholar]
- Leung, D. W., E. Chan and D. W. Goeddel, 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Techniques 1: 11–15. [Google Scholar]
- Lew, D. J., and S. I. Reed, 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liku, M. E., V. Q. Nguyen, A. W. Rosales, K. Irie and J. J. Li, 2005. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol. Biol. Cell 16: 5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho, E., and H. Bousbaa, 2008. Kinetochore-microtubule interactions “in check” by Bub1, Bub3 and BubR1: the dual task of attaching and signalling. Cell Cycle 7: 1763–1768. [DOI] [PubMed] [Google Scholar]
- Loog, M., and D. O. Morgan, 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108. [DOI] [PubMed] [Google Scholar]
- Louvion, J. F., B. Havaux-Copf and D. Picard, 1993. Fusion of GAL4–VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 131: 129–134. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., 2003. Targets of the cyclin dependent kinase Cdk1. Nature 425: 859–864. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22: 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., 1993. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr. Opin. Cell Biol. 5: 166–179. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co, K. Irie and J. J. Li, 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr. Biol. 10: 195–205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co and J. J. Li, 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073. [DOI] [PubMed] [Google Scholar]
- Niepel, M., C. Strambio-de-Castillia, J. Fasolo, B. T. Chait and M. P. Rout, 2005. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170: 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden, C., M. Mendoza, J. Dobbelaere, C. V. Kotwaliwale, S. Biggins et al., 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125: 85–98. [DOI] [PubMed] [Google Scholar]
- Picard, D., 2000. Posttranslational regulation of proteins by fusions to steroid-binding domains. Methods Enzymol. 327: 385–401. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., and S. Biggins, 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15: 486–493. [DOI] [PubMed] [Google Scholar]
- Rahal, R., and A. Amon, 2008. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 22: 1534–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauter, H., and Y. Barral, 2006. Cytokinesis goes polo. Dev. Cell 11: 136–137. [DOI] [PubMed] [Google Scholar]
- Richardson, H., D. J. Lew, M. Henze, K. Sugimoto and S. I. Reed, 1992. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6: 2021–2034. [DOI] [PubMed] [Google Scholar]
- Ruchaud, S., M. Carmena and W. C. Earnshaw, 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8: 798–812. [DOI] [PubMed] [Google Scholar]
- Schulman, B. A., D. L. Lindstrom and E. Harlow, 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95: 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., and K. Nasmyth, 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7: 1160–1175. [DOI] [PubMed] [Google Scholar]
- Segal, M., D. J. Clarke and S. I. Reed, 1998. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J. Cell Biol. 143: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, M., D. J. Clarke, P. Maddox, E. D. Salmon, K. Bloom et al., 2000. Coordinated spindle assembly and orientation requires Clb5p-dependent kinase in budding yeast. J. Cell Biol. 148: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., A. Toth, M. Galova and K. Nasmyth, 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402: 203–207. [DOI] [PubMed] [Google Scholar]
- Surana, U., H. Robitsch, C. Price, T. Schuster, I. Fitch et al., 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65: 145–161. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Warren, C. D., D. M. Brady, R. C. Johnston, J. S. Hanna, K. G. Hardwick et al., 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13: 3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasch, R., and F. R. Cross, 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418: 556–562. [DOI] [PubMed] [Google Scholar]
- Widlund, P. O., J. S. Lyssand, S. Anderson, S. Niessen, J. R. Yates, III et al., 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell 17: 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes, G. M., V. Archambault, R. J. Austin, M. D. Jacobson, S. P. Bell et al., 2004. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Woodbury, E. L., and D. O. Morgan, 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9: 106–112. [DOI] [PubMed] [Google Scholar]
- Yeong, F. M., H. H. Lim, C. G. Padmashree and U. Surana, 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell 5: 501–511. [DOI] [PubMed] [Google Scholar]