Abstract
Background
Oral squamous cell carcinoma (OSCC) is one of the most common forms of cancer associated with the presence of precancerous oral leukoplakia. Given the poor prognosis associated with oral leukoplakia, and the difficulties in distinguishing it from cancer lesions, there is an urgent need to elucidate the molecular determinants and critical signal pathways underlying the malignant transformation of precancerous to cancerous tissue, and thus to identify novel diagnostic and therapeutic target.
Results
We have utilized two dimensional electrophoresis (2-DE) followed by ESI-Q-TOF-LC-MS/MS to identify proteins differentially expressed in six pairs of oral leukoplakia tissues with dysplasia and oral squamous cancer tissues, each pair was collected from a single patient. Approximately 85 differentially and constantly expressed proteins (> two-fold change, P < 0.05) were identified, including 52 up-regulated and 33 down-regulated. Gene ontological methods were employed to identify the biological processes that were over-represented in this carcinogenic stage. Biological networks were also constructed to reveal the potential links between those protein candidates. Among them, three homologs of proteosome activator PA28 a, b and g were shown to have up-regulated mRNA levels in OSCC cells relative to oral keratinocytes.
Conclusion
Varying levels of differentially expressed proteins were possibly involved in the malignant transformation of oral leukoplakia. Their expression levels, bioprocess, and interaction networks were analyzed using a bioinformatics approach. This study shows that the three homologs of PA28 may play an important role in malignant transformation and is an example of a systematic biology study, in which functional proteomics were constructed to help to elucidate mechanistic aspects and potential involvement of proteins. Our results provide new insights into the pathogenesis of oral cancer. These differentially expressed proteins may have utility as useful candidate markers of OSCC.
Backgound
Oral, head, and neck squamous cellular carcinoma is one of the most common forms of cancer associated with the presence of precancerous lesions. It is now believed that OSCC follows a similar pattern in its development, and thus is preceded by precancerous lesions, among which oral leukoplakia (OLK) is the most common type. The World Health Organization (WHO) first defined oral leukoplakia as a white plaque that could not be characterized clinically or pathologically as any other disease in oral mucosa. The malignant potential of oral leukoplakia was evidenced by the progression from metaplasia without dysplasia to low grade dysplasia, high grade dysplasia, and ultimately to invasive carcinoma [1]. The risk of developing malignancies is 8–10 times higher in people who have oral leukoplakia than people who do not [2]. The risk is also increasing with the series of dysplasia stages [3]. There is an urgent need to elucidate the molecular determinants and key signal pathways underlying the malignant transformation from precancerous to cancerous tissue, and thus to identify novel diagnostic and therapeutic targets.
Proteomics is an established molecular profiling technology that may significantly accelerate human cancer research. Recently, a lot of progress has been made in oral cancer proteomics generating some potential applications in this emerging field. This technology platform has been utilized to discover highly sensitive and specific protein markers for oral cancer diagnosis and prognosis by comparing the protein profiles of cancer cells [4,5], tissues [6], plasma [7], and saliva[8,9] with appropriate controls. However, there are fewer reports about discrimination of protein expression profiles between tumor and precancerous lesion with different stages of dysplasia.
The development of bioinformatics tools has allowed the compilation of searchable genomic and proteomic databases accessible via the Internet. Among them, the application of Gene Ontology (GO) and the pathway analysis was considered as a powerful tool in systematic biology for elucidating the complexity of expression profiles in cellular processes. The term of GO describes the role of a given gene in a biological process, its molecular function and cellular component. Each gene is provided with different levels of GO terms, ranging from high-level, broadly descriptive terms to very low-level, highly specific terms [10]. Thus, profiling the expression data based on GO will provide another dimension for understanding the key regulatory processes in oral cancer. The application of the pathway analysis reveals the interactions between the proteins, thus quickly generating new insights into potential complex molecular mechanisms underlying disease related processes [11].
In this study, we have evaluated protein expression differences to identify potential biomarkers of disease progress from oral leukoplakia to OSCC in order to gain further insight into potential mechanisms underlying these transformations. Six pairs of protein lysates were obtained from six patients. The tissues were analyzed by two-dimensional gel electrophoresis, followed by ESI-Q-TOF tandem mass spectrometry. GO analysis was applied to identify biological processes over-represented in the carcinogenesis. Biological networks were also constructed to reveal the potential links between the protein candidates. By using this approach, new therapeutic targets or protein markers can possibly be identified to improve patient survival.
Results
2-DE profiling of OSCC and the oral leukoplakia tissues
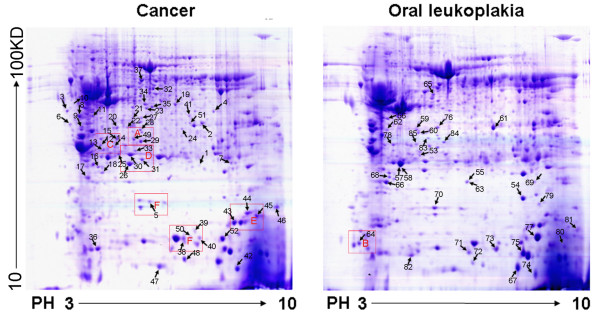
A total of 6 pairs of OSCC tumor tissues and the oral leukoplakia control tissues were obtained from 6 patients. Figure 1 showed their representative clinical photos and HE-staining histographs. 2-DE with immobilized pH gradients was performed to study the expression patterns of proteins extracted from both tissues, and each sample was analyzed twice to ensure the reproducibility. Figure 2 showed representative 2-DE patterns obtained from the paired tissues. After automatic spot detection, background subtraction, and volume normalization, 859 ± 68 protein spots in OSCC tissue, and 844 ± 56 protein spots in control tissues were detected. Of these spots, 730 (85%) in OSCC tissues and 708 (84%) in control tissues were reproducibly detected in all of the twelve runs, and only the reproducibly detected spots were subjected to statistical analysis(p < 0.05). As a result, 85 protein spots showed more than two fold changes in at least 4 of the twelve repeats (marked in figure 2) while 68 proteins (80%) repeated in more than 8 of 12 pairs.11 proteins were selected as examples (boxed in figure 2) showing the consistent expression changes in enlarged form (see additional file 1).
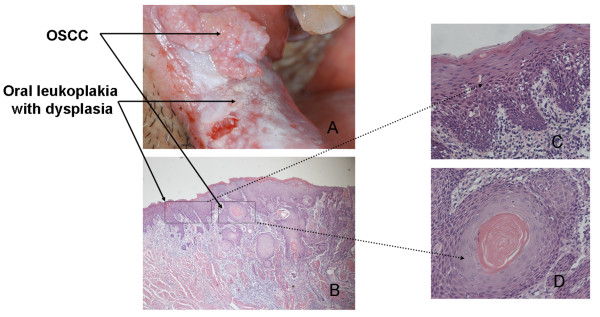
Figure 1.
Representative clinical and histological pictures. Representative clinical (A) and histological pictures (B, C, D) showing a pair of OSCC and oral leukoplakia tissues with dysplasia obtained from the same patient.
Figure 2.
Representative two-dimensional maps marked altered proteins. Representative two-dimensional maps of a pair of OSCC cancer tissue and precancerous oral leukoplakia tissue from the same patient. The proteins were separated on a pH 3–10 nonlinear IPG strip, followed by a 12% SDS-polyacrylamide gel, as stated under Methods. The gel was Coomassie-blue stained and the spots were analyzed by ESI-Q-TOF-LC-MS/MS. Arrows indicate identified protein spots significantly and consistently altered between carcinoma tissue and control tissue. 11 boxed proteins were selected as examples showing the consistent expression changes in enlarged form in additional file 1.
Identification of putative OSCC biomarkers
As shown in Table 1, a total of 85 differentially expressed proteins, including 52 up-regulated and 33 down-regulated proteins, were identified. Proteasome activator complex PA28 a and b were chosen for further validation. They both exhibited a high expression level in cancer tissues (4–6 fold increase) when compared with the precancerous OLK tissues. The mass spectra for both were shown in Figure 3.
Table 1.
Summary of protein alterations
| Spot numbera | Swissprot No.b | Protein name | Mrc | PId | Scoree | Peptidesf | Mean foldg | Repeath |
| 1 | *Q5T2P8 | Annexin A8-like protein 1 | 37086 | 5.45 | 221 | 13 | -3.09 ± 0.24 | 8 |
| 2 | P13928 | Annexin A8 | 36842 | 5.56 | 269 | 11 | -2.98 ± 0.26 | 8 |
| 3 | Q01105 | Protein SET | 33469 | 4.23 | 107 | 5 | 5.89 ± 0.52 | 6 |
| 5 | Q99497 | Protein DJ-1 | 20,050 | 6.33 | 187 | 5 | 3.22 ± 0.43 | 8 |
| 4 | P52907 | F-actin-capping protein subunit alpha-1 | 32902 | 5.45 | 118 | 6 | 2.23 ± 0.31 | |
| 6 | Q13765 | Nascent polypeptide-associated complex subunit alpha | 23370 | 4.52 | 451 | 10 | 4.37 ± 0.51 | 8 |
| 7 | *P24534 | Elongation factor 1-beta | 24748 | 4.5 | 45 | 1 | 2.42 ± 0.3 | 8 |
| 8 | *P02533 | Keratin, type I cytoskeletal 14 | 51589 | 5.09 | 432 | 17 | 4.32 ± 0.56 | 8 |
| 9 | P09493 | Tropomyosin alpha-1 chain | 32746 | 4.69 | 162 | 15 | 6.27 ± 0.38 | 8 |
| 10 | P08107 | Heat shock 70 kDa protein 1 | 70009 | 5.48 | 234 | 8 | 2.33 ± 0.28 | 8 |
| 11 | *P07910 | Heterogeneous nuclear ribonucleoproteins C1/C2 | 33650 | 4.95 | 140 | 9 | 2.95 ± 0.35 | 6 |
| 12 | P10809 | 60 kDa heat shock protein, mitochondrial precursor | 61016 | 5.7 | 103 | 7 | -4.32 ± 0.58 | 10 |
| 13 | P12004 | Proliferating cell nuclear antigen | 28750 | 4.57 | 113 | 4 | 7.95 ± 0.38 | 12 |
| 14 | P40261 | Nicotinamide N-methyltransferase | 30011 | 5.56 | 59 | 10 | -4.06 ± 0.58 | 8 |
| 15 | Q9UL46 | PA28 b | 27515 | 5.44 | 759 | 28 | 5.88 ± 0.68 | 12 |
| 16 | P52566 | Rho GDP-dissociation inhibitor 2 | 23031 | 5.1 | 248 | 11 | 3.14 ± 0.28 | 12 |
| 17 | * P02679 | Fibrinogen gamma chain precursor | 51479 | 5.37 | 326 | 18 | -3.05 ± 0.42 | 8 |
| 18 | P52565 | Rho GDP-dissociation inhibitor 1 | 23193 | 5.02 | 211 | 9 | 3.44 ± 0.32 | 12 |
| 19 | * P05112 | Interleukin-4 precursor | 17481 | 9.17 | 81 | 1 | 4.12 ± 0.62 | 4 |
| 20 | *P68363 | Tubulin alpha-1B chain | 50120 | 4.94 | 325 | 16 | 3.86 ± 0.31 | 10 |
| 21 | P05388 | 60S acidic ribosomal protein P0 | 34252 | 5.71 | 68 | 6 | 5.24 ± 0.71 | 6 |
| 22 | Q07021 | Complement component 1 Q | 31742 | 4.74 | 309 | 6 | 2.12 ± 0.49 | 8 |
| 23 | *P02675 | Fibrinogen beta chain precursor | 56577 | 8.54 | 236 | 18 | 13.65 ± 1.44 | 8 |
| 24 | *P01009 | Alpha-1-antitrypsin precursor | 46707 | 5.37 | 99 | 2 | 4.22 ± 0.65 | 4 |
| 25 | P09936 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 25151 | 5.33 | 326 | 22 | 11.5 ± 0.84 | 4 |
| 26 | P09211 | Glutathione S-transferase P | 23341 | 5.43 | 468 | 16 | 5.44 ± 0.24 | 12 |
| 27 | P07437 | Tubulin beta chain | 50095 | 4.78 | 197 | 12 | 5.67 ± 1.02 | 10 |
| 28 | P14618 | Pyruvate kinase isozymes M1/M2 | 58470 | 7.96 | 285 | 18 | 5.22 ± 0.29 | 12 |
| 29 | Q06323 | PA28 a | 28705 | 5.78 | 147 | 9 | 4.44 ± 0.25 | 12 |
| 30 | P04792 | heat shock 27 kDa protein 1 | 22768 | 5.98 | 565 | 42 | 5.56 ± 0.37 | 10 |
| 32 | P30048 | peroxiredoxin 3 | 27675 | 7.67 | 367 | 28 | 3.04 ± 0.4 | 10 |
| 31 | *P11142 | Heat shock cognate 71 kDa protein | 70854 | 5.37 | 134 | 7 | 5.26 ± 0.79 | 8 |
| 33 | Q13162 | Peroxiredoxin-4 | 30521 | 5.86 | 95 | 8 | 4.74 ± 0.22 | 12 |
| 34 | P36952 | Serpin B5 precursor | 42111 | 5.72 | 324 | 19 | 5.26 ± 0.47 | 12 |
| 35 | Q15019 | Septin-2 | 41461 | 6.15 | 191 | 6 | 3.88 ± 0.65 | 8 |
| 36 | Q01469 | Fatty acid-binding protein, epidermal | 15155 | 6.6 | 41 | 3 | 6.24 ± 0.25 | 12 |
| 37 | *P19013 | Keratin, type II cytoskeletal 4 | 57649 | 6.25 | 1023 | 28 | 3.12 ± 0.70 | 4 |
| 38 | P60709 | Actin, cytoplasmic 1 | 42052 | 5.29 | 806 | 28 | 5.88 ± 1.02 | 6 |
| 39 | Q969H8 | Uncharacterized protein C19orf10 | 18897 | 6.2 | 167 | 11 | 3.23 ± 0.56 | 8 |
| 40 | P47929 | Galectin-7 | 15066 | 7.03 | 730 | 22 | 6.44 ± 0.26 | 12 |
| 41 | * P06733 | Alpha-enolase | 47139 | 7.01 | 1000 | 27 | 4.33 ± 0.49 | 8 |
| 42 | P60903 | Protein S100-A10 | 11196 | 6.82 | 149 | 18 | 5.45 ± 0.62 | 6 |
| 43 | P62937 | Peptidyl-prolyl cis-trans isomerase A | 18229 | 7.68 | 2056 | 75 | 5.68 ± 0.85 | 6 |
| 44 | P23528 | Cofilin-1 | 18491 | 8.22 | 1677 | 35 | 3.11 ± 0.42 | 8 |
| 45 | *P22392 | Nucleoside diphosphate kinase B | 17287 | 8.52 | 235 | 20 | 4.44 ± 0.23 | 12 |
| 46 | Q01995 | Transgelin | 22596 | 8.87 | 238 | 16 | 3.28 ± 0.19 | 12 |
| 47 | P01834 | Ig kappa chain C region | 11602 | 5.58 | 228 | 24 | 3.44 ± 0.65 | 10 |
| 48 | P31949 | Protein S100-A11 | 10173 | 6.56 | 185 | 8 | 5.56 ± 0.75 | 8 |
| 49 | P62736 | Actin, aortic smooth muscle | 41982 | 5.23 | 119 | 6 | 3.67 ± 0.55 | 8 |
| 50 | Q64133 | Amine oxidase [flavin-containing] A | 60157 | 7.94 | 111 | 1 | 7.56 ± 0.8 | 6 |
| 51 | P47755 | F-actin-capping protein subunit alpha-2 | 32929 | 5.57 | 119 | 11 | 3.24 ± 0.45 | 8 |
| 52 | Q96A32 | Myosin regulatory light chain 2, skeletal muscle isoform | 19116 | 4.91 | 517 | 33 | 5.73 ± 0.35 | 12 |
| 53 | *P02743 | Serum amyloid P-component | 25485 | 6.1 | 140 | 5 | -2.34 ± 0.45 | 6 |
| 54 | P02511 | Alpha-crystallin B chain | 20146 | 6.76 | 874 | 34 | -9.5 ± 1.05 | 10 |
| 55 | P61088 | Ubiquitin-conjugating enzyme E2 N | 17184 | 6.1 | 3 | 86 | -3.23 ± 0.45 | 10 |
| 56 | P08758 | Annexin A5 | 35914 | 4.94 | 496 | 10 | -5.74 ± 0.28 | 12 |
| 57 | P62942 | FK506-binding protein 1A | 11943 | 7.88 | 51 | 2 | -2.56 ± 0.45 | 12 |
| 58 | P68871 | Hemoglobin subunit beta | 16102 | 6.75 | 996 | 45 | -2.02 ± 0.52 | 8 |
| 59 | Q14152 | Eukaryotic translation initiation factor 3 subunit I | 36479 | 5.38 | 190 | 15 | -2.96 ± 0.47 | 10 |
| 60 | P06702 | Protein S100-A9 | 13234 | 5.71 | 548 | 23 | -6.42 ± 0.58 | 12 |
| 61 | P04083 | Annexin A1 | 38690 | 6.57 | 2528 | 60 | -5.03 ± 0.28 | 12 |
| 62 | P55083 | Microfibril-associated glycoprotein 4 | 28972 | 5.38 | 95 | 1 | -3.69 ± 0.55 | 8 |
| 63 | P40925 | Malate dehydrogenase cytoplasmic | 36,403 | 6.91 | 4 | 86 | -2.87 ± 0.62 | 8 |
| 64 | P27482 | Calmodulin-like protein 3 | 16937 | 4.3 | 1562 | 55 | -4.22 ± 0.35 | 10 |
| 65 | * P61158 | Actin-related protein 3 | 47341 | 5.61 | 93 | 9 | -2.14 ± 0.45 | 8 |
| 66 | *P02768 | Serum albumin precursor | 71317 | 5.92 | 856 | 52 | -7.67 ± 0.85 | 10 |
| 67 | P05109 | Protein S100-A8 | 10828 | 6.51 | 136 | 13 | -3.34 ± 0.55 | 12 |
| 68 | *P05976 | Myosin light chain 1, skeletal muscle isoform | 21132 | 4.97 | 881 | 49 | -13.5 ± 0.85 | 12 |
| 69 | P30086 | Phosphatidylethanolamine-binding protein 1 | 21044 | 7.01 | 210 | 5 | -3.78 ± 0.65 | 6 |
| 70 | P11177 | Pyruvate dehydrogenase E1 component subunit beta, | 39208 | 6.2 | 64 | 5 | -5.35 ± 0.25 | 12 |
| 71 | P31151 | Protein S100-A7 | 11564 | 6.28 | 131 | 7 | -6.23 ± 0.44 | 12 |
| 72 | Q96FQ6 | Protein S100-A16 | 11794 | 6.28 | 68 | 4 | -7.14 ± 0.86 | 8 |
| 73 | P49773 | Histidine triad nucleotide-binding protein 1 | 13793 | 6.43 | 40 | 3 | -3.23 ± 0.52 | 8 |
| 74 | P69905 | Hemoglobin subunit alpha | 15305 | 8.72 | 769 | 43 | -7.68 ± 1.04 | 8 |
| 75 | P04080 | Cystatin-B | 11133 | 6.96 | 378 | 28 | -3.89 ± 0.18 | 12 |
| 76 | P07195 | L-lactate dehydrogenase B chain | 36615 | 5.71 | 246 | 12 | -2.69 ± 0.55 | 6 |
| 77 | P02144 | Myoglobin | 17173 | 7.14 | 2284 | 53 | -19.22 ± 1.38 | 12 |
| 78 | O00299 | Chloride intracellular channel protein | 26906 | 5.09 | 405 | 16 | -2.23 ± 0.48 | 6 |
| 79 | P07737 | Profilin-1 | 15216 | 8.44 | 1252 | 53 | -5.69 ± 0.35 | 12 |
| 80 | O95994 | Anterior gradient protein 2 homolog | 19967 | 9.03 | 292 | 10 | -2.87 ± 0.52 | 8 |
| 81 | *P62988 | Ubiquitin | 18560 | 6.56 | 112 | 5 | -5.22 ± 0.43 | 6 |
| 82 | P45378 | Troponin T, fast skeletal muscle | 31805 | 5.71 | 233 | 10 | -2.99 ± 0.45 | 8 |
| 83 | P12429 | Annexin A3 | 36353 | 5.63 | 169 | 10 | -4.41 ± 0.28 | 12 |
| 84 | P09525 | Annexin A4 | 35860 | 5.84 | 694 | 20 | -2.88 ± 0.56 | 8 |
| 85 | P35232 | Prohibitin | 29843 | 5.57 | 214 | 14 | -3.24 ± 0.45 | 8 |
a The spot number correspond to those on 2-DE images shown in Fig. 2
b Swiss-Prot accession number.
c Theoretical molecular mass (kDa) and pI from the ExPASy database.
d Theoretical pI from the ExPASy database.e The number of unique peptides identified by MS/MS sequencing.
f Probability-based MOWSE (molecular weight search) scores. g Expression change level in OSCC tumor tissue compared with control (+, increase in tumor;-, decrease in tumor). Data were represented as mean ± SD
h frequency of the up-regulation (or down-regulation) of the 85 proteins identified in the six sample pairs.
* new proteins obtained by analysis of expression pattern through web-based tool (GOTree)
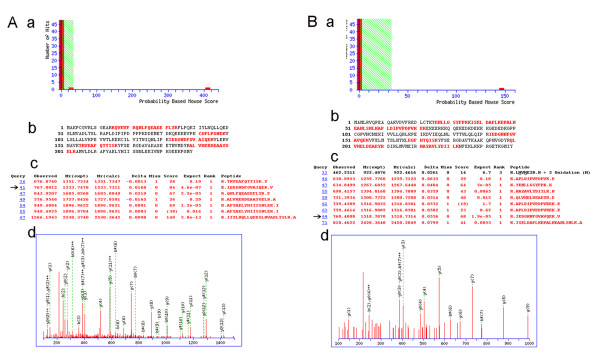
Figure 3.
Results of PA28 b and a as the representative protein identified using ESI-Q-TOF-MS/MS. A a b, output of the database searching by the MASCOT program using MS/MS data used in the identification of PA28 b. The matched peptides were shown in bold red. A c d, MS/MS spectrum of parent ions with m/z values of 1533.7311(arrow marked). B a b, output of the database searching by the MASCOT program using MS/MS data used in the identification of PA28 a. B c d, MS/MS spectrum of parent ions with m/z values of 1518.7314(arrow marked).
Finding functional enrichment in transformation from oral leukoplakia to OSCC through gene ontology
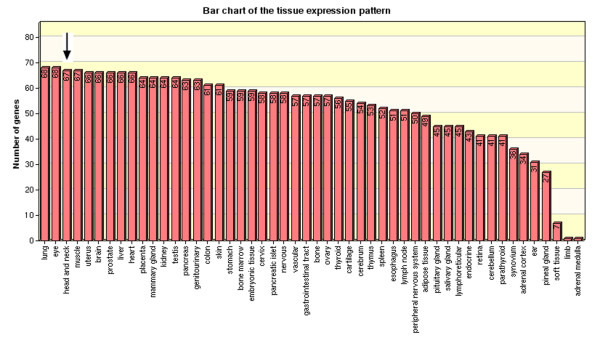
Ontological methods were employed to structure the biological processes that were over-represented in the carcinogenic stage from oral leukoplakia to infiltrative oral cancer. A web-based tool (GOTree) was employed in which the biological process of the proteins encoded by the genes is scored. Those with a level higher than 4 were highlighted. The observed versus the expected number of genes in the GO categories using a Homo sapiens reference data set was analyzed. 18 novel proteins, each marked with an asterisk in Table 1, were demonstrated to be expressed in head and neck by the analysis of tissue expression profile using GOTree, and their bar chart is shown in Figure 4. X-axis represented the different tissue. The Y-axis means the proteins has been reported in corresponding tissue. Figure 5 shows the tree-like structure with their respective molecular functions, cellular components and biological processes. Taken the biological processes as an example, it involved six processes, the processes responsive to stimulus (including heat shock 70 kDa, 60 kDa and 27 kDa proteins, tubulin, heat shock 70 kDa protein 8, annexin A5, proliferating cell nuclear antigen, S100A7,8,9, annexin A1 and A8, and interleukin 4); physiological response(including serpin peptidase inhibitor, Rho GDP dissociation inhibitor beta, fibrinogen, PA28 a and b); negative regulation of biological process (including capping protein muscle, annexin A4, SET translocation, glutathione S-transferase, S100A11, annexin A1, non-metastatic cells 2 protein, and prohibitin); locomotion proteins (including annexin A1, SERPINB5, Rho GDP dissociation inhibitor (GDI) alpha, tropomyosin 1, and heat shock 27 kDa protein 1); cell death process (including heat shock 27 and 70 kDa protein, annexin A4 annexin A5, galectin 7, and prohibitin); and coagulation (including annexin A4, A5, A8, and fibrinogen).
Figure 4.
Expression Profiling Diagram. The diagram was constructed with the use of the Ingenuity Pathway Analysis software as described in Materials and Methods and in Results. 18 novel proteins were shown to be firstly expressed in head and neck.
Figure 5.
Directed acyclic graph (DAG) view of the enriched GO categories in the transformation process from precancerous oral leukoplakia to OSCC. The enriched GO categories were brought together and visualized as a DAG. Categories in red were enriched, while those in black were non-enriched parents http://bioinfo.vanderbilt.edu/gotm/. A) biological process, B) physiological process C) response to stimulus D) molecular function E) cellular component.
Link of the Proteins to Biological Pathways
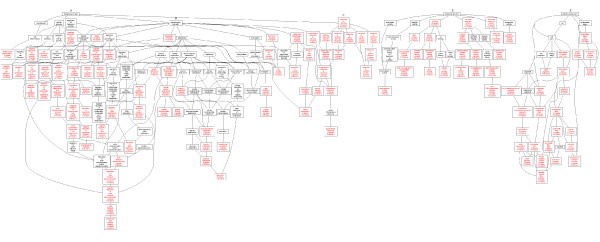
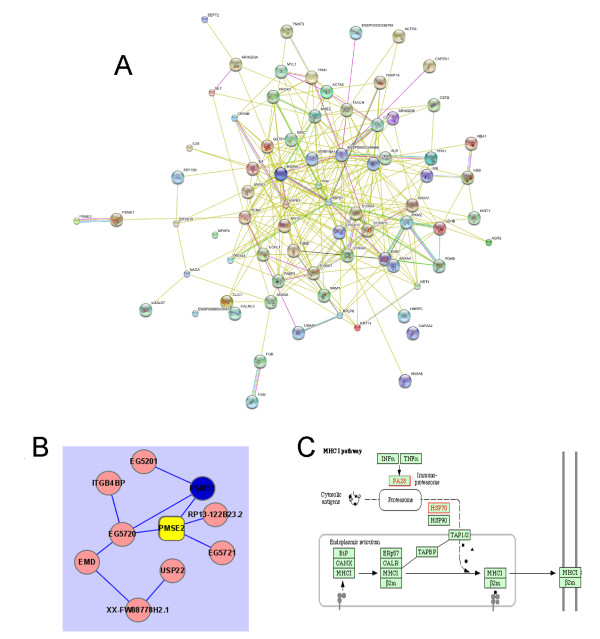
To establish an overview of the interactions among differentially regulated proteins, and to prioritize proteins and pathways for further evaluation, we used the Pathway Studio software to explore the associations between differentially regulated proteins, essentially based on the available knowledge about eukaryotic molecular interactions documented in the ResNet database. Notably, central nodes of proteins of the biological network, generated by the Pathway Architect assembly of GO designations for the most prominently over-represented genes, were shared in oral leukoplakia tissues and tumor tissues including S100 family (A7, 8, 9, 10, and 11), HSP family (HSPB1 and HSPA8), ANX family (A1, 3, 4, and 5), tumor metastasis suppressor NME2 and Rho GDP dissociation inhibitor alpha and beta (ARHGDIA and B), pyruvate kinase (PKM2), transgelin (TAGLN), glutathione S-transferase (GSTP1), SERPINA1, PCNA, and so on (Figure 6A). Those proteins and their interactive pathways merit further study for their roles in oral carcinogenesis.
Figure 6.
Organization of dysregulated protein genes into common pathways in the transformation process from precancerous oral leukoplakia to OSCC by Pathway Studio. Proteins with their SWISS-PROT number were loaded into Pathway Studio software http://www.ariadnegenomics.com/, and analyzed using the Pathway Studio program with the ResNet 2.5 database. (A) 85 protein genes with a high confidence index of interactions were included in the pathway layout graph. (B) The indirect interaction of eight proteins with PA28 a and b (gene name: PSME1 and 2) as reported in the cancer-related literature. (C) The key proteins in MHC antigen presenting pathway, among which PA28, proteosome, HSP70 and HSP90 have been detected up-regulated in our study. Each node represents either a protein entity or a control mechanism of the interaction. Connecting lines between the protein symbols indicate interactions; different types of interactions are denoted by symbols on the lines. Green square indicates regulation; purple square, binding; blue square, expression; orange circle, protein modification; red diamond, metabolism; green circle, promoter binding; yellow triangle, transport; "+" in gray circle, positive effect; and "-" in gray circle, negative effect.
Biological Pathways related to PA28
Pathway analysis was also used to reveal the protein interactions and potential pathways of PA28. The result showed that some proteins interacted with PA28 either directly or indirectly, which was not identified by the proteomics approach due to their low abundance (Figures 6B). It was generally accepted that PA28ab contributes to Class I presentation in immune tissues. Our results also reinforced the connection between PA28ab and cellular immunity as the proteins involved in the MHC-I antigen presenting pathway like PA28, proteosome, HSP70 and HSP90 have been detected to be up-regulated (Table 1, Figures 6C).
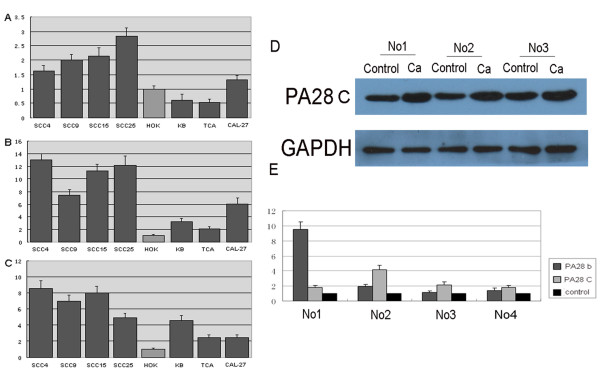
Overexpression of PA28 in OSCC Cancer Cells and Cancer Tissues
Evaluation of PA28 expression in seven OSCC cell lines relative to the human immortalized oral keratinocytes (HOK16E6E7) and of PA28 expression in OSCC tumor tissues versus oral leukoplakia tissues was performed by real time RT-PCR and Western blotting. Consistent with observations from 2-DE, expression of PA28 was markedly increased at both the mRNA and protein levels in OSCC cells and tumor tissues compared with normal keratinocytes and oral leukoplakia tissues (Figure 7).
Figure 7.
Validation of PA28 in OSCC tissues and cells relative to control oral leukoplakia tissue and normal keratinocytes. (A, B, C) The mRNA expression evaluated by real-time PCR in seven OSCC cell lines when compared to the oral keratinocytes. The mRNA level of the three PA28 homologs, a, b and g were shown in A, B, and C, respectively. Except PA28 a in KB and Tca8113 cell lines, the mRNA expression in OSCC cells were reproducibly increased. (D) Crops of images of quantitative measurement of PA 28 g protein in 3 pairs of OSCC tissues (Ca) and oral leukoplakia tissues (control) by Western blotting analysis using PA 28 g-specific antibody. (E) The mRNA expression evaluated by real-time PCR in 4 pairs of OSCC tissues and oral leukoplakia tissues.
Discussion
Only a few studies were done on proteomic analysis of the malignant transformation mechanism from precancerous lesion with different stage of dysplasia into invasive cancer. We have performed a comparative proteomic analysis to profile differentially expressed proteins in the transformation process. Using GO analysis, we further analyzed the biological process and pathway network of these proteins, which can generate a new insight into systemic biology in carcinogenesis.
Oral squamous carcinoma, like esophageal adenocarcinoma, has been associated with the presence of precancerous lesion with different stage of dysplasia, thus providing a good model to elucidate every stage of carcinogenesis in more detail. In our study, each pair of precancerous and cancer tissues was from the same patient, which provides an opportunity to eliminate or at least reduce heterogeneity. Using the proteomic approach, we have identified 85 differently expressed gene products and found some proteins are related to apoptosis, response to stimulus, metabolic regulation and etc. We can thus conclude that these proteins may play an important role in malignant transformation process. Our most significant finding was that several proteins in the same protein families and homologs were identified in this transformation process, such as peroxiredoxins (Peroxiredoxin-3 and 4), Annexin family (A1, A 3, A 4, A 5, A 8), Rho GDP-dissociation inhibitor 1 and 2, Heat shock protein family (70 kDa protein 1, 71 kDa protein, and Heat shock protein beta-1), PA28 homolog (PA28 a and b), Protein S100 family (A7, A8, A9, A10, A11, and A16). Among which, the annexins and S100 are two super-families of closely related calcium and membrane-binding proteins and their relationship with carcinogenesis has been widely studied. They have a diverse range of cellular functions including vesicle trafficking, cell division, apoptosis, calcium signaling and growth regulation.
Many studies have revealed the annexins to be among the genes whose expression is differentially altered in neoplasia. Some annexins showed increased expression in specific types of tumors, while others displayed loss of expression. In our report, the expression level of annexin A1, 3, 4, 5, 8 were all decreased while annexin A8 showed increased expression. Annexin A1 has been extensively studied in vitro and in vivo. The loss of expression of annexin A1 in our study confirmed previous findings in head and neck squamous carcinomas[12]. Expression of annexin A3 has only been studied in a limited number of tumor types with only one report regarding its expression in head and neck cancer [13]. For annexin A4, a few studies reported its increased expression in clear cell renal cancer and colorectal cancer by using a combination of proteomics tools [14,15]. The change of Annexin A5 was also observed in our study, which has been considered as one of the signals on the surface of the apoptotic cells and has been used as a probe for apoptosis [16]. Annexin A8 has been shown to be consistently over-expressed in acute promyelocytic leukaemia, breast cancers, pancreatic cancer by a combination of gene expression microarrays and immunohistochemistry. The expression of annexin A4, 5, and 8 in head and neck cancer has been reported in this study for the first time, which was consistent with the results obtained in other studies concerning their expression in other cancers [17,18].
The S100 proteins are a multi-gene calcium-binding family of proteins comprising 20 known human members. There has been growing interest in the S100 protein family and their relationship with different cancers. While the precise role of S100 proteins in the development and promotion of cancer remains unclear, it is evident that the S100 proteins have a variety of intracellular and extracellular roles, and that disruption of any one of these functions may contribute to carcinogenesis. There is evidence that these proteins play a major role in tumor metastasis by interacting with a number of different proteins, including matrix metalloproteinase, cytoskeletal proteins, p53, Jab1, Cox-2 and BRCA1. In this study, we have identified a series of members including S100A7, 8, 9, 10, 11, and 16 with differential expression in OSCC tissues. S100A7 (psoriasin) was a member first characterized as being highly expressed in psoriatic keratinocytes [19]. There is accumulating evidence that S100A7 is up-regulated in bladder cancer skin tumors and some invasive carcinomas. Its expression is associated with a poorer prognosis and reduced survival [20-22]. On the contrary, other reports in OSCC showed its expression was associated with a better prognosis based on the finding that S100A7 is highly expressed in pre-invasive, well-differentiated and early staged OSCC, but little or no expression was found in poorly differentiated, later-staged invasive tumors [23]. Other reports showed that S100A7 inhibits both OSCC cell proliferation in vitro and tumor growth/invasion in vivo [24]. These results were echoed by our study, in which S100A7 was identified to be down-regulated in the transformation process form precancerous dysplasia to invasive cancer. Therefore, unlike in other tumors, our data suggests S100A7 to be a tumor suppressor in OSCC. The detailed function should be further elucidated. S100A8 and S100A9 which form a heterodimer complex 90 are up-regulated in many cancers and have been implicated in the metastatic process including gastric cancer, prostate cancer, colorectal cancer, and breast cancer [25-27]. In OSCC, one study reported there was more than a 10-fold over-expression of S100 A8 in HPV18+ OSCC [28]. For S100A11, its function has been somewhat controversial. In bladder carcinoma and renal carcinoma, its expression is related to tumor suppression, and decreased expression of S100A11 has been associated with an increase in histopathological grade, poorer prognosis and decreased survival [29]. However, in prostate cancer and breast cancer it is thought to be a tumor promoter. Its increased expression in prostate cancers has been shown to be associated with advanced pathological stage [30]. There is only one report about the gene expression of S100A11 related to its diverse functions [31]. S100A16 protein, a new and unique member of the EF-hand Ca (2+)-binding proteinswas found to accumulate within nucleoli and be translocated to the cytoplasm in response to Ca (2+) stimulation [32]. Here we report for the first time the expression of S100A16 protein in carcinogenesis from precancerous dysplasia to OSCC. It is possible that each S100 protein may play multiple roles in tumourigenesis and metastasis. This highlights the need for an improved understanding of the S100 family, before the design of S100 protein-targeted therapies can be achieved.
Proteasomes are large complexes that carry out crucial roles in many cellular pathways by degrading proteins in the cytosol and nucleus of eukaryotic cells [33]. Proteasomes are activated by protein complexes that bind to the end rings of subunits. PA28 (also known as 11S or REG) has been shown to bind specifically to and activate 20S proteasomes against model peptide substrates [34]. The biological roles of PA28 are less well understood. There are three PA28 homologs, called a, b and g. Although PA28a and b subunits are expressed in many organs, they are particularly abundant in immune tissues and are virtually absent from the brain. By the late 1990s, PA28ab was found to contribute to Class I presentation, based on the high levels of PA28ab in immune tissues, the IFNg induction of PA28ab and many components of the class I pathway, and the direct production of some Class I epitopes by PA28ab-proteasome complexes[35,36]. Our results also reinforce the connection between PA28ab and cellular immunity by showing that the key proteins in the MHC I antigen presenting pathway like PA28, proteosome, HSP70 and HSP90 have been detected up-regulated in OSCC tissues. An earlier study showed that PA28g expression correlated with cell proliferation. Recently, some researchers have gained more insight into the role of PA28g in apoptosis [37]. These findings were paralleled by studies suggesting that PA28g functions in cell cycle progression and has an immune role [38]. Two-hybrid screens have identified several proteins that interact with PA28g as well. Interestingly, all these findings suggest that PA28g is an anti-apoptotic factor. Less is known about how PA28g may suppress apoptosis in oral carcinesogenesis. In our validation study, Three homologs were all included. The results have confirmed the up-regulation of PA28 in carcinogenesis by comparison between several OSCC cell lines and oral keratinocytes. In our further studies, we would valuate the PA28 ab complex and PA28 g immunostaining pattern in different stage of tissue samples from normal, precancerous to infiltrative OSCC. Moreover, the relationships of immunostaning with survival rate and recurrence will be analyzed.
Conclusion
In summary, we have applied proteomic technologies to analyze the malignant transformation from precancerous oral leukoplakia to oral cancer from 6 patients, and we have identified 85 different proteins with altered expression levels in OSCC in the transformation, of which 53 were up-regulated. Previous characterizations regarding their functions and possible interactions with other proteins and in particular the pathways involved were also evaluated. As a key factor in tumor metastasis, PA was chosen for transformation at first. The proteosome activator PA28 was studied for their expression and interactive networks correlated with oral malignancy. We have being started the further research on other potential biomakers like peroxiredoxins, annexin family and S100 family. This is an example of a systems biology study, in which functional proteomics was constructed to help to elucidate mechanical aspects and potential involvement of proteins of interest in biological pathways.
Methods
Cell Culture
Seven oral squamous cell carcinoma cell lines (SSC4, SCC9, SCC15, SCC25, Cal 27, KB, Tca8113) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum (Invitrogen), 100 units/liter penicillin, and 10 mg/liter streptomycin. HOK16E6E7 cells, a human immortalized oral keratinocyte cell line, was cultured in keratinocyte growth medium containing 0.15 Mm calcium and supplemented with epidermal growth factor (Invitrogen). All cell lines were maintained at 37°C in an atmosphere containing 5% CO2.
Tissue collection and sample preparation
Six pairs of tumors and oral leukoplakia tissues with dysplasia were obtained from six patients in West China Stomatological Hospital, Sichuan University. The specimens were examined histologically by hematoxylin and eosin (HE) staining, and the clinicopathologic stage was determined according to the TNM classification system of the International Union against Cancer [39]. Patients receiving previous chemotherapy or radiation treatment were excluded. All the tissue specimens were snap-frozen in liquid nitrogen for proteomic analysis. Hematoxylin-stained 5 μm frozen sections were reviewed by a Board-Certified pathologist (Y. Chen) for tumor cellularity (oral squamous carcinoma) or oral leukoplakia mucosa (moderate-high grade dysplasia). Informed consent was obtained from all patients or their relatives for the experimental use of their tissues. Medical records were reviewed and data were coded to protect patient confidentiality. The project was approved by the Scientific and Ethics Committee of Sichuan University.
Two-Dimensional Electrophoresis
Two-dimensional gel electrophoresis was performed essentially as previously described [40]. The protein concentration of the supernatants was determined using a Bio-Rad protein kit. All the paired samples were quantitatively analyzed in group. Samples of 1 mg protein were applied on immobilized pH 3–10 nonlinear gradient strips in sample cups at their basic and acidic ends. Focusing started at 200 V and the voltage was gradually increased to 8000 at 4 V/min and kept constant for a further 3 h (approximately 150 000 Vh totally). The second dimensional separation was performed in 12% SDS-polyacrylamide gels. The gels (180 × 200 × 1.5 mm3) were run at 40 mA/gel. After protein fixation in 50% methanol containing 5% phosphoric acid for 2 h, the gels were stained with Coomassie Brilliant Blue R-250 (Merck, Germany) for 12 h and the protein spots were visualized. Each experiment was performed twice to ensure the accuracy of analyses. The images were scanned using a Bio-Rad high quality white light GS-800 scanner (400–750 nm). The differentially expressed proteins were identified using the PD-Quest 2DE analysis software (Bio-Rad, USA). The quantity of each spot in a gel was normalized as a percentage of the total quantity of all spots in that gel and evaluated in terms of O.D. The student's t-test was applied to compare the spot relative volume between two groups. Significant spots that showed changed consistently and at least 2.0-fold difference (p < 0.05) were selected for tandem mass spectrometry (MS/MS) analysis.
Protein identification by nano-HPLC-ESI-Q-TOF-MS/MS
The protein spots were excised manually and digested using sequence grade trypsin (V511A, Promega). The protein samples were reduced, alkylated, and then digested with trypsin using standard protocols as previously described. The digests were analyzed using a nano-HPLC system coupled to Q-TOF Primer mass spectrometer (Q-TOF, Micromass, Micromass, Manchester, UK) equipped with an electrospray ionization source. Spectra were accumulated until a satisfactory signal/noise ratio had been obtained. Only double, or more than double, charge peaks, in the mass range from 400 to 1600 m/z, were considered for MS/MS. Ions exhibiting a detection intensity exceeding 10 counts/second were selected for production of ion spectra by Collision Induced Dissociation (CID). Trypsin autolysis products and keratin-derived precursor ions were automatically excluded. Three MS/MS ions were selected for each survey scan. All data used to extract peak information, which was used to create the MS/MS peak list, were generated from one combined spectrum. The tandem mass spectrometry (MS/MS) data, "pkl list (pkl) " files acquired by the software of ProteinLynx 2.2.5 (Waters), included the mass values, the intensity and the charge of the precursor ions (parent ions with +2 or +3 charge in this study). The pkl files were analyzed using MASCOT search engine http://www.matrixscience.com against SWISS-PROT protein database. Proteins were identified on the basis of two or more peptides whose ion scores both exceeded the threshold, P < 0.05, which indicated the 95% confidence level for these matched peptides [41].
Functional genome (Gene) ontology analysis
To translate sets of differentially regulated genes into functional profiles, we applied GOTree Machine http://bioinfo.vanderbilt.edu/gotm/ for data analysis. GOTree Machine generates a GOTree, a tree-like structure to navigate the Gene Ontology-directed acyclic graph for input protein sets, and to provide navigational data and visualization. The software categorizes gene products based on the location of the protein within cellular components and suggests possible biochemical, biological, and molecular functions. Statistical analysis was performed to identify the most important GO categories for the input protein sets and to suggest their potential biological importance in each of the categories. The tissue expression pattern diagram was constructed with the software too.
Biological association network
Changes in gene/protein expression are not isolated events. Therefore, we hypothesize that differentially regulated genes/proteins that have interacted with others may be more likely to play crucial roles in oncogenesis. To identify those key factors, we explored the biological associations among the differentially expressed proteins using String http://string.embl.de/. Differentially expressed proteins each identified with a SWISS-PROT number were uploaded into String. The proteins were integrated into biological association networks based on protein interactions documented in a curated database
Real-Time Quantitative PCR
Real-time PCR was performed on a LightCycler (Roche Diagnostics) platform as previously described [42]. The following forward and reverse primers were selected by using PRIMER EXPRESS software and were synthesized by TaKaRa Co. Ltd. PA28 a F: 5'-GCC AAC TTG AGC AAT CTG A-3'; PA28 a R:5'-ACA GGG AGG ACC TTT GTC-3'; PA28 a TM: 5'-FAM-AGAAAGAGGAGCGGAAGAAACAGC-ECLIPSE-3'; PA28 b F: 5'-TAG CGA CTG AAG CAG CAT G-3'; PA28 b: 5'-GGA AGT CAA GTC AGC CAC AT-3'; PA28 b TM: 5'-FAM-ATT CCT CTA CAG ATT CTT GCC ACA GA-ECLIPSE-3'; PA28 g F: 5'-AGA AGA CTT GGT GGC AAA T-3'; PA28 g R: 5'-TCC AGT CCA TCA TGG CTA T-3'; PA28 g TM: 5'-FAM-CAT GAA TCT CCC AGT CCC TGA CCC-ECLIPSE-3'. A SYBR green PCR kit (Applied Biosystems) was used by following the manufacturer's instructions, and the analyses were performed in duplicate or triplicate. Target mRNA values were normalized by using β-actin mRNA as an internal control.
Western Blotting
Protein extracts were prepared using a lysis buffer containing protease inhibitor mixture 8340. For Western blotting analysis, 20 ug of protein were separated by 12% SDS-PAGE, then transferred to a PVDF membrane (Millipore), and probed with monoclonal mouse anti-PA28 g antibody (1:1000; Cell signaling Technology). The blots were labeled with horseradish peroxidase-conjugated secondary antibodies (1:10,000) and visualized with an ECL detection system (Pierce).
Statistical Analysis
All samples were processed twice by 2-D gels and real-time PCR. The results of the individual analyses correlated well with each other without unexpected deviations. The comparison was done by unpaired Student's t test. Significance was set a p < 0.05. Computations were performed using the SPSS version 11.5 software package.
Authors' contributions
ZW and XDF contribute equally to conducting the experiments and writing the paper, XYL contributed to the genome (Gene) ontology analysis and network analysis, LJ and XZ contributed to expression validation, NJ and JL contributed to Two-Dimensional Electrophoresis and trypsin digestion, QMC and LJL provided the original concept of the study, supervised the study and contributed to writing the paper. All authors read and approved the final manuscript.
Supplementary Material
The enlarged and cropped images of 11 selected protein spots in 2D Gel. 11 proteins were selected as examples showing the consistent expression changes in enlarged form. The images of each changed protein spot were compared with the control. (A) 2-DE gel images of 11 selected protein indicated by arrows in the panels. Each panel shows an enlarged view of the gel spots from Figure 2. (B) Volume density analysis graphs: the data were expressed as mean ± SD of twelve repeats.
Acknowledgments
Acknowledgements
We would like to thank Yi jia (National Jewish Health) and Professor David Wong (UCLA, School of Dentistry) and Professor A. Ian Smith (Medicine Nursing and Health Sciences, Monash University) for helpful comments on the paper. This work was supported by grants from the National Science Funds for Talented Professionals (No.30725041), the 973 National Basic Research Program of China (2008CB517307), the National Natural Science Foundation of China (No. 30300387, 30471891, 30672323, 30801294), Specialized Research Fund for the Doctoral Program of Higher Education (200806101110).
Contributor Information
Zhi Wang, Email: wangzhi0506@126.com.
Xiaodong Feng, Email: fengxiaodong84@126.com.
Xinyu Liu, Email: xinyuliu33@126.com.
Lu Jiang, Email: jianglu20002.student@sina.com.
Xin Zeng, Email: zengxin22@163.com.
Ning Ji, Email: jiningcoming@sohu.com.
Jing Li, Email: liJing19840108@126.com.
Longjiang Li, Email: muzili63@163.com.
Qianming Chen, Email: qmchen@scu.edu.cn.
References
- Saranath D, (Ed) Contemporary Issues in Oral Cancer. Oxford University Press, New Delhi; 2000. pp. 30–71. [Google Scholar]
- Lee JJ, Hong WK, Hittelman WN, Mao L, Lotan R, Shin DM, Benner SE, Xu XC, Lee JS, Papadimitrakopoulou VM, Geyer C, Perez C, Martin JW, El-Naggar AK, Lippman SM. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res. 2000;6:1702–1710. [PubMed] [Google Scholar]
- Jaber MA, Porter SR, Speight P, Eveson JW, Scully C. Oral epithelial dysplasia: clinical characteristics of western European residents. Oral Oncol. 2003;39:589–96. doi: 10.1016/S1368-8375(03)00045-9. [DOI] [PubMed] [Google Scholar]
- Koike H, Uzawa K, Nakashima D, Shimada K, Kato Y, Higo M, Kouzu Y, Endo Y, Kasamatsu A, Tanzawa H. Identification of differentially expressed proteins in oral squamous cell carcinoma using a global proteomic approach. Int J Oncol. 2005;27:59–67. [PubMed] [Google Scholar]
- Kato H, Uzawa K, Onda T, Kato Y, Saito K, Nakashima D, Ogawara K, Bukawa H, Yokoe H, Tanzawa H. Down-regulation of 1D-myo-inositol 1,4,5-trisphosphate 3-kinase A protein expression in oral squamous cell carcinoma. Int J Oncol. 2006;28:873–81. [PubMed] [Google Scholar]
- He QY, Chen J, Kung HF, Yuen AP, Chiu JF. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4:271–8. doi: 10.1002/pmic.200300550. [DOI] [PubMed] [Google Scholar]
- Balys R, Alaoui-Jamali M, Hier M, Black M, Domanowski G, Rochon L, Jie S. Clinically relevant oral cancer model for serum proteomic eavesdropping on the tumour microenvironment. J Otolaryngol. 2006;35:157–166. [PubMed] [Google Scholar]
- Xie H, Onsongo G, Popko J, de Jong EP, Cao J, Carlis JV, Griffin RJ, Rhodus NL, Griffin TJ. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–498. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- Hu S, Arellano M, Boontheung P. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RJ, Cramer R. PIGOK: Linking protein identity to gene ontology and function. J Proteome Res. 2006;5:3429–3432. doi: 10.1021/pr0601537. [DOI] [PubMed] [Google Scholar]
- Koehn J, Krapfenbauer K, Huber S, Stein E, Sutter W, Watzinger F, Erovic BM, Thurnher D, Schindler T, Fountoulakis M, Turhani D. Potential involvement of MYC- and p53-related pathways in tumorigenesis in human oral squamous cell carcinoma revealed by proteomic analysis. J Proteome Res. 2008;7:3818–3829. doi: 10.1021/pr800077a. [DOI] [PubMed] [Google Scholar]
- Garcia Pedrero JM, Fernandez MP, Morgan RO, Herrero Zapatero A, Gonzalez MV, Suarez Nieto C, Rodrigo JP. Annexin A1 downregulation in head and neck cancer is associated with epithelial differentiation status. Am J Pathol. 2004;164:73–79. doi: 10.1016/S0002-9440(10)63098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu A, Uzawa K, Nakashima D, Kouzu Y, Endo Y, Koike H, Yokoe H, Harada K, Sato M, Tanzawa H. A proteomics approach to characterizing human submandibular gland cell lines by fluorescent two-dimensional differential in-gel electrophoresis. Int J Mol Med. 2006;17:253–260. [PubMed] [Google Scholar]
- Zimmermann U, Balabanov S, Giebel J, Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B, Walther R. Increased expression and altered location of annexin IV in renal clear cell carcinoma: a possible role in tumour dissemination. Cancer Lett. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98:426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsten MF, Hofstra L, Narula J, Reutelingsperger CP. Counting heads in the war against cancer: defining the role of annexin A5 imaging in cancer treatment and surveillance. Cancer Res. 2006;66:1255–1260. doi: 10.1158/0008-5472.CAN-05-3000. [DOI] [PubMed] [Google Scholar]
- Stein T, Price KN, Morris JS, Heath VJ, Ferrier RK, Bell AK, Pringle MA, Villadsen R, Petersen OW, Sauter G, Bryson G, Mallon EA, Gusterson BA. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin Cancer Res. 2005;11:6872–6879. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Illei PB, Ashfaq R, Infante JR, Murphy K, Pandey A, Schulick R, Winter J, Sharma R, Maitra A, Goggins M, Hruban RH. New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol. 2008;32:188–96. doi: 10.1097/PAS.0b013e31815701f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Leffers H, Honoré B, Celis JE. Molecular cloning and expression of a novel keratinocyte protein (psoriasisassociated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acidbinding proteins. J Invest Dermatol. 1992;99:299–305. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- Celis JE, Rasmussen HH, Vorum H, Madsen P, Honoré B, Wolf H, Orntoft TF. Bladder squamous cell carcinomas express psoriasin and externalize it to the urine. J Urol. 1996;155:2105–12. doi: 10.1016/S0022-5347(01)66118-4. [DOI] [PubMed] [Google Scholar]
- Moubayed N, Weichenthal M, Harder J, Wandel E, Sticherling M, Gläser R. Psoriasin (S100A7) is significantly up-regulated in human epithelial skin tumours. J Cancer Res Clin Oncol. 2007;133:253–261. doi: 10.1007/s00432-006-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley ED, Niu Y, Njue C, Kliewer EV, Murphy LC, Watson PH. Psoriasin (S100A7) expression is associated with poor outcome in estrogen receptor-negative invasive breast cancer. Clin Cancer Res. 2003;9:2627–2631. [PubMed] [Google Scholar]
- Kesting MR, Sudhoff H, Hasler RJ, Nieberler M, Pautke C, Wolff KD, Wagenpfeil S, Al-Benna S, Jacobsen F, Steinstraesser L. Psoriasin (S100A7) up-regulation in oral squamous cell carcinoma and its relation to clinicopathologic features. Oral Oncol. 2009;45:731–6. doi: 10.1016/j.oraloncology.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Zhou G, Xie TX, Zhao M, Jasser SA, Younes MN, Sano D, Lin J, Kupferman ME, Santillan AA, Patel V, Gutkind JS, Ei-Naggar AK, Emberley ED, Watson PH, Matsuzawa SI, Reed JC, Myers JN. Reciprocal negative regulation between S100A7/psoriasin and beta-catenin signaling plays an important role in tumor progression of squamous cell carcinoma of oral cavity. Oncogene. 2008;27:3527–3538. doi: 10.1038/sj.onc.1211015. [DOI] [PubMed] [Google Scholar]
- Hermani A, De Servi B, Medunjanin S, Tessier PA, Mayer D. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res. 2006;312:184–197. doi: 10.1016/j.yexcr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Stulík J, Koupilova K, Osterreicher J, Knízek J, Macela A, Bures J, Jandík P, Langr F, Dedic K, Jungblut PR. Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis. 1999;20:3638–3646. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3638::AID-ELPS3638>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Arai K, Takano S, Teratani T, Ito Y, Yamada T, Nozawa R. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr Cancer Drug Targets. 2008;8:243–52. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- Lo WY, Lai CC, Hua CH, Tsai MH, Huang SY, Tsai CH, Tsai FJ. S100A8 is identified as a biomarker of HPV18-infected oral squamous cell carcinomas by suppression subtraction hybridization, clinical proteomics analysis, and immunohistochemistry staining. J Proteome Res. 2007;6:2143–2151. doi: 10.1021/pr060551+. [DOI] [PubMed] [Google Scholar]
- Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- Rehman I, Azzouzi AR, Cross SS, Deloulme JC, Catto JW, Wylde N, Larre S, Champigneuille J, Hamdy FC. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol. 2004;35:1385–1391. doi: 10.1016/j.humpath.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Kornberg LJ, Villaret D, Popp M, Lui L, McLaren R, Brown H, Cohen D, Yun J, McFadden M. Gene expression profiling in squamous cell carcinoma of the oral cavity shows abnormalities in several signaling pathways. Laryngoscope. 2005;115:690–698. doi: 10.1097/01.mlg.0000161333.67977.93. [DOI] [PubMed] [Google Scholar]
- Sturchler E, Cox JA, Durussel I, Weibel M, Heizmann CW. S100A16, a novel calcium-binding protein of the EF-hand superfamily. J Biol Chem. 2006;281:38905–38917. doi: 10.1074/jbc.M605798200. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediatedproteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hill CP, Masters EI, Whitby FG. The 11S regulators of 20S proteasome activity. Curr Top Microbiol Immunol. 2002;268:73–89. doi: 10.1007/978-3-642-59414-4_4. [DOI] [PubMed] [Google Scholar]
- Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol. 2004;5:685–692. doi: 10.1038/ni1088. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immuno. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Shimada K, Nishida Y, Lee RS, Pardee AB, Nishizuka Y. Loss in transformed cells of cell cycle regulation of expression of a nuclear protein recognized by SLE patient antisera. Exp Cell Res. 1989;182:284–289. doi: 10.1016/0014-4827(89)90299-1. [DOI] [PubMed] [Google Scholar]
- Barton LF, Runnels HA, Schell TD, Cho Y, Gibbons R, Tevethia SS, Deepe GS, Jr, Monaco JJ. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J Immunol. 2004;172:3948–54. doi: 10.4049/jimmunol.172.6.3948. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Wittekind Ch, (eds) UICC TNM Classification of Malignant Tumors. 6. John Wiley and Sons, Inc., NewYork; 2002. [Google Scholar]
- Wang Z, Jiang L, Huang C, Li Z, Chen L, Gou L, Chen P, Tong A, Tang M, Gao F, Shen J, Zhang Y, Bai J, Zhou M, Miao D, Chen Q. Comparative proteomics approach to screening of potential diagnostic and therapeutic targets for oral squamous cell carcinoma. Mol Cell Proteomics. 2008;7:1639–1650. doi: 10.1074/mcp.M700520-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence database using mass spectrometric data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kornberg LJ, Villaret D, Popp M, Lui L, McLaren R, Brown H, Cohen D, Yun J, McFadden M. Gene expression profiling in squamous cell carcinoma of the oral cavity shows abnormalities in several signaling pathways. Laryngoscope. 2005;115:690–8. doi: 10.1097/01.mlg.0000161333.67977.93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The enlarged and cropped images of 11 selected protein spots in 2D Gel. 11 proteins were selected as examples showing the consistent expression changes in enlarged form. The images of each changed protein spot were compared with the control. (A) 2-DE gel images of 11 selected protein indicated by arrows in the panels. Each panel shows an enlarged view of the gel spots from Figure 2. (B) Volume density analysis graphs: the data were expressed as mean ± SD of twelve repeats.