Abstract
The mechanism of muscle weakness was investigated in an Australian family with a M9R mutation in TPM3 (α-tropomyosinslow). Detailed protein analyses of five muscle samples from two patients showed that nemaline bodies are restricted to atrophied type 1 (slow) fibers in which the TPM3 gene is expressed. Developmental expression studies showed that α-tropomyosinslow is not expressed at significant levels until after birth, thereby likely explaining the childhood (rather than congenital) disease onset in TPM3 nemaline myopathy. Isoelectric focusing demonstrated that α-tropomyosinslow dimers, comprised of equal ratios of wild-type and M9R-α-tropomyosinslow, are the dominant tropomyosin species in three separate muscle groups from an affected patient. These findings suggest that myopathy-related slow fiber predominance likely contributes to the severity of weakness in TPM3 nemaline myopathy because of increased proportions of fibers that express the mutant protein. Using recombinant proteins and far Western blot we demonstrated a higher affinity of tropomodulin for α-tropomyosinslow compared to β-tropomyosin; the M9R substitution within α-tropomyosinslow greatly reduced this interaction. Finally, transfection of the M9R mutated and wild-type α-tropomyosinslow into myoblasts revealed reduced incorporation into stress fibers and disruption of the filamentous actin network by the mutant protein. Collectively, these results provide insights into the clinical features and pathogenesis of M9R-TPM3 nemaline myopathy.
Keywords: Disease severity, Nemaline myopathy, Skeletal muscle, Tropomodulin, Tropomyosin
INTRODUCTION
Nemaline myopathy (NM) is an inherited neuromuscular disease characterized by general muscle weakness and rod shaped structures in skeletal muscle. The disorder ranges in severity from severe (lethal) congenital to childhood- and adult-onset forms. NM results from mutations in genes encoding thin filament proteins, including α-skeletal actin, nebulin, β-tropomyosin, troponin T, α-tropomyosinslow (1) and the actin regulator protein cofilin-2 (2). The first disease causing mutation (M9R) was identified over a decade ago, in the α-tropomyosinslow gene (TPM3) in a large Australian autosomal dominant family with childhood onset NM (3). Two additional mutations in TPM3 with recessive inheritance have been identified in patients with severe congenital (4) and intermediate (5) forms of NM. An R167H mutation in TPM3 has been identified in a sporadic (6) case and in a large family with NM (7). Every NM patient who harbors a TPM3 mutation contains rods only within type 1 muscle fibers (4–8). In addition, dominant mutations in TPM3 have recently been identified in 11 patients from six families with congenital fiber type disproportion in whom the muscle pathology is characterized by small type 1 fibers but with absence of nemaline bodies (9).
The identification of the M9R mutation in TPM3 paved the way for research into understanding the molecular mechanisms of NM. A transgenic mouse model of the M9R α-tropomyosinslow mutation has a late onset of muscle weakness, nemaline bodies within its skeletal muscle fibers, a predominance of atrophied oxidative (type 1) fibers and hypertrophied glycolytic (type 2) fibers (10); features that are also observed in patients with the M9R mutation in TPM3 (8). Isolated muscles from M9R transgenic mice do not show any overt differences in the maximal force generated when muscle fibers are stimulated at optimum sarcomere lengths, but they exhibit impaired force generation at shorter (i.e. below optimum) sarcomere lengths (11).
Cell biology studies have highlighted functional deficits of the M9R substitution within TPM3 that may contribute to the hypotonia and sarcomeric disruption in NM patients possessing the M9R mutation. Studies with recombinant proteins have shown that introduction of the M9R substitution in TPM3 reduces the affinity of recombinant M9R α-tropomyosin for F-actin by as much as 100-fold (12, 13). Circular dichroism studies showed that the binding of tropomodulin to an N-terminal tropomyosin fragment containing the M9R substitution was abolished (14). Furthermore, substitution of M9R within the human α-tropomyosinfast gene resulted in reduced sensitivity of isometric force production to activating calcium in adenovirally transduced rat cardiac myocytes (15).
Previous analysis of skeletal muscle from two related patients bearing the M9R mutation revealed a highly specific pathological finding of atrophied type 1 fibers containing nemaline rods and hypertrophied type-2 fibers without rods (8). We have since obtained three muscle samples from one of these individuals with M9R TPM3 NM and have shown that this pathology is not consistent in all four muscle samples, thereby providing valuable information with respect to the diagnosis of primary TPM3 mutations in patients with NM. In the present study, we performed a thorough analysis of four muscle samples from a single patient bearing the M9R mutation, determining the mutant protein load in each muscle and the effect on endogenous tropomyosin isoform regulation. We show that slow muscle fibers in TPM3 NM muscle express α-tropomyosinslow dimers as the dominant sarcomeric tropomyosin species, with roughly equal levels of wild-type and mutant M9R proteins detected within the myofibrillar fraction. We also demonstrate that altered tropomyosin dimer populations may affect fiber-type specific troponin expression and we examine the localization and binding of tropomodulin within patient muscle expressing a roughly equal ratio of wild-type and mutant M9R α-tropomyosinslow.
MATERIALS AND METHODS
Skeletal Muscle Samples
We studied archival frozen human fetal skeletal muscle biopsies from fetuses delivered at 14, 16, 19, and 22 weeks gestation. All specimens were de-identified in accordance with the ethical guidelines of the Children's Hospital at Westmead. We also studied skeletal muscle biopsies from individuals of the following ages: 25 and 36 weeks gestation, 8 days, 5 months, 5 years, and 28 years. All of the samples were from patients with no known form of neuromuscular disease and showed normal muscle histology. All muscle samples from patient 1 were obtained at autopsy (at age 46 years); the controls (28 and 38 years) and Patient 2 muscle samples (22 years) were obtained at biopsy.
Immunohistochemistry Analysis
Indirect immunofluorescence was performed as previously described (16) with the following exceptions: 1) For troponin Islow, muscle cryosections were fixed with 3% paraformaldehyde for 3 minutes, washed three times in phosphate buffered saline (PBS), then extracted with –20°C methanol for 5 minutes. 2) For tropomodulin, immunohistochemistry (IHC) was performed as previously described (17). Antibody sources and dilutions are listed in the Table. Images were captured as previously described (16). Muscle morphometry was performed on at least 200 fibers using ImageQuant software.
Table.
Antibodies Used for Immunohistochemical Analysis*
| Antibody | mAb or pAb | Dilution | Source |
|---|---|---|---|
| Anti-skeletal slow myosin | mAb | 1:250 | Chemicon International |
| Anti-skeletal fast myosin | mAb | 1:200 | Sigma |
| Anti-α-actinin 2 | pAb | 1:500 | Gift from Prof Alan Beggs, Boston Children's Hospital (Boston, MA) |
| Anti-troponin T slow | mAb | 1:30 | Developed by Lin JC obtained from DSHB** |
| Anti-troponin I fast | mAb | 1:100 | Dr Takeshi Nakamura, Kitasato University, Kanagawa, Japan |
| Troponin I slow | mAb | 1:10 | Dr Takeshi Nakamura, Kitasato University |
| Anti-tropomodulin #1749 | pAb | 1:185 | Dr Velia Fowler, Scripps Institute, La Jolla University (La Jolla, CA) |
| CY3-conjugated anti-mouse IgG secondary | mAb | 1:250 | Jackson Laboratories |
| CY3-conjugated anti-rabbit IgG secondary | pAb | 1:250 | Jackson Laboratories |
| Alexa488 conjugated anti-mouse IgG | pAb | 1:200 | Molecular Probes |
| Rhodamine-phalloidin-TRITC conjugated | Not an antibody, but a phallotoxin that binds to filamentous actin | 1:400 | Sigma |
Abbreviations: mAb = monoclonal antibody, pAb= polyclonal antibody, DSHB = Developmental Studies Hybridoma Bank.
The anti-troponin T slow antibody was developed by JC Lin and obtained from the DSHB under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, The University of Iowa.
Protein purification
Recombinant human Tmod1 protein was expressed in E. coli and purified as described (18). Tropomyosins were purified as in (13).
Far Western Blots
Increasing amounts of purified Tmod1 or TPM isoforms were separated by 12% or 15% SDS-PAGE. Parallel gels were stained with Coomassie Brilliant Blue or transferred onto nitrocellulose and Tmod1 or TPM binding was assessed by a blot overlay technique (19). The transferred proteins were bound to the membrane followed by two 5-minute washes at 4°C in OverWash Buffer (20 mM HEPES, pH 7.3, 80 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2% Triton X-100). The blots were overlayed with 3 μg of purified protein incubated for 12 to 18 hours at 4°C and excess protein was removed by five 5-minute washes with OverWash. Primary antibodies (anti-Tmod1 monoclonal mAb95 [20] or anti-TPM polyclonal antibodies [17]) were added and incubated for 12 to 18 hours at 4°C. The blots were rinsed with two consecutive OverWash rinses followed by five 10-minute shaking washes. Peroxidase-conjugated secondary antibody was added for 1 hour in Blotto (50 mM Tris, pH 7.5, 185 mM NaCl, 0.05% Tween-20, 3% powdered skim milk). The blots were subsequently washed twice in Blotto and twice in Otto (Blotto minus milk) followed by ECL detection. Western blots were performed as above without the protein overlay step. Gels and blots were scanned and quantitated using ImageJ (NIH). The percent of Tmod1 binding to each TPM is the average of the Far Western blot signal divided by the corresponding Coomassie signal for each isoform set with the Tmod1:wild-type α-tropomyosinslow interaction set to 100%. The Far Western analysis was repeated multiple times with representative blots shown.
Western Blot Analysis of Tropomyosin and Tropomodulin
Samples were extracted into myofibrillar (insoluble) and soluble protein pools as previously described (16). Tropomyosin isoforms were separated as outlined in (21) and immunoblotted as described in (22) using anti-tropomyosin antibody (TM311, 1:10,000 dilution, Sigma). Separation of tropomodulin was performed using 4% to 12% Bis-Tris NuPAGE™ pre-cast gels (Invitrogen, Carlsbad, CA). Immunoblotting of tropomodulin was performed as previously described (17).
Isoelectric Focusing
Isoelectric focusing was performed using a Multiphor II system (GE Healthcare, Little Chalfont, Buckinghamshire, UK) according to methods described in (16) using 18-cm immobiline dry strips (pH 4.5-5.5, GE Healthcare) and focused for a total of 60,000 V/hours. The isoelectric point (pI) for proteins was calculated using the pI analysis tool available at the web site http://www.expasy.org/
Denaturation of Muscle Thin Filament with Urea
An insoluble myofibrillar fraction was obtained from frozen muscle sections after two washes in 1 ml PBS containing 0.5% triton X-100 and protease inhibitors (1:500, Sigma). The pellet was resuspended in 250 μl of PBS containing protease inhibitors and aliquots were incubated with increasing concentrations of urea (prepared in PBS). The samples were incubated for exactly 15 minutes on ice then centrifuged at 18,000 g for 10 minutes at 4°C. Forty μl of the supernatant were removed and solubilized in 2X SDS loading buffer (4% SDS, 40% glycerol, 100 mM Tris pH 6.8, 100 mM DTT), heat inactivated at 94° C for 3 minutes and analyzed by SDS-PAGE.
Expression Constructs
The wild-type TPM3 cDNA sequence was amplified from reverse transcribed human skeletal muscle RNA (Applied Biosystems, Foster City, CA) using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and the following primers (forward 5’-cgtgtacggtgggaggtct-3’; reverse 5’-atcctggtcgagctggacg-3’, SigmaGenosys, Sydney, Australia), which introduced an XhoI and EcoR1 restriction site respectively. The resultant polymerase chain reaction (PCR) product was cloned into the TOPO (blunt) plasmid (Invitrogen) and clones were sequenced to confirm that no PCR-induced errors were present. The M9R mutation was introduced into an error-free WT-TPM3TOPO clone through site-directed mutagenesis (QuickChange Site-Directed Mutagenesis kit, Stratagene). Both the WT-TPM3 and M9R-TPM3 cDNA sequences were released from the TOPO (blunt) vector using XhoI and EcoRI (New England Biolabs, Ipswich, MA), gel purified (Qiagen, Hilden, Germany) and cloned into a similarly digested pEGFP-N1 vector (Clontech, Mountain View, CA). Clones were sequenced to confirm that the WT-TPM3EGFP sequence was still error-free and that the M9R-TPM3 EGFP had the appropriate mutation.
Cell Culture
C2C12 cell culture and transfection was performed as previously described (16).
RESULTS
Clinical Patient Data
This study examines two patients (Patients 1 and 2) from a large autosomal dominant Australian family with NM and a M9R mutation in TPM3 (3). Interestingly, Patients 1 and 2 presented with predominantly distal weakness and foot drop, reminiscent of a peripheral neuropathy. Patient 1 was confined to a wheelchair in her late thirties and died at age 46 years from adult respiratory distress syndrome. Her cousin, Patient 2 presented in mid-childhood with foot drop and predominantly distal muscle weakness. She has had a relatively non-progressive disease course and is ambulant and active in her mid-twenties.
Patients with a M9R Mutation in TPM3 Show Variable Fiber Type Distribution with Nemaline Bodies Detected Only in Type 1 Fibers
Our previous characterization of pathological findings in NM due to a M9R mutation within TPM3 was performed on single muscle samples from Patient 1 (biceps) and Patient 2 (quadriceps) (8) (Fig. 1). Histological analysis highlighted a striking pathology associated with this mutation. The muscle samples consisted of a mixed population of rod-containing atrophied type 1 fibers and hypertrophied type 2 fibers that do not contain rods (Fig. 1C, G). We have since obtained three additional muscle samples from Patient 1 (quadriceps, deltoid, and adductor pollicis). In contrast to our findings in the biceps, the quadriceps, deltoid, and adductor pollicis exhibit almost complete type 1 myofiber predominance, with rods in almost all fibers (Fig. 1E, F). Closer examination of fiber size reveals these rod-containing slow fibers are atrophic (quadriceps mean diameter 32.0 μm ± 5.4, n = 200 fibers), compared to the mean fiber diameter from a healthy adult female quadriceps muscle (51.3 μ ± 7.9 [23]). Thus, different muscle groups from a single individual affected with M9R TPM3 NM respond differently to expression of the mutant M9R α-tropomyosinslow isoform, with respect to slow fiber predominance. Importantly, in all muscles examined from both patients bearing this mutation, the abnormalities remain restricted to slow fibers in which the TPM3 gene is expressed. Thus, slow fiber predominance likely contributes to the severity of weakness and slow fiber conversion may contribute to disease progression.
Figure 1.
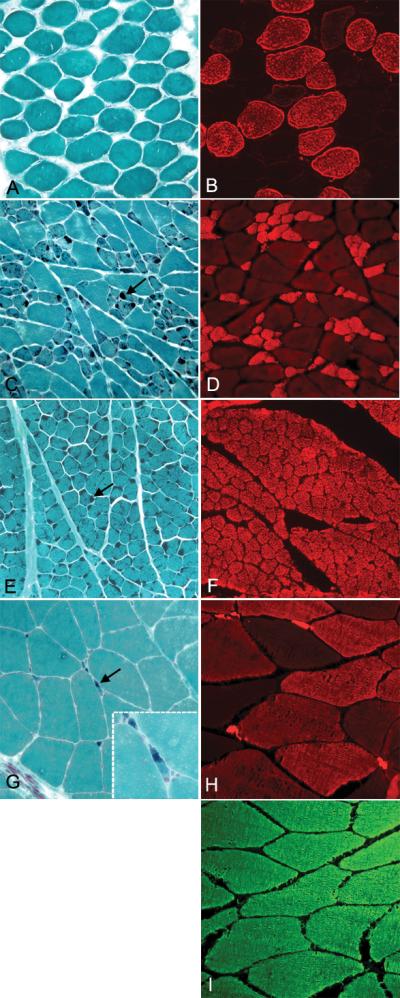
Rods are localized to atrophied type 1 fibers in patients with the M9R mutation in TPM3. (A, C, E, G) Frozen muscle sections stained using the modified Gomori Trichrome method (31) in control (A) (quadriceps muscle biopsy from a 35-year-old female with no known neuromuscular disease), Patient 1 (bicep, C and quadriceps E, 46 years of age) and Patient 2 (quadriceps, G, 22 years of age). (B, D, F, H) Panels show immunolabeling with anti-myosin heavy chain slow (anti-MHCslow) in control (B), Patient 1 (bicep, D and quadriceps F) and Patient 2 (quadriceps, H). (I) Panel shows immunolabeling with anti-myosin heavy chain fast (anti-MHCfast) in Patient 2. Rods are dark blue in all fibers of the quadriceps (E) muscle in Patient 1 (arrow). They are evident in atrophied fibers in the biceps muscle from Patient 1 (C, arrows) and in Patient 2 (G, arrows). Immunolabeling with anti-myosin heavy chain slow (anti-MHCslow) shows that the rods are restricted to slow fibers (D, F, H). Staining of control muscle for anti-MHCslow shows labeling of ~50% of fibers (B). Patient 2 exhibits a complete type 2 fiber predominance (I, anti-MHCfast) with ~50% of these fibers co-expressing anti-MHCslow (H). All images were originally taken at 20x magnification except for inset of (G), which is 100x magnification.
Late Developmental Expression of α-Tropomyosinslow Explains the Childhood Onset Form of Patients with a M9R Mutation in TPM3
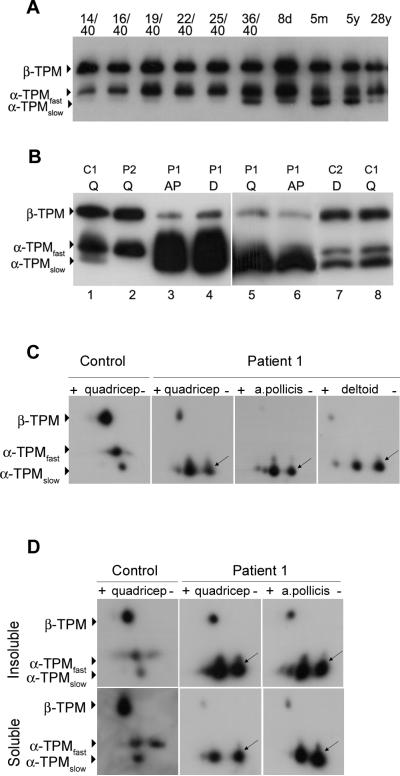
All patients with the M9R mutation in TPM3 have the childhood onset form of NM. α-tropomyosinslow and α-tropomyosinfast are expressed in slow and fast skeletal muscle fibers respectively. During human skeletal muscle development, fast fibers predominate and complete fiber typing profile is not established until approximately nine months of age, as determined using myosin heavy chain fiber type specific antibodies (24). The developmental expression of tropomyosin isoforms in human skeletal muscle has not previously been defined at the protein level. By examining the expression of tropomyosin isoforms during human skeletal muscle development, we determined that β-tropomyosin and α-tropomyosinfast are the predominant isoforms during development (Fig. 2A). In contrast, α-tropomyosinslow is expressed at extremely low levels during development and is not expressed at significant levels until around birth. The lack of α-tropomyosinslow expression during development likely accounts for the childhood (rather than congenital) onset of clinical disease in patients with NM due to a M9R mutation in TPM3.
Figure 2.
Expression of M9R mutant α-tropomyosinslow suggests a dominant negative effect. Frozen muscle samples were separated by SDS-PAGE using 3.4 M urea containing acrylamide gels. The PVDF membranes were probed with a tropomyosin antibody that recognizes both muscle and non-muscle isoforms of tropomyosin (anti-TM311, Sigma). (A) Frozen skeletal muscle samples from controls ranging in age from 14 weeks gestation (14/40) to 28 years (y) were used to assess the expression of tropomyosin isoforms throughout human development. β-Tropomyosin (β-TPM) and α-tropomyosinfast (α-TPMfast) are expressed during embryonic development, the neonatal period and throughout adulthood. α-Tropomyosinslow (α-TPMslow) is expressed at very low levels during embryonic development (14–25 weeks gestation) and is upregulated after birth (36/40) and throughout adulthood. (B) Three tropomyosin isoforms are expressed in adult control (C1 [deltoid 38 years] and C2 [quadriceps 28 years]) muscle: β-tropomyosin (β-TPM), α-tropomyosinfast and α-tropomyosinslow. The quadriceps (Q), deltoid (d) and adductor pollicis (AP) muscles from Patient 1 (P1) show increased levels of α-tropomyosinslow and reduced levels of β-tropomyosin compared to controls. Patient 2 (P2) does not express α-tropomyosinslow due to the predominant expression of fast MHC in the majority of large fibers. (C) Isoelectric focusing of an age-matched control using whole muscle lysates shows three spots corresponding to wild-type β-tropomyosin (β-TPM), α-tropomyosinfast (α-TPMfast) and α-tropomyosinslow (α-TPMslow). The predicted pI of α-TPMslow is 4.68. In contrast, two spots are clearly evident in all Patient 1 muscle samples examined. M9R mutant α-tropomyosinslow (predicted pI of 4.71, arrows) is slightly more basic and is expressed at roughly equal levels to wild-type α-TPMslow in these samples. α-TPMfast is not present in the patient muscle due the complete type 1 fiber predominance. In all of the patient muscle samples examined β-TPM is reduced relative to α-TPMslow. (D) Extraction of muscle into insoluble and soluble protein pools shows that M9R mutant α-tropomyosinslow (arrows) localizes to the insoluble protein pool in appreciable quantities, suggesting that it contributes to the sarcomeric thin filaments and supporting a dominant negative (poison-protein) effect rather than haploinsufficiency (shortage of normal tropomyosin protein).
2D PAGE Demonstrates Expression of Mutant M9R α-Tropomyosinslow in Patient Muscle, Suggesting a Dominant Negative Mode of Pathogenesis
Western blot analysis of tropomyosin isoform expression in Patient 1 shows upregulation of α-tropomyosinslow protein and reduced levels of β-tropomyosin in the quadriceps muscle (Fig. 2B, lanes 3–6), confirming findings previously described for the deltoid and adductor pollicis (16, 25). α-Tropomyosinfast was markedly reduced or absent in the quadriceps and adductor pollicis muscle from Patient 1, consistent with complete type 1 fiber predominance as indicated by positive staining of all fibers with MHCslow and absent labeling with MHCfast (shown for adductor pollicis muscle, see Fig. 1). Normal levels of β-tropomyosin and α-tropomyosinfast were, however, observed in Patient 2, with absent α-tropomyosinslow (Fig. 2B, lane 2). These results likely reflect tropomyosin expression within healthy type 2 fibers in this patient due to extreme atrophy of the type 1 rod-containing fibers and their low relative contribution to the whole muscle lysate (Fig. 1G).
Mutant M9R α-tropomyosinslow protein is more basic (predicted pI of 4.71) compared to wild-type α-tropomyosinslow (predicted pI of 4.68) and is expressed in roughly equal proportions, with slight variations in the ratio of mutant to wild-type α-tropomyosinslow. We exploited the net charge change resulting from the M9R substitution to examine the levels of mutant and wild-type α-tropomyosinslow in the three muscle samples from Patient 1 that exhibited complete type 1 fiber predominance. In whole muscle lysates from control muscle, three spots corresponding to β-tropomyosin, α-tropomyosinfast and α-tropomyosinslow were detected using the anti-TM311 antibody (Fig. 2C). In all three muscles from Patient 1 we confirmed the marked downregulation of β-tropomyosin, absent α-tropomyosinfast, and two distinct spots for α-tropomyosinslow.
To confirm that mutant M9R α-tropomyosinslow was present within the insoluble myofibrillar fraction we first extracted muscle samples into insoluble and soluble fractions with 0.5% triton X-100 and repeated 2D-PAGE analysis. In both the quadriceps and adductor pollicis (insufficient material was available for the deltoid), mutant M9R type α-tropomyosinslow was detected in the insoluble myofibrillar protein fraction, strongly supporting its localization within the thin filament (Fig. 2D). Densitometric analysis showed a slight predominance of the relative levels of myofibrillar WT tropomyosin (quadriceps 57%, a. pollicis 58%) compared with M9R tropomyosin (quadriceps 43%, a. pollicis 42%); these experiments could be performed only once. Thus, mutant M9R and WT α-tropomyosinslow proteins are equally represented within the insoluble protein fraction that would include proteins of the thin filament, and potentially also proteins sequestered within insoluble nemaline bodies.
In summary, Figure 2 demonstrates three separate muscle groups from Patient 1, each consisting of only type 1 fibers, which show marked reduction in levels of β-tropomyosin and dominant expression of α-tropomyosinslow, comprised of roughly equal amounts of M9R and WT α-tropomyosinslow proteins. We have previously shown that recombinant M9R- α-tropomyosinslow can effectively dimerize with itself, WT-α–tropomyosinslow and with β-tropomyosin, although shows a preference for α:α dimers rather than the usual α:β heterodimer pairing (25). Thus, our observation that both M9R and WT α-tropomyosinslow are equally represented within the insoluble myofibrillar fraction, together with previous evidence that M9R can effectively form dimers in vitro (25), collectively support the notion that M9R tropomyosin is incorporated into the skeletal muscle thin filament and exerts a dominant negative effect on skeletal muscle function. In addition, the marked skewing in the tropomyosin dimer population, from α/β heterodimers to α/α homodimers (some containing the mutant M9R protein), may also induce a detrimental effect on contractile function.
M9R TPM3 Patient Muscle with Complete type 1 Fiber Predominance Shows Abnormal Expression of Troponin Ifast in type 1 Fibers Expressing Only Slow Myosin
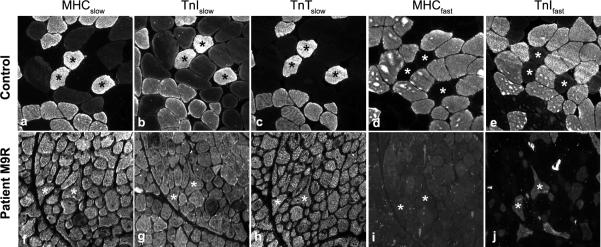
Due to the altered ratio of tropomyosin isoform expression observed in Patient 1, we queried whether altered tropomyosin dimers affected troponin isoform regulation and recruitment to the sarcomere (Fig. 3). Troponin Islow, and troponin Tslow labeled all fibers of the quadriceps, adductor pollicis and deltoid muscles, consistent with complete type 1 fiber predominance (Fig. 3g, h). Interestingly, troponin Ifast was detected in type 1 fibers positive only for MHCslow (negative for MHCfast, actinin 3, developmental myosin [data not shown]) in all three muscles examined (Fig. 3j, shown for adductor pollicus). Despite positive labeling of troponin Ifast in a subset of muscle fibers by IHC, only a very faint band was detected for troponin Ifast by Western blotting (Fig. 4B). This suggests that IHC is a more sensitive tool than Western blotting and that the level of expression of troponin Ifast is below the level of detection by Western analysis. Dysregulation of troponin Ifast in slow fibers was not observed in four other NM patients with type 1 fiber predominance (3 patients with a defined ACTA1 mutation, V163M and I136M and 1 patient with unknown genetic cause in whom mutations in ACTA1 and TPM3 have been excluded (data not shown). This suggests that altered regulation of troponin Ifast is not a common feature observed in NM and may be specific to Patient 1 (M9R TPM3), perhaps as a consequence of altered tropomyosin isoform regulation.
Figure 3.
Inappropriate expression of troponin I fast is specific to a patient with a M9R mutation in TPM3. IHC was performed on an age-matched control (deltoid, 38 years) and Patient 1 (M9R mutation, a. pollicis) samples. Sequential sections were used to track individual fibers in multiple stains (asterisks). Appropriate expression of fiber types is evident in control muscle using myosin heavy chain (MHC)- and troponin (Tn)-specific antibodies (a–e). In contrast, the patient with the M9R mutation in TPM3 (f–j) expresses Troponin I fast (j) in a subset of fibers despite complete type 1 fiber predominance in this muscle, as shown by MHCslow (f). All images taken at 20x magnification.
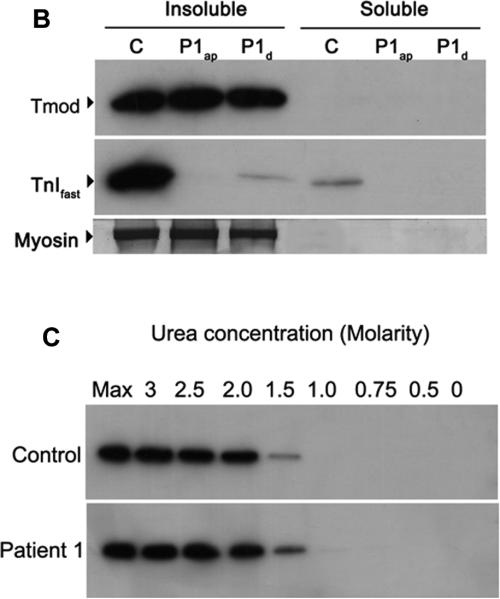
Figure 4.
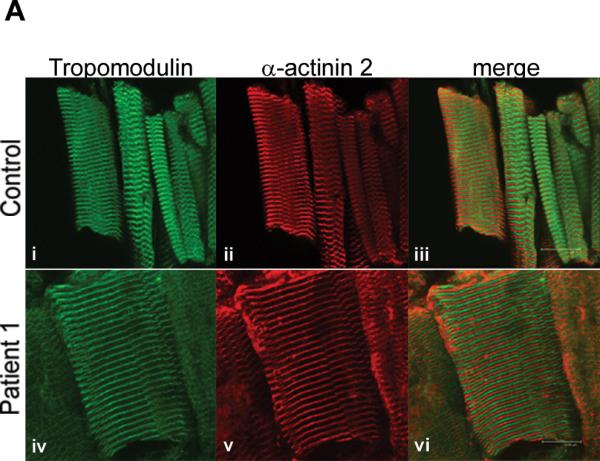
Tropomodulin is normally expressed and bound to the thin filament in a patient with a M9R mutation in TPM3. (A) Immunohistochemistry with tropomodulin shows a striated pattern in control (i, quadriceps muscle) and in Patient 1 (iv, adductor pollicis muscle). Labeling with α-actinin 2 also shows a striated pattern indicating its localization to the Z-line (ii) and (v). Tropomodulin does not colocalize with α-actinin 2 (iii and vi) due to its localization at the pointed end of the thin filament. The images were taken on a Leica SP2 confocal microscope at 60x magnification and 2.5x zoom. (B) Frozen muscle sections from Patient 1 (M9Rap = adductor pollicis, M9Rd = deltoid) and an age-matched control (quadriceps) were extracted into insoluble and soluble protein pools using 0.5% triton X-100 and separated by SDS-PAGE. Tropomodulin is expressed in the control and in both of the patient muscle samples in the insoluble pool. Troponin Ifast is detected at high levels in the control with only low levels in the patient samples. The PVDF membrane was stained with Coomassie blue myosin is shown to illustrate equal protein loading for each sample. (C) Myofibrillar proteins were extracted from frozen muscle sections from Patient 1 (adductor pollicis muscle) and an age matched control using 0.5% triton X-100 and denatured using varying concentrations of urea (0.5–3 M). The samples were solubilized, separated by SDS-PAGE and immunoblotted using anti-tropomodulin (Tmod#1749). In both the patient and the control samples the majority of tropomodulin (~80%) is denatured at 1.5 M urea, suggesting that there is no difference in the binding dynamics of tropomodulin to the thin filament.
The M9R Mutation Abolishes the Interaction or Tmod1 with α-Tropomyosinslow but Does not Disrupt Tmod1 Localization to Thin Filament Pointed Ends
In vitro binding studies using circular dichroism had shown that binding of a tropomodulin peptide to an N-terminal fragment of α-tropomyosinfast containing the M9R substitution is abolished (14). Therefore, we explored the expression, localization and binding of tropomodulin to the thin filament in muscle from the patient with a M9R mutation. Co-labeling of tropomodulin (Fig. 4A, i and iv) with α-actinin 2 to mark the Z-line (Fig. 4A, ii and v) showed a clear striated staining pattern in the quadriceps and adductor pollicis muscles from Patient 1, identical to that of the control (Fig. 4A, iii and vi). Partial overlay was observed following co-labeling for tropomodulin and phalloidin (not shown), consistent with normal localization of tropomodulin at the pointed end of the thin filament (Fig. 4A).
To examine the strength of the association of tropomodulin to the thin filament further, we used partial denaturation with increasing concentrations of urea to compare dissociation of tropomodulin from a muscle myofibrillar fraction prepared from Patient 1 and an age-matched control. Figure 4B demonstrates that tropomodulin localizes exclusively to the insoluble myofibrillar fraction following crude extraction in 0.5% triton X-100, in both Patient 1 and control muscle. Denaturation of this myofibrillar preparation with increasing concentrations of urea shows an identical profile of tropomodulin dissociation in both patient and control (Fig. 4C). Thus, our data suggest that tropomodulin is normally localized to the pointed end of the thin filament in patient muscle containing a roughly equal ratio of M9R:WT α-tropomyosinslow as the dominant tropomyosin species; in addition, there is no major difference in the tenacity of this binding as determined by denaturation of myofibrillar protein with urea.
Tmod Far Western Analysis
Purified recombinant tropomyosin proteins (β, αslow and M9Rαslow) were separated by SDS-PAGE, transferred to PVDF membranes and incubated with recombinant Tmod1 in the overlay solution. Figure 5 demonstrates that Tmod1 binds to both wild-type α-tropomyosinslow and β-tropomyosin, but shows a greatly decreased interaction with the mutant M9R α-tropomyosinslow. Tmod1 also shows a distinct preference for binding to α-tropomyosinslow over β-tropomyosin, as indicated by an increase in Tmod1 binding (Fig. 5A). Quantification of the relative binding of Tmod1 to the TPM isoforms demonstrated that the interaction of Tmod1 with the mutant M9R-α-tropomyosinslow was approximately 10% of the level of binding to wild-type α-tropomyosinslow whereas binding of Tmod1 to α-tropomyosin was approximately 50% of the level of binding to wild-type α-tropomyosinslow (Fig. 5C). Next, we compared Tmod1-TPM interactions using the TPMs in the overlay solution and Tmod1 on the blots (Fig. 5B). This approach also showed that binding of M9R-α-tropomyosinslow to Tmod1 could not be detected. (Note that in this approach, binding of α-tropomyosinslow to Tmod1 cannot be compared directly to binding of β-tropomyosin to Tmod1 because exposures for the two blots were different).
Figure 5.
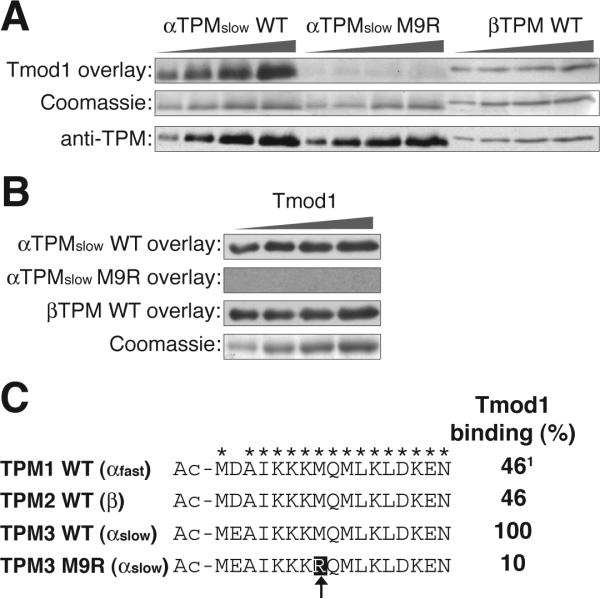
Tmod1 has a decreased interaction with the M9R α-tropomyosinslow mutant. (A) Increasing amounts of TPM isoforms were separated by SDS-PAGE and the interaction with Tmod1 (upper panel) was monitored by Far Western blotting for Tmod1. Parallel gels were Coomassie-stained (middle panel) to demonstrate equivalent loading of the gel and Western blotted with an anti-TPM polyclonal antibody (bottom panel) to demonstrate that the anti-TPM antibody used in B recognizes all of the purified TPMs. (B) Increasing amounts of Tmod1 were separated by SDS-PAGE on parallel gels and the interaction with wild-type α-tropomyosinslow (upper panel), M9R α-tropomyosinslow mutant (upper middle panel), and wild-type β-tropomyosin (bottom middle panel) was monitored by Far Western blotting for TPM. A parallel gel was run and Coomassie stained (bottom panel) to monitor loading of the gel. (C) The N-terminal Tmod1-binding domains of the human TPM genes are identical except for an aspartic acid [D] to glutamic acid [E] substitution (asterisks represent conserved residues). The position of the M9R mutation in α-tropomyosinslow is highlighted and marked by an arrow. The relative interaction affinity between Tmod1 and each TPM isoform (determined from A) is indicated on the right where the Tmod1:wild-type α-tropomyosinslow interaction is set to 100%. α-tropomyosinfast binding to Tmod1 was not tested; the Tmod1-binding quotient was deduced to be similar to β-tropomyosin as their Tmod1-interaction domains at the N-terminus are identical.
Comparison of the Tmod1 binding N-terminal sequences in exon 1a from the TPM1, TPM2, and TPM3 genes coding for α-tropomyosinfast, β-tropomyosin, and α-tropomyosinslow, respectively, demonstrates that the αfast and the β sequences are identical, while the αslow sequence differs only by a conserved glutamic acid [E] for aspartic acid [D] substitution (Fig. 5C). Yet, this amino acid change leads to about a two-fold increase in Tmod1 binding to the α-tropomyosinslow compared to β-tropomyosin (equivalent to α-tropomyosinfast). Nevertheless, the M9R substitution in the α-tropomyosinslow leads to a 10-fold reduction in binding of Tmod1 (our data), similar to the effect of the M9R substitution in the α-tropomyosinfast sequence (14). Thus, small changes within the Tmod1-binding site of TPMs can drastically alter the interaction between these proteins (Fig. 5C). Additionally, our data suggest that the compensatory upregulation of the stronger Tmod1-binding α-tropomyosinslow expression (Fig. 2) is responsible for localizing normal levels of Tmod1 to pointed ends in the patients’ skeletal muscles.
The M9R Mutant Tropomyosin Disrupts the Cytoarchitecture and Morphology of C2C12 Myoblasts
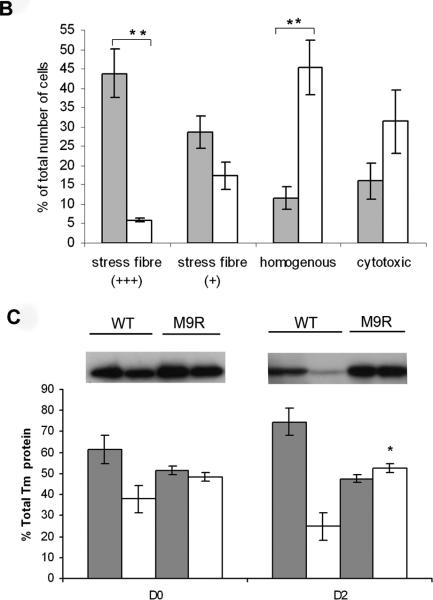
To examine the localization of the M9R mutant tropomyosin in C2C12 myoblasts we generated a C-terminal tagged tropomyosin-enhanced green fluorescent protein (EGFP) fusion construct. Transfection with wild-type α-tropomyosinslow-EGFP (WT-TPMEGFP) into C2C12 myoblasts resulted in stress fiber staining in the majority of cells (Fig. 6A, i–iii), diffuse cytoplasmic staining in a subset of cells, and evidence of cytotoxicity in a few cells. By contrast, C2C12 myoblasts transfected with M9R-TPMEGFP did not typically localize to stress fibers but predominantly showed a homogenous distribution of staining (Fig. 6A, iv-vi), with many cells appearing cytopathic, as assessed by membrane blebbing and rounding (Fig. 6A, iv). In many, but not all cases, C2C12 myoblasts transfected with M9R-TPMEGFP also showed reduced labeling of phalloidin to stress fibers (Fig. 6A, v), suggesting that expression of the mutant TPM isoform disrupts endogenous actin filaments. By semiquantitative assessment (Fig. 6B) 1,179 WT-transfected cells were scored as showing moderate and high stress fibers (+++/+, 36 ± 13%), homogenous staining (12 ± 6%,) and cytotoxicity (16 ± 9%) (see Fig. 6 legend for details of morphological scoring). For M9R-TPMEGFP–transfected cells, 1,270 were scored as moderate and high stress fibers (+++/+, 12 ± 8%, p = 0.0006 vs. WT-transfected for clearly defined stress fibers [+++]), homogenous staining 45 ± 14% (p = 0.011 vs. WT-transfected), cytotoxicity = 31 ± 16% (p = 0.16 vs. WT-transfected).
Figure 6.
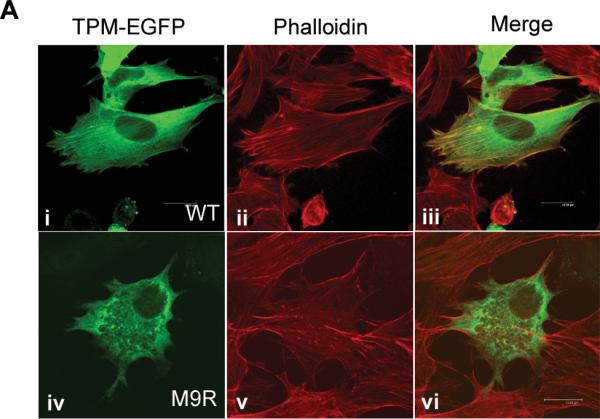
M9R mutant α-tropomyosinslow protein does not incorporate efficiently into stress fibers and disrupts the cellular architecture. (A) WT-TPMEGFP predominantly localizes to stress fibers (i) in transfected C2C12 cells, as shown by co-labeling for phalloidin (ii), which specifically recognizes filamentous actin. Colocalization is evident in (iii). In contrast, C2C12 cells transfected with M9R-TPMEGFP generally results in homogenous cytoplasmic staining (iv) in the majority of transfected cells. In many M9R-TPMEGFP transfected cells, phalloidin stress-fiber labeling appeared less defined (v), suggesting disruption of the endogenous actin cytoskeleton; there is no colocalization (vi). Images were captured on a SP2 Leica confocal microscope at 63x magnification and 2x zoom. (B) Semiquantitative analysis of stress fiber localization in WT- and M9R-TPMEGFP-transfected cells was performed in two separate experiments in which replicate cover slips were independently assessed by three blinded reviewers, and ~300 cells scored as follows: +++ clearly defined stress fibers, + predominantly homogenous staining but some stress fibers evident, - homogenous staining, 0 cytotoxicity (as assessed by membrane blebbing and cell rounding). Shaded bars are data from WT-transfected and open bars are M9R-TPMEGFP-transfected cells. Asterisks indicate significant differences (see Results). (C) C2C12 cells transfected with WT-TPMEGFP and M9R-TPMEGFP were separated into insoluble (grey bars) and soluble (white bars) protein pools after 24 hours of transfection (D0) or following two days of differentiation (D2). SDS-PAGE was performed using anti-TPM311 and a band at ~60 kDa was detected for the tropomyosin/EGFP fusion protein. Representative western blots images are shown from 5 independent experiments performed in duplicate. There is significantly more soluble protein in M9R-transfected compared to WT-transfected in D2 myotubes (p < 0.05, using a Mann-Whitney non-parametric test).
These immunocytochemical observations are supported by centrifugation studies from transfected C2C12 cells in which M9R-TPMEGFP showed a significant impairment in its ability to contribute to insoluble filaments isolated from transfected C2C12 myoblasts (Fig. 6C, Day 0: insoluble fraction, WT = 61 ± 5%; M9R = 52 ± 4%), and myotubes (Fig. 6C, Day 2: insoluble fraction, WT= 75± 7%, M9R = 47± 3%, p < 0.05).
DISCUSSION
Since the discovery of the M9R mutation in TPM3, a number of different model systems have been established to improve understanding of the etiology of muscle weakness in NM and to determine the functional consequences of this mutation. Here, we examine five different muscle samples obtained from two related patients with the M9R mutation in TPM3 to investigate proposed mechanisms of disease pathogenesis. Analysis of the developmental expression of the sarcomeric tropomyosin isoforms revealed that α-tropomyosinslow is not expressed at robust levels until after birth, thereby likely explaining the childhood, rather than congenital, onset in patients with TPM3-related disease.
Nemaline bodies were evident only in type 1 fibers; therefore, the pathological phenotype is chiefly observed in fibers that express mutant α-tropomyosinslow protein. M9R Patient 1 showed a complete type 1 fiber predominance in three of the four muscles examined, resulting in expression of mutant protein in all fibers. Type 1 myofiber predominance is a common feature in many inherited myopathies and it has been postulated to be a compensatory mechanism for conserving energy because of the more efficient oxidative nature of slow muscle. In this case, predominance of type 1 fibers (due to fiber-conversion, or, perhaps altered maturation) results in whole muscle groups in which all fibers express mutant tropomyosin and thereby likely contributing to the severity of the phenotype. Differences in fiber composition may explain the more severe phenotype of Patient 1 (wheelchair confinement, death at age 46 due to respiratory distress), compared to that of Patient 2 (moderate weakness, remaining ambulatory in her twenties). In the single quadriceps biopsy examined from Patient 2, rod-containing type-1 fibers are extremely atrophied and the bulk of muscle cross-sectional area consists of mainly type 2 fibers of which some are hybrid fibers that co-express fast and slow MHC.
Fiber atrophy of rod-containing type-1 fibers is a consistent pathological feature of all muscles examined from both patients possessing the M9R mutation in TPM3. Fiber atrophy is a common feature in NM (8) and has been observed in other NM patients with mutations in ACTA1 (26). Fiber atrophy may be a primary manifestation of the disease process due to expression of the mutant protein; in both Patients 1 and 2 it was most pronounced in type 1 fibers that contained rods. This concept is supported by the recent identification of mutations in TPM3 as a cause of congenital fiber type disproportion (9) in which the major pathology is small type 1 fibers without rods. Alternatively, atrophy of slow fibers might be a compensatory mechanism and/or be exacerbated by muscle disuse. Patient 1 became wheelchair-bound in her late thirties following a period of prolonged immobilization due to a broken leg. Thus, fiber atrophy observed in the quadriceps muscle might be due in part to disuse in that case. Furthermore, anecdotal evidence from this and other patients with NM suggests that immobilization results in accelerated muscle atrophy and acute chronic deterioration in muscle strength (27). Interestingly, the biceps muscle of Patient 1 was the only one of four muscles examined to contain a subset of hypertrophied type 2 fibers. This patient likely would have maintained higher levels of activity in her upper limbs, including pushing her wheel-chair and this might in part explain the variation in fiber size in this muscle.
Analysis of tropomyosin isoform expression in three different muscles from Patient 1 using 2-D gel electrophoresis shows that α-tropomyosinslow species dominate, with roughly equal proportions of mutant and wild-type α-tropomyosinslow, and marked downregulation of β-tropomyosin. Since these three muscle groups showed complete type-1 fiber predominance, we can conclude that type 1 fibers in Patient 1 express α-tropomyosin dimers as the dominant tropomyosin species. We also observed abnormal expression of troponin Ifast in slow fibers (positive for slow myosin heavy chain in the absence of fast myosin expression) in Patient 1. This may be due either to dysregulation of troponin isoform expression secondary to altered expression of tropomyosin isoforms and/or residual troponin Ifast expression following fast-slow fiber conversion. Altered fiber-specific expression of troponin Ifast has not, however, been observed in skeletal muscle patients with other forms of NM with complete Type 1 fiber predominance.
The M9R mutant α-tropomyosinslow isoform localized to the insoluble protein pool, which consists of the insoluble cytoskeleton and the sarcomeric apparatus. This suggests that the mutant α-tropomyosin protein is incorporated into the thin filament and has a dominant negative (poisoning) effect on sarcomere thin filament function. We cannot exclude the possibility that the insoluble fraction of M9R mutant α-tropomyosinslow is present in the rods, however. A dominant-negative mode of disease pathogenesis has also been shown for four patients with NM due to mutations in ACTA1 (16, 28, 29).
Here, we provide evidence that tropomodulin localizes normally to the pointed end of the thin filament in M9R patient muscle. These results contrast with in vitro binding results reported by Greenfield and Fowler, which showed loss of binding of synthetic M9R mutant peptide to tropomodulin (14). This discrepancy may reflect the different tropomyosin isoform (α-tropomyosinfast, which is not expressed at high levels in affected muscles [Fig. 2]), employed in their studies (14), Indeed, we show that Tmod1 binds more strongly to α–tropomyosinslow than to β-tropomyosin, which has a Tmod1-binding site equivalent to α-tropomyosinfast. Thus, the upregulation of α-tropomyosinslow, to which Tmod1 binds better than β–tropomyosin, may account for the wild-type localization of Tmod1 in the presence of M9R mutant protein in patient muscle. Tmod1 may bind to heterodimers of the M9R mutant and wild-type α–tropomyosinslow at sarcomeric pointed ends, but this has not been tested. Alternatively, wild-type α–tropomyosinslow homodimers may be located at the majority of the sarcomeric thin filament pointed ends, whereas M9R mutant α-tropomyosinslow could localize along the remainder of the thin filament or be sequestered into nemaline rods. In this scenario, Tmod1 would be able to interact with wild-type α-tropomyosinslow at or near the sarcomeric pointed ends. Additionally, in human muscle there are likely to be other proteins that link tropomyosin and tropomodulin directly or indirectly to sarcomeric pointed ends in order to maintain and regulate thin filament length. Nebulin is known to bind directly to tropomodulin (30) and plays a supportive role in promoting the thin filament stability and length, and thus may contribute to the normal expression, localization, and binding of tropomodulin observed in the patient muscle.
Transfection studies in C2C12 myoblasts showed that M9R mutant α-tropomyosinslow incorporates less efficiently into filamentous structures such as stress fibers, results in cellular toxicity and may disrupt or destabilize the endogenous actin cytoskeleton of transfected cells. In patient muscle, myofiber formation is completed prior to expression of significant levels of the mutant M9R-tropomyosin (i.e. around birth). Thus, defective myotube formation is unlikely to contribute to disease pathogenesis in M9R TPM3 NM but a detrimental effect of mutant M9R-tropomyosin on actin filament stability or formation could influence muscle fiber growth, and may well contribute to the fiber atrophy/hypotrophy consistently observed in slow fibers expressing mutant protein in M9R TPM3 NM.
In summary, we present a detailed pathological and biochemical analysis of NM associated with a M9R mutation in TPM3 and provide insights into the disease mechanism. Patients affected with TPM3-NM may have mixed fiber muscle groups or complete slow fiber predominance. In all cases, slow fibers are atrophied, and nemaline bodies are localized to type 1 fibers in which the gene is expressed. Atrophy of type 1 myofibers is likely a primary consequence of expression of the mutant M9Rα-tropomyosinslow protein and of the resulting dysfunction this mutant protein imparts, although slow fiber atrophy may also be exacerbated by muscle disuse. The M9R mutation alters the expression of other tropomyosin isoforms and results in a switch from normal α/β tropomyosin heterodimers to a predominance of α/α dimers, of which some likely contain the mutant M9R protein. This alteration in TPM dimer populations may also directly contribute to muscle dysfunction.
Support
Sandra T. Cooper was supported by a USA MDA Development grant and an Australian National Health and Medical Research Council (NH&MRC) grant 301946. Biljana Ilkovski, Kristen J. Nowak and Kathryn N. North were supported by NH&MRC Grant 40394. Velia M. Fowler and Raymond, A. Lewis were supported by a National Institute of Health grant to Velia M. Fowler (HL083464). Nigel G. Laing was supported by an NH&MRC Fellowship 403904.
REFERENCES
- 1.Sanoudou D, Beggs AH. Clinical and genetic heterogeneity in nemaline myopathy--a disease of skeletal muscle thin filaments. Trends Mol Med. 2001;7:362–68. doi: 10.1016/s1471-4914(01)02089-5. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal PB, Greenleaf RS, Tomczak KK, et al. Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am J Hum Gen. 2007;80:162–67. doi: 10.1086/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laing NG, Wilton SD, Akkari PA, et al. A mutation in the alpha tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy NEM1. Nat Genet. 1995e;10:249. doi: 10.1038/ng0695-249. [DOI] [PubMed] [Google Scholar]
- 4.Tan P, Briner J, Boltshauser E, et al. Homozygosity for a nonsense mutation in the alpha-tropomyosin slow gene TPM3 in a patient with severe infantile nemaline myopathy. Neuromuscul Disord. 1999;9:573–79. doi: 10.1016/s0960-8966(99)00053-x. [DOI] [PubMed] [Google Scholar]
- 5.Wattanasirichaigoon D, Swoboda KJ, Takada F, et al. Mutations of the slow muscle alpha-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology. 2002;59:613–17. doi: 10.1212/wnl.59.4.613. [DOI] [PubMed] [Google Scholar]
- 6.Durling HJ, Reilich P, Muller-Hocker J, et al. De novo missense mutation in a constitutively expressed exon of the slow alpha-tropomyosin gene TPM3 associated with an atypical, sporadic case of nemaline myopathy. Neuromuscul Disord. 2002;12:947–51. doi: 10.1016/s0960-8966(02)00182-7. [DOI] [PubMed] [Google Scholar]
- 7.Penisson-Besnier I, Monnier N, Toutain A, et al. A second pedigree with autosomal dominant nemaline myopathy caused by TPM3 mutation: A clinical and pathological study. Neuromuscul Disord. 2007;17:330–37. doi: 10.1016/j.nmd.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Ryan MM, Ilkovski B, Strickland CD, et al. Clinical course correlates poorly with muscle pathology in nemaline myopathy. Neurology. 2003;60:665–73. doi: 10.1212/01.wnl.0000046585.81304.bc. [DOI] [PubMed] [Google Scholar]
- 9.Clarke FN, Kolski H, Dye ED, et al. Mutations in TPM3 are a common cause of congenital fibre type disproportion. Ann Neurol. 2008;63:329–37. doi: 10.1002/ana.21308. [DOI] [PubMed] [Google Scholar]
- 10.Corbett MA, Robinson CS, Dunglison GF, et al. A mutation in alpha-tropomyosin(slow) affects muscle strength, maturation and hypertrophy in a mouse model for nemaline myopathy. Hum Mol Genet. 2001;10:317–28. doi: 10.1093/hmg/10.4.317. [DOI] [PubMed] [Google Scholar]
- 11.de Haan A, van der Vliet MR, Gommans IM, et al. Skeletal muscle of mice with a mutation in slow alpha-tropomyosin is weaker at lower lengths. Neuromuscul Disord. 2002;12:952–57. doi: 10.1016/s0960-8966(02)00138-4. [DOI] [PubMed] [Google Scholar]
- 12.Moraczewska J, Greenfield NJ, Liu Y, et al. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophys J. 2000;79:3217–25. doi: 10.1016/S0006-3495(00)76554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akkari PA, Song Y, Hitchcock-DeGregori S, et al. Expression and biological activity of Baculovirus generated wild-type human slow alpha tropomyosin and the Met9Arg mutant responsible for a dominant form of nemaline myopathy. Biochem Biophys Res Comm. 2002;296:300–304. doi: 10.1016/s0006-291x(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 14.Greenfield NJ, Fowler VM. Tropomyosin requires an intact N-terminal coiled coil to interact with tropomodulin. Biophys J. 2002;82:2580–91. doi: 10.1016/S0006-3495(02)75600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michele DE, Albayya FP, Metzger JM. A nemaline myopathy mutation in alpha-tropomyosin causes defective regulation of striated muscle force production. J Clin Invest. 1999;104:1575–81. doi: 10.1172/JCI7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilkovski B, Nowak KJ, Domazetovska A, et al. Evidence for a dominant-negative effect in ACTA1 nemaline myopathy caused by abnormal folding, aggregation and altered polymerization of mutant actin isoforms. Hum Mol Genet. 2004;13:1727–43. doi: 10.1093/hmg/ddh185. [DOI] [PubMed] [Google Scholar]
- 17.Ursitti JA, Fowler VM. Immunolocalization of tropomodulin, tropomyosin and actin in spread human erythrocyte skeletons. J Cell Sci. 1994;107:1633–39. doi: 10.1242/jcs.107.6.1633. [DOI] [PubMed] [Google Scholar]
- 18.Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J Biol Chem. 2003;278:40000–9. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 19.Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. J Biol Chem. 1987;262:12792–12800. [PubMed] [Google Scholar]
- 20.Almenar-Queralt A, Lee A, Conley CA, et al. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. J Biol Chem. 1999;274:28466–75. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- 21.Pieples K, Wieczorek DF. Tropomyosin 3 increases striated muscle isoform diversity. Biochemistry. 2000;39:8291–97. doi: 10.1021/bi000047x. [DOI] [PubMed] [Google Scholar]
- 22.Cooper ST, Lo HP, North KN. Single section Western blot: Improving the molecular diagnosis of the muscular dystrophies. Neurology. 2003;61:93–97. doi: 10.1212/01.wnl.0000069460.53438.38. [DOI] [PubMed] [Google Scholar]
- 23.Doriguzzi C, Mongini T, Palmucci L, et al. Quantitative analysis of quadriceps muscle biopsy. Results in 30 healthy females. J Neurol Sci. 1984;66:319–26. doi: 10.1016/0022-510x(84)90020-0. [DOI] [PubMed] [Google Scholar]
- 24.Compton AG, Cooper ST, Hill PM, et al. The syntrophin-dystrobrevin subcomplex in human neuromuscular disorders. J Neuropathol Exp Neurol. 2005;64:350–61. doi: 10.1093/jnen/64.4.350. [DOI] [PubMed] [Google Scholar]
- 25.Corbett MA, Akkari PA, Domazetovska A, et al. An alphaTropomyosin mutation alters dimer preference in nemaline myopathy. Ann Neurol. 2005;57:42–49. doi: 10.1002/ana.20305. [DOI] [PubMed] [Google Scholar]
- 26.Ilkovski B, Cooper ST, Nowak K, et al. Nemaline myopathy caused by mutations in the muscle alpha-skeletal-actin gene. Am J Hum Genet. 2001;68:1333–43. doi: 10.1086/320605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan MM, Schnell C, Strickland CD, et al. Nemaline myopathy: A clinical study of 143 cases. Ann Neurol. 2001;50:312–20. doi: 10.1002/ana.1080. [DOI] [PubMed] [Google Scholar]
- 28.Wallefeld W, Krause S, Nowak KJ, et al. Severe nemaline myopathy caused by mutations of the stop codon of the skeletal muscle alpha actin gene (ACTA1). Neuromuscul Disord. 2006;16:541–47. doi: 10.1016/j.nmd.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 29.D'Amico A, Graziano C, Pacileo G, et al. Fatal hypertrophic cardiomyopathy and nemaline myopathy associated with ACTA1 K336E mutation. Neuromuscul Disord. 2006;16:548–52. doi: 10.1016/j.nmd.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 30.McElhinny AS, Kolmerer B, Fowler VM, et al. The N-terminal end of nebulin interacts with tropomodulin at the pointed ends of the thin filaments. J Biol Chem. 2001;276:583–92. doi: 10.1074/jbc.M005693200. [DOI] [PubMed] [Google Scholar]
- 31.Engel WK, Cunningham GG. Rapid examination of muscle tissue: An improved trichrome method for fresh-frozen biopsy sections. Neurology. 1963;13:919–23. doi: 10.1212/wnl.13.11.919. [DOI] [PubMed] [Google Scholar]