Abstract
A general method for isotopic labeling of the purine base moiety of nucleotides and RNA has been developed through biochemical pathway engineering in vitro. A synthetic scheme was designed and implemented utilizing recombinant enzymes from the pentose phosphate and de novo purine synthesis pathways, with regeneration of folate, aspartate, glutamine, ATP, and NADPH cofactors, in a single-pot reaction. Syntheses proceeded quickly and efficiently in comparison to chemical methods with isolated yields up to 66% for 13C, 15N enriched ATP and GTP. The scheme is robust and flexible, requiring only serine, NH4+, glucose and CO2 as stoichiometric precursors in labeled form. Using this approach, U-13C- GTP, U-13C,15N- GTP, 13C2,8- ATP and U-15N- GTP were synthesized on a millimole scale, and the utility of the isotope labeling is illustrated in NMR spectra of HIV-2 transactivation region (TAR) RNA containing 13C 2,8-adenosine and 15N-1,3,7,9,2-guanosine. Pathway engineering in vitro permits complex synthetic cascades to be effected expanding the applicability of enzymatic synthesis.
INTRODUCTION
The expanding field of biocatalysis has emerged as a viable alternative to the chemical synthesis of many compounds. Enzymatic synthesis offers distinct advantages over chemical methods, including reduced reaction time, increased product yield, increased product specificity, reduced cost, and reduced environmental impact (1–3). For these reasons, many basic enzymatic syntheses have been adapted for the efficient production of a wide variety of compounds. Examples include: vanillin (4), amylose (5), β-lactam antibiotics (6), glycopeptides (7), iminocyclitols (8), and chiral amino acids (9).
Through pathway engineering, it is possible to expand enzymatic synthesis to include a combination of biochemical pathways, increasing the complexity of the accessible targets significantly, yet continuing to maintain the same advantages enjoyed by simple enzymatic syntheses. An elegant example of a complex multistep enzymatic reaction was the synthesis of vitamin B12 from simple precursors, pioneered by A. Ian Scott, using cloned enzymes from different organisms (10–12). A bioactive heparan sulfate pentasaccharide was synthesized in vitro using eight enzymes from its biosynthetic pathway in the Golgi apparatus with a faster reaction time, increased yield, and using one-tenth of the steps required for total chemical synthesis (13). In addition, multistep enzymatic synthesis has recently been used to synthesize enterocin polyketides (14). Multi-step enzymatic synthesis has also been used for efficient preparative synthesis of 13C- and 2H-labeled ribonucleotides for NMR studies of RNA structure (15–17).
Uniformly 13C, 15N- labeled nucleotides used in NMR studies of RNA structure and dynamics are obtained from bacteria grown on a minimal medium with 15NH4Cl as the sole nitrogen source and/or 13C6-glucose or 13C-methanol as the carbon source (18–20). Using this method, labeled nucleotides can be produced economically in gram quantities, but specific isotope labeling patterns are not possible. Specific incorporation of 13C, 15N can be used to reduce spectral crowding, as a spectral filter or to simplify the dipolar network for relaxation studies (16, 21–23).
Selective incorporation of isotope labels into nucleotides can be effected by chemical and/or enzymatic synthesis. Using 13C,2H-labeled glucose and unlabeled bases, efficient enzymatic synthesis of a variety of specific ribose labeled nucleotides was carried out, facilitating the study of RNA molecules of increasing size (15–17, 24). Specific labeling of the ribose moiety permits assignment of sugar-sugar and intermolecular NOE correlations by reducing ambiguity and crowding of the spectra. The two most important protons for NMR structural analysis, H1′ and H2′, can be selectively observed, and the variety of available labeling patterns for the glucose starting material provides a great degree of flexibility for labeling of the ribose moiety (17).
The next logical step is to expand specific isotope labeling to the base. The base moiety of RNA and DNA molecules contains key structural information in local interactions including: hydrogen bonding, protonation, ligand interactions, and stacking (22, 23, 25). Chemical synthesis has been the main source for specific 13C, 15N base labeling to date (26, 27). Several groups have incorporated 13C, 15N at specific positions in the base of purine nucleotides using a combination of chemical and enzymatic synthesis. Starting from 4,5,6-triaminopyrimidine or 2,4,5,6-tetraaminopyrimidine, 13C8-labeled adenine or 2,6-diaminopurine were chemically synthesized, followed by combined chemical and enzymatic conversion to ATP or GTP, with overall yields of 37% and 29%, respectively (23). Similarly, starting from imidazole-4,5-dicarboxylic acid, hypoxanthine and 2-methylthio-hypoxanthine were chemically synthesized, followed by a combination of enzymatic and chemical synthesis to introduce specific labels at 13C2 or 15N1,3, amino, giving 35% overall yield for adenosine and 40% for guanosine (28). Starting from inosine, 13C215N1, amino-guanosine was synthesized in 32% overall yield (29). The 2-amino group of GTP has also been labeled with 15N through enzymatic interconversion of NMP’s (30). These synthetic schemes and starting materials vary widely in implementation and difficulty depending on the desired nucleotide and labeling pattern. In general, uniformly or specifically labeled purine bases are not commercially available and chemical synthesis of labeled purines is complex (22, 26, 27,31). A general, flexible and efficient enzymatic synthesis of base labeled purine nucleotides would have many applications in biochemical and biophysical studies of nucleic acid structure and function.
In the present work, de novo purine biosynthesis is coupled with phosphoribosyl-pyrophosphate (PRPP) synthesis to effect a total enzymatic synthesis of the purine nucleotides ATP and GTP. The synthesis is carried out with the precursors glucose, glutamine, serine, ammonia, and CO2 that are available in a variety of isotopically labeled forms. A total of 28 biosynthetic enzymes were used for the efficient synthesis of ATP and GTP with yields of up to 66% in a single-pot reaction. The reaction design relies heavily on NTP and NAD(P)H cofactor regeneration schemes (32, 33), and in addition, novel recycling reactions for folate, fumarate, and glutamine cofactor pools were implemented. Due to the flexibility afforded by selection of labeled starting materials, a wide variety of purine isotope labeling patterns are possible. Using this approach, U-13C- GTP, U-13C,15N- GTP, 13C2,8- ATP and U-15N- GTP were synthesized on a millimolar scale, and the utility of these labeled nucleotides is illustrated in NMR spectra of an RNA oligomer containing 13C 2,8-adenosine and 15N-1,3,7,9,2- guanosine.
RESULTS AND DISCUSSION
Design of the enzymatic synthesis
The well-established metabolic pathway for de novo purine biosynthesis is outlined in Figure 1a,b. Enzymes of the pentose phosphate pathway convert glucose to PRPP, which enters into the linear cascade of reactions that assemble the purine ring to produce IMP, which is the precursor for both ATP and GTP. The net reaction for biosynthesis of ATP and GTP is shown in Figure 2a,b, including the precursor molecules as well as the numerous cofactors that are consumed in the reaction cascade. The atoms in the purine ring are metabolically derived from glycine, CO2, aspartate, glutamine, and 10-formyl THF as shown in Figure 2c,d, and this set of precursors defines the possible isotope labeling patterns that can be achieved using in vitro biosynthesis. The goal of the present work is to recapitulate the de novo purine biosynthesis cascade in vitro, using purified recombinant enzymes and a defined set of labeled precursors to generate specifically isotopically labeled nucleotides.
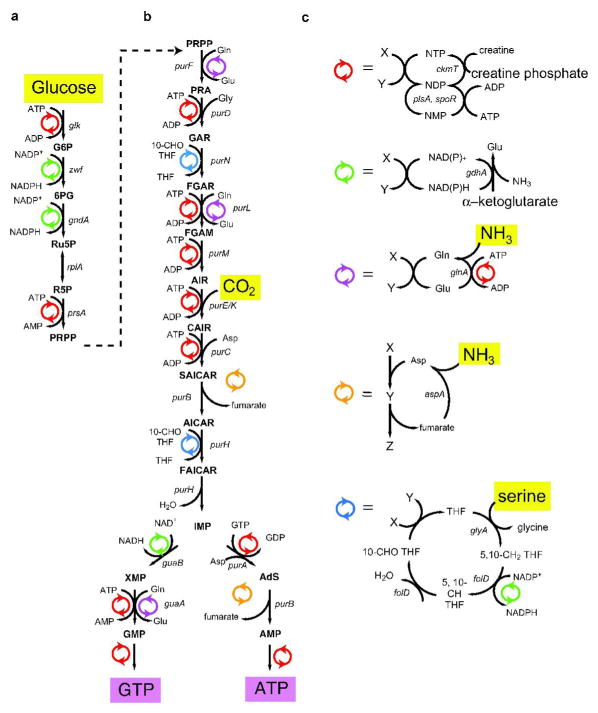
Figure 1. Scheme for Enzymatic Synthesis of Purine Nucleotides.
a) Conversion of Glucose to PRPP. Glucose and ATP are converted to glucose-6-phosphate (G6P) by the action of glucokinase (glk) with the production of ADP. G6P and NADP+ is converted to 6-phosphogluconate (6PG) by glucose-6-phosphate dehydrogenase (zwf) with the production of NADPH. 6PG and NADP+ is converted to ribulose-5-phosphate (Ru5P) by the action of 6-phosphogluconate dehydrogenase (gndA) with the production of NADPH. Phosphoriboisomerase (rpiA) interconverts Ru5P and ribose-5-phosphate (R5P). R5P and ATP are converted to phosphoribose-pyrophosphate (PRPP) by the action of PRPP synthase (prsA) with the production of AMP. b) Conversion of PRPP to ATP or GTP. PRPP and glutamine are converted to phosphoribose-amine (PRA) by the action of amidophosphoribosyl-transferase (purF) with the production of glutamine. PRA, glycine and ATP are converted to phosphoribosyl-glycinamide (GAR) by the action of phosphoribosylamine-glycine ligase (purD) with the production of ADP. GAR and 10-formyl-tetrahydrofolate are converted to phosphoribosyl-formylglycinamide (FGAR) by the action of phosphoribosyl-glycine amide formyltransferase (purN) with the production of tetrahydrofolate (THF). FGAR, glutamine and ATP are converted to phosphoribosyl-formylglycinamidine (FGAM) by the action of FGAM synthase (purL) with the production of glutamate and ADP. FGAM and ATP are converted to phosphoribosyl-aminoimidazole (AIR) by phosphoribosyl-formyl glycinamidine cyclo-ligase (purM) with the production of ADP. AIR, CO2 and ATP (47) are converted to phosphoribosyl-amino-carboxy-imidazole (CAIR) by the action of phosphoribosyl-amino-imidazole carboxylase (purE, purK) with the production of ADP. CAIR, aspartate and ATP are converted to phosphoribosyl-amino-succinocarboxamide-imidazole (SAICAR) by phosphoriobsyl-aminoimidazole-succino-carboxamide synthase (purC) with the production of ADP. SAICAR is converted to phosphoribosyl-amino-imidazole carboxamide (AICAR) by adenylosuccinate lyase (purB) with the production of fumarate. AICAR and 10-formyl-THF are converted to phosphoribosyl-formylamido-imidazole carboxamide (FAICAR) by the action of phophoribosylamino-imidazole-carboxamide formyltransferase (purH) with the production of THF. FAICAR is converted to inosine monophosphate (IMP) by IMP cyclohydrolase (purH) with the production of H2O. For ATP: IMP and GTP are converted to adenylosuccinate (AdS) by adenylosuccinate synthase (purA) with the production of GDP. AdS is converted to AMP by the action of purB with the production of fumarate. For GTP: IMP and NAD+ are converted to xanthosine monophosphate (XMP) by the action of IMP dehydrogenase (guaB) with the production of NADH. XMP, glutamine and ATP are converted to GMP by the action of GMP synthase (guaA) with the production of AMP and glutamate. c) Cofactor Regeneration Schemes. NTP Regeneration (Red). AMP and ATP are converted to ADP by the action of adenylate kinase (plsA). GMP and ATP are converted to GDP by the action of guanylate kinase (spoR) with the production of ADP. NDP’s and creatine phosphate are converted to NTP’s by the action of creatine phosphokinase (ckmT) with the production of creatine. Nicotinamide Adenine Dinucleotide Regeneration (Green). NAD(P)H, α-ketoglutarate (α-KG) and NH3 are converted to NAD(P)+ by the action of glutamate dehydrogenase (gdhA) with the production of glutamate. Glutamine Recycling (Purple). Glutamate, ATP and NH3 are converted to glutamine by the action of glutamine synthase (glnA) with the production of ADP. Aspartate Recycling (Orange). Fumarate and NH3 are converted to aspartate by the action of aspartate ammonia-lyase (aspA). Folate Regeneration (Blue). THF and serine are converted to 5,10-CH2-THF by the action of glycine hydroxymethyl-transferase (glyA) with the production of glycine. 5,10-CH2-THF and NADP+ is converted to 5,10-CH-THF by the action of methylene-THF dehydrogenase (folD) with the production of NADPH. 5,10-CH-THF is converted to 10-CHO (formyl) THF by the action of methenyl-THF cyclohydrolase (folD) with the production of H2O.
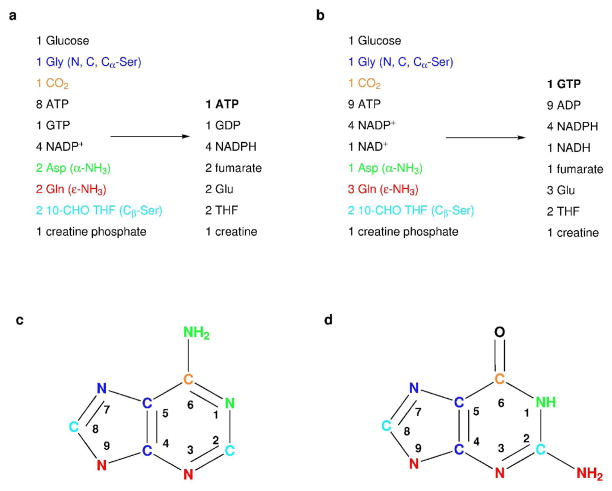
Figure 2.
Substrates and products of the biosynthesis for: (a) ATP, (b) GTP. Metabolic origin of purine atoms for: (c) adenine, (d) guanine. The substrates and the metabolic source for each purine atom is color coded: Glycine (blue), CO2 (orange), Aspartate (green), Glutamine (red), 10-formyl-THF (cyan)
For efficient enzymatic synthesis in vitro, there are three major considerations. First, isotopically labeled amino acids are expensive starting materials. 13C-labeled 10-formyl THF is not commercially available, and it is necessary to generate these labeled precursor components in situ. Second, the numerous ATP and GTP equivalents required cannot be supplied in stoichiometric amounts to avoid isotopic dilution of the desired nucleotide products, and it is essential to employ enzymatic cofactor regeneration schemes. Third, it is important to provide a driving force for the overall reaction to ensure high yields of the desired labeled ATP and GTP. All of these considerations can be taken into account to design and effect an efficient and cost-effective enzymatic synthesis scheme.
The overall strategy for de novo purine nucleotide synthesis builds on the previously implemented syntheses of nucleotides from glucose and bases, via the PRPP intermediate, as shown in Figure 1a (15–17). The cofactor recycling schemes for regeneration of NTPs from NMPs or NDPs and regeneration of NAD(P)+ from NAD(P)H are well-established (34, 35), shown in Figure 1c. Creatine phosphate is a suitable phosphate source for re-phosphorylation of NDPs by the action of creatine kinase. NAD(P)+regeneration is accomplished by the action of glutamate dehydrogenase on α-ketoglutarate and ammonia, to produce glutamate. Since neither glutamate nor creatine directly supply labeled atoms to the final product, these reagents may be used in excess to provide a driving force for the overall reaction.
During the ATP synthesis, the enzymes continue to utilize and regenerate labeled ATP as it is produced. To initiate the reaction, it is necessary to add a catalytic amount of unlabeled ATP (1%). In addition, a catalytic amount of unlabeled GTP (1%) is added as a required cofactor for conversion of IMP to adenylosuccinate by adenylosuccinate synthase (purA). The labeled ATP that is synthesized will unavoidably contain 1% unlabeled ATP and 1% unlabeled GTP, but for preparation of labeled RNAs for NMR studies, this has a negligible effect on the overall labeling.
GTP synthesis is equally dependent on ATP, but addition of 1% catalytic ATP is not sufficient to initiate the reaction. Although increasing the ATP concentration is effective, the subsequent separation of labeled GTP from unlabeled ATP involves a difficult chromatography step. Fortunately, the ATP requirement for the GTP synthesis reaction can be met by the addition of 10% dATP that is readily separated from the final product GTP during the boronate affinity chromatography step. Several enzymes including hexokinase, PRPP synthase (prsA), both monophosphate kinases (plsA, spoR) and glutamine synthase (glnA) utilize dATP with slightly lower efficiency than ATP. However, GMP synthase (guaA), will not accept dATP as a substrate, and it is still necessary to add a catalytic amount of ATP (1%) during GTP synthesis in the presence of dATP. Both ATP and dATP are regenerated by creatine phosphokinase, efficiently fueling the GTP reaction. In this way, dATP provides the critical phosphate source in the GTP synthesis reaction without contributing to isotopic dilution or mixing of the nucleotide pool.
To implement the purine biosynthesis cascade, shown in Figure 1b, several additional cofactor recycling schemes needed to be developed. Four of the nitrogen atoms in ATP or GTP are derived from the α-amino group of aspartate or the ε-amino group of glutamine, and both of these positions can be enzymatically labeled, in situ from 15N-ammonium ions, as shown in Figure 1c. Generation of ε-15N-glutamine can be accomplished in situ from glutamate using glutamine synthase, while generation of α-15N-aspartate can be accomplished in situ from fumarate using aspartate-ammonia lyase (aspA). Glutamine is used in three reactions for the incorporation of the purine N3 and N9 atoms and the guanine N2 atom, releasing glutamate, which can be recycled to form ε-15N-glutamine by the action of glnA, as shown in Figure 1b,c. Aspartate is used in two similar two-step reaction sequences for incorporation of the purine N1 atom and the adenine N6 atom, both reactions releasing fumarate, which can be recycled to form α-15N-aspartate by the action of aspA, as shown in Figure 1b,c. In this reaction scheme, glutamate is generated in situ from recycling of NAD(P)H, and only catalytic amounts of fumarate must be added. The action of glnA and aspA effectively recycles the pools of glutamine and aspartate cofactors.
The purine C2 and C8 positions are derived from 10-formyl-THF, however 13C-labeled folates are not commercially available. It is possible to generate 13C-10-formyl-THF in situ and recycle catalytic amounts of tetrahydrofolate (THF) as a cofactor, using the scheme shown in Figure 1c. The β-carbon of serine is incorporated into 10-formyl-THF by the sequential action of serine hydroxymethyl transferase (glyA) and the multifunctional enzyme folD, which provides for efficient recycling of the folate cofactor pool during nucleotide synthesis.
The recycling schemes shown in Figure 1c for the NTP, NAD(P)H, glutamate, fumarate, and folate pools, make the overall synthesis of ATP and GTP, shown in Figure 1a,b, streamlined and efficient. Almost every step of the linear sequence from glucose to ATP/GTP is facilitated by recycling of one of these cofactor pools. The net reaction for the enzymatic scheme shown in Figure 1a,b including the effects of cofactor recycling, is given in Figure 3a,b for ATP and GTP synthesis, respectively. The set of starting materials for the overall reaction shown in Figure 2a,b is simplified to a small number of reagents that fall into three classes. First, there is a set of stoichiometric reagents that are ultimately incorporated into the nucleotide, which includes glucose, CO2, ammonia, and serine. Second, the large number of phosphate and reducing equivalents are supplied with creatine phosphate and α-ketoglutarate, respectively. These two reagents are supplied in excess to provide a driving force that fuels the overall reactions. Third, a set of cofactors is supplied in catalytic amounts. The complex cascade of reactions in Figure 1a,b is effectively reduced to a net transformation of glucose, CO2, ammonia, and serine into ATP/GTP, with one byproduct molecule of glycine in addition to the creatine and glutamate byproducts from cofactor regeneration.
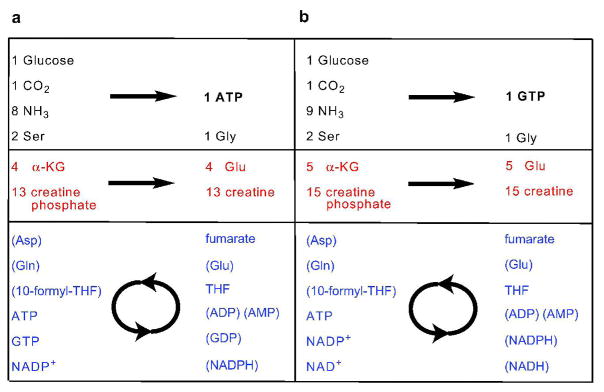
Figure 3.
Pathway engineered synthesis scheme for: (a) ATP and (b) GTP. Reagents are color coded as: Stoichiometric isotopically labeled reagents (black), Phosphate and oxidizing equivalents as the driving force (red), and Recycled cofactors (blue). Cofactors are added in catalytic amounts, and intermediates that are generated in situ are shown in parentheses.
In the net reaction, carbons of the purine ring originate from only serine (C2,C4,C5,C8) and CO2 (C6), while the nitrogens of the purine ring are derived from serine (N7) and NH4+ ions (N1,N2,N3,N6,N9). There is a tremendous flexibility available generating specific labeling patterns by the suitable choice of labeled starting materials. Due to the availability of specifically labeled serine, all the purine base carbons can be separately labeled except for C2 and C8. For nitrogen, it is possible to separate the N3, N9 from N1 by using labeled aspartate directly, and N7 is separately labeled from serine.
There is one restriction placed on possible labeling patterns due to the production of CO2 by decarboxylation of 6-phosphogluconate to ribulose-5-phosphate during PRPP generation. This step effectively couples the isotope composition of the C1 of glucose to C6 in the purine base via the CO2 pool, and care must be taken to avoid isotopic scrambling or isotopic dilution due to atmospheric CO2 in certain cases. If 13C-ribose and 13C6-purine labeling are both desired, then degassing and inert atmosphere are sufficient to prevent isotopic dilution (see Supplementary Figure 9). If 13C6 purine labeling is desired without ribose labeling, then PRPP can be generated enzymatically directly from unlabeled ribose (15). If 13C-ribose labeling from 13C-glucose is desired without 13C6-purine labeling, then an excess of 12CO2 can be supplied. For synthesis of two different labeled nucleotides, 13C labeling of the base and the ribose moieties was successfully combined.
Implementation of the enzymatic synthesis
There are 28 enzymes required to implement the de novo enzymatic synthesis of GTP and ATP shown in Figure 1, and few of these are commercially available. Given our previous success with E. coli phosphoribosyltransferases (15, 16, 36, 37) and the extensive characterization of the purine biosynthesis pathway (38), the entire set of E. coli enzymes for the reactions in Figure 1 was cloned, expressed, and purified. Many of the intermediates in Figure 1 are not commercially available, and it was not possible to perform activity assays on many of the expressed enzymes. In these cases, the value for the specific activity was assumed to be 1 U/mg. Fortunately, all of the selected enzymes were catalytically active, and de novo synthesis of ATP was observed in the first model reaction. The reactions described below were not extensively optimized, and it appears that this enzymatic synthesis scheme is very stable and robust. To illustrate the flexibility and utility of the pathway engineered nucleotide synthesis, four different labeled nucleotides were synthesized, as shown in Figure 4.
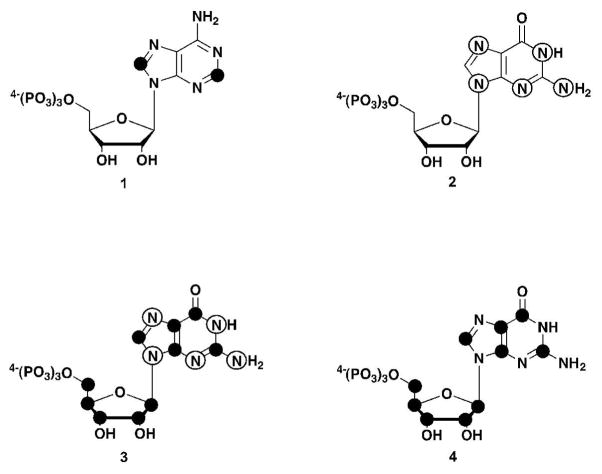
Figure 4.
Molecular structures of compounds synthesized: (1) 13C-C2,8-ATP, (2) U-15N-GTP, (3) U- 13C,15N-GTP, (4) U-13C-GTP. 13C labels are indicated by black circles and 15N labels are indicated by open circles.
Synthesis of 13C2,8-ATP
A 1.2 mmole scale ATP synthesis was performed with labeling of the C8 and C2 of adenine using β-13C-serine. The labeled serine was combined with stoichiometric substrates (glucose, NH4Cl, glutamine, KHCO3), fuel reagents (α-ketoglutarate, creatine phosphate), catalytic cofactors (ATP, GTP, NADP+, fumarate, THF) and the 23 enzymes listed in Table 1(1) in a 120 ml buffered reaction incubated at 37°C. ATP formation was monitored by HPLC over the course of two days, when the reaction ceased to progress. Affinity purification afforded a 57% isolated yield of 13C2,8-ATP, based on input glucose, which compares favorably to yields for ATP synthesis from adenine and glucose (16, 17).
Table 1.
Complete list of enzymes for de novo enzymatic synthesis of ATP and GTP. The amounts of each enzyme used for synthesis of compounds 1–4 in Figure 4 is provided as either Units of enzyme or mg of enzyme.
| Enzyme | Gene | EC | Source | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|
| Hexokinase | hxk1/2 | 2.7.1.1 | BY | 5.6 U | 5.6 U | --- | --- |
| Glucokinase | glk | 2.7.1.2 | RE | --- | --- | 2.4 U | 2.4 U |
| Glucose-6-phosphate dehydrogenase | zwf1 | 1.1.1.49 | BY | 6.2 U | 6.35 U | --- | --- |
| Glucose-6-phosphate dehydrogenase | zwf | 1.1.1.49 | RE | --- | --- | 2.6 U | 2.6 U |
| Phosphogluconate dehydrogenase | gndA | 1.1.1.44 | RE | 6.8 U | 6.85 U | 2.9 U | 2.9 U |
| Ribose-5-phosphate isomerase | rpiA | 5.3.1.6 | RE | 7.5 U | 7.5 U | 3.2 U | 3.2 U |
| Ribose-phosphate diphosphate kinase | prsA | 2.7.6.1 | RE | 8.2 U | 8.2 U | 3.5 U | 3.5 U |
| Amidophosphoribosyl-transferase | purF | 2.4.2.14 | RE | 10.5 mg | 15.2 mg | 4.0 mg | 4.0 mg |
| Phosphoribosylamine-glycine ligase | purD | 6.3.4.13 | RE | 14.4 mg | 12.5 mg | 4.3 mg | 4.3 mg |
| Phosphoribosylglycinamide formyltransferase | purN | 2.1.2.2 | RE | 4.6 mg | 7.5 mg | 4.7 mg | 4.7 mg |
| Phosphoribosylformylglycinamidine synthase | purL | 6.3.5.3 | RE | 25.6 mg | 25.2 mg | 5.4 mg | 5.4 mg |
| Phosphoribosylformylglycinamidine cyclo-ligase | purM | 6.3.3.1 | RE | 11.7 mg | 7.8 mg | 5.7 mg | 5.7 mg |
| Phosphoribosylamino-imidazole carboxylase (catalytic subunit) | purE | 4.1.1.21 | RE | 18.7 mg | 35.4 mg | 6.5 mg | 6.5 mg |
| Phosphoribosylamino-imidazole carboxylase (ATPase subunit) | purK | 4.1.1.21 | RE | 6.0 mg | 8.5 mg | 6.2 mg | 6.2 mg |
| Phosphoribosylamino-imidazole-succinocarboxamide synthase | purC | 6.3.2.6 | RE | 3.4 mg | 6.9 mg | 7.0 mg | 7.0 mg |
| Adenylosuccinate lyase | purB | 4.3.2.2 | RE | 56 U | 28 U | 10 U | 10 U |
| Phosphoribosylamino-imidazole-carboxamide formyltransferase | purH | 2.1.2.3 | RE | 14 mg | 31 mg | 18 mg | 18 mg |
| Inosine-monophosphate cyclohydrolase | purH | 3.5.4.10 | RE | ||||
| Adenylosuccinate synthase | purA | 6.3.4.4 | RE | 10 U | --- | --- | --- |
| Inosine-monophosphate dehydrogenase | guaB | 1.1.1.205 | RE | --- | 25.4 U | 15 U | 15 U |
| Guanosine-monophosphate synthase | guaA | 6.3.5.2 | RE | --- | 25.8 U | 16.5 U | 16.5 U |
| Adenylate kinase | ak5 | 2.7.4.3 | CM | 200 U | --- | --- | --- |
| Adenylate kinase | plsA | 2.7.4.3 | RE | --- | 47 U | 9 U | 9 U |
| Creatine phosphokinase | ckmT | 2.7.3.2 | RM | 250 U | 118 U | --- | --- |
| Creatine phosphokinase | ckmT | 2.7.3.2 | RE | --- | --- | 125 U | 125 U |
| Guanylate kinase | spoR | 2.7.4.8 | RE | 1 U | 55 U | 18 U | 18 U |
| Glycine hydroxymethyltransferase | glyA | 2.1.2.1 | RE | 22.5 mg | 25 mg | 20.2 mg | 20.2 mg |
| Methylene-tetrahydrofolate dehydrogenase | folD | 1.5.1.5 | RE | 10.5 mg | 23.4 mg | 20.2 mg | 20.2 mg |
| Methenyl-tetrahydrofolate cyclohydrolase | folD | 3.5.4.9 | RE | ||||
| Aspartate ammonia-lyase | aspA | 4.3.1.1 | RE | 92 U | 32 U | 20 U | 20 U |
| Glutamate dehydrogenase | glud1 | 1.4.1.3 | BL | 94 U | 141 U | --- | --- |
| Glutamate dehydrogenase | gdhA | 1.4.1.4 | RE | --- | --- | 39 U | 39 U |
| Glutamine synthase | glnA | 6.3.1.2 | RE | --- | --- | 38 U | --- |
| Inorganic diphosphatase | ppa | 3.6.1.1 | RE | 23.4 U | 24 U | 26 U | 26 U |
Source Key: RE = Recombinant from E. coli, BY = Baker’s yeast, CM = Chicken muscle, RM = Rabbit muscle, BL = Bovine liver
Synthesis of U-15N-GTP
1.2 mmole scale GTP synthesis was performed to produce uniform 15N-labeling by combining stoichiometric substrates (glucose, 15NH4Cl, 15Nε-glutamine, serine, KHCO3), fuel reagents (α-ketoglutarate, creatine phosphate), catalytic cofactors (ATP, dATP, NADP+, fumarate, THF) and the 24 enzymes listed in Table 1(2) in a 120 ml buffered reaction incubated at 37°C. GTP formation was monitored by HPLC and after approximately 2 days, the reaction ceased to progress, and U-15N-GTP was obtained in 24% yield after purification. The lower yield is likely due to the instability of one or more of the de novo purine biosynthesis enzymes. Without commercially available substrates, it is difficult to determine which enzymes suffered loss of activity over the months that elapsed after the initial synthesis of 13C2,8-ATP. In earlier pilot scale models, the GTP synthesis proceeded similarly to the ATP synthesis, and subsequent syntheses with fresh enzymes were more efficient.
Synthesis of U-13C-15N-GTP
A 0.5 mmole scale GTP synthesis was carried out starting with stoichiometric labeled reagents (13C-glucose, 15NH4Cl, 13C/15N-serine, NaH13CO3), fuel reagents and catalytic cofactors as described above. Fresh preparations of the 27 recombinant enzymes listed in Table 1(3) were used, and the enzymatic recycling of glutamine was included to permit use of 15NH4Cl as the primary nitrogen source. Increased consumption of creatine phosphate was observed due to the addition of (d)ATP dependent glnA. The reaction was monitored by HPLC, and after 40 hours it was complete. An isolated yield of 42% U-13C-15N-GTP was obtained after purification.
Synthesis of U-13C-GTP
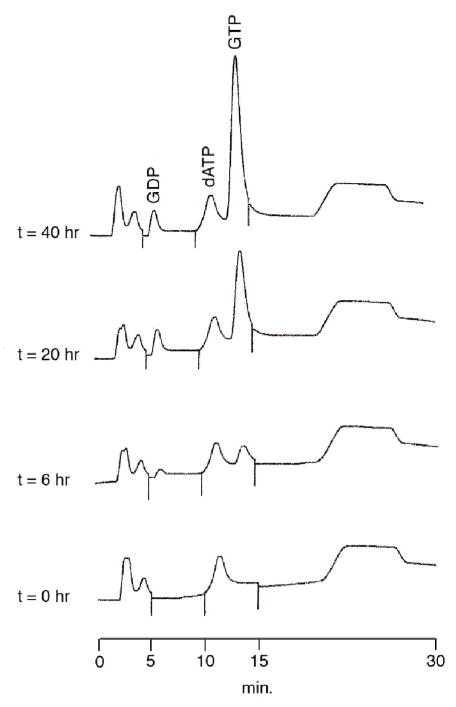
A 0.5 mmole scale GTP synthesis was performed starting with stoichiometric reagents (13C-glucose, NH4Cl, glutamine, 13C-serine, NaH13CO3) and fuel reagents and recycled cofactors as described above. The 26 recombinant enzymes listed in Table 1(4) were added, and the time course of the reaction was monitored by HPLC over the course of 40 hours, as shown in Figure 5. An isolated yield of 66% U-13C-GTP was obtained after purification.
Figure 5.
HPLC chromatograms showing the timecourse of the U-13C GTP-forming reaction. The early-eluting peaks are from the NAD(P)+ and THF cofactors.
Incorporation of labeled nucleotides into RNA
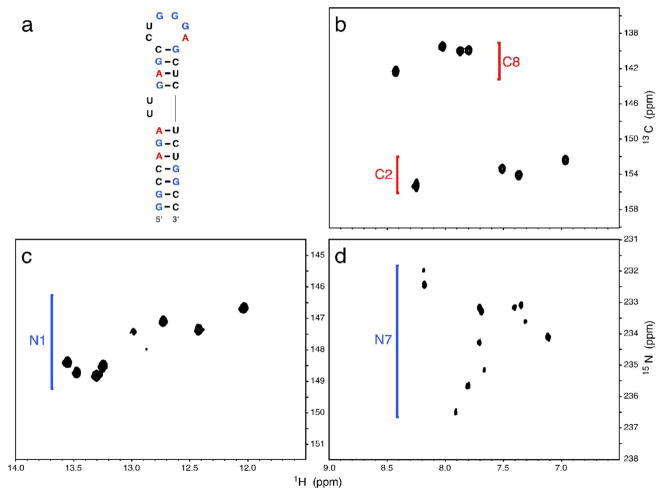
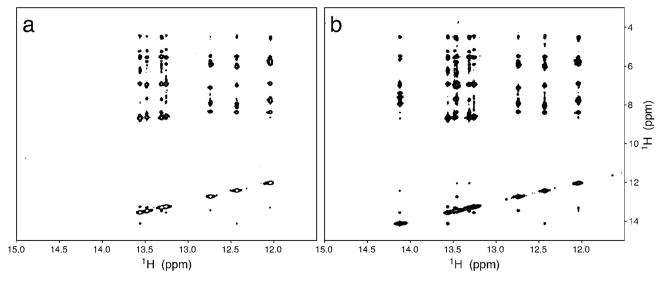
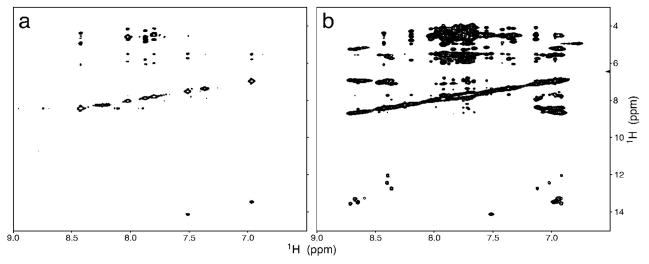
To demonstrate the utility of the labeled nucleotides, HIV-2 TAR RNA was transcribed in vitro using unlabeled CTP and UTP and the U-15N-GTP and 13C2,8-ATP prepared as described above. The H2 and H8 resonances of adenine can be selectively observed in a 1H,13C HSQC spectrum, shown in Figure 6b. Similarly, correlations between the guanine H8 and N7 atoms are selectively observed in the 1H,15N-HSQC spectrum shown in Figure 6c,d. NOESY spectra were also acquired on the RNA, showing selection for specific proton interactions edited by both the 15N and 13C labels shown in Figures 7, 8 respectively. This labeling pattern imparts the ability to discern specific base interactions and reduce crowding and overlap seen with unlabeled or uniformly labeled samples.
Figure 6.
1H-15N and 11H-13C HSQC correlation spectra at 10°C for TAR RNA uniformly labeled with 15N-labeled guanosine and adenosine labeled with 13C at positions 2 and 8. (a) Sequence of the partially labeled TAR-2 construct. (b) One bond correlations between protons and the labeled carbons in the four adenosine bases. (c) One bond correlations between N1 the eight hydrogen-bonded imino protons. (d) Two bond correlations between the non-exchangeable H8 proton and the N7 atom.
Figure 7.
15N-editted (a) and homonuclear (b) NOESY (Tmix = 250 ms) spectra of TAR RNA uniformly labeled with 15N-labeled guanosine and adenosine labeled with 13C at positions 2 and 8.
Figure 8.
13C-editted (a) and homonuclear (b) NOESY (Tmix = 250 ms) spectra of TAR RNA uniformly labeled with 15N-labeled guanosine and adenosine labeled with 13C at positions 2 and 8.
CONCLUSIONS
Using simple starting materials that are readily available in isotopic form, both ATP and GTP can be synthesized de novo with a variety of isotopic labeling patterns in comparable yields to syntheses with a preformed base. Enzymes originating from different biochemical pathways combined with several cofactor regeneration schemes, including a portion of the folate cycle, worked together elegantly in one pot. This approach provides isotopically labeled nucleotides in superior efficiency to both chemical synthesis and biomass extraction. With all of these factors included in our pathway design, we have been successful in implementing de novo purine biosynthesis in vitro to effect a flexible, robust, and cost effective synthesis of isotopically labeled purine nucleotides. It should be possible to extend this approach to labeled pyrimidine nucleotide synthesis, as well as synthesis of labeled amino acids and cofactors.
Through pathway engineering, the efficiency and elegance of synthesis in nature can be employed in vitro. The interdependence of biosynthetic pathways, the complex flux of metabolites, and the cost and limited availability of biosynthetic precursors provide significant challenges when excising a pathway from its cellular context. Selective fusion of metabolic pathways, regeneration of expensive cofactors and in situ labeling can be accomplished while simultaneously providing a driving force for the desired synthesis from a simple set of starting materials. Based on all of these factors and the complexity of the purine nucleotide synthetic scheme, we suggest that this work expands the limits of the capabilities for in vitro enzymatic synthesis and the scope of its applicability for synthesis of high value biochemicals.
EXPERIMENTAL
Materials
Chemicals were purchased from Sigma. L-serine (13C3, 99%), L-serine (13C1,2,3, 98%), L-serine (15N, 98%), L-serine (15N, 98%; 13C 1,2,3, 98%), L-glutamine (15Namide, >98%), NaHCO3 (13C, 99%), D-glucose (13C1,2,3,4,5,6, 99%) and NH4Cl (15N, 99%) were purchased from Cambridge Isotope Laboratories. Hexokinase from baker’s yeast, glucose-6-phosphate dehydrogenase from baker’s yeast, creatine phosphokinase from rabbit muscle, adenylate kinase from chicken muscle, and glutamic dehydrogenase from bovine liver were purchased from Sigma. PrsA prepared as described previously was used for synthesis of compounds 1 and 2 (39), and the new recombinant prsA was used for synthesis of compounds 3 and 4.
Enzyme Cloning
The genes for all recombinant E. coli enzymes in Table 1 were cloned from the E. coli K12 MRE600 genome, based on the reported gene sequences in Genbank using standard procedures. Each gene was amplified from genomic DNA using PCR with gene specific primers containing compatible restriction sites of either BamHI, HindIII, EcoRI, or XhoI for cloning into pET22-HT (derived from pET22b, Novagen) encoding a N-terminal hexahistidine tag. E. coli strain DH5α was used for cloning and plasmid maintenance. All constructs were verified by DNA sequencing. Rabbit muscle creatine phosphokinase was amplified from pET17b-RMCK, a gift from George Kenyon, University of Michigan (40). A single point mutation, Y397F was introduced to glutamine synthase enhancing specific activity as previously shown (41).
Enzyme Purification
Plasmids were transformed into E. coli BL21(DE3) cells for protein overexpression. Overnight 5 mL cultures in LB with 100 μg/mL ampicillin were diluted 100 fold and grown at 37°C with shaking at 280 rpm until the OD600 was ~0.6. Protein expression was induced by addition of 1 mM IPTG, followed by growth for an additional 4 hours. Cells were harvested by centrifugation at 10000 g for 30 minutes. All steps after cell growth were carried out at 4°C. The cell pellets from 6 L of growth were gently resuspended in 250 mL of 50 mM Tris-HCl (pH 7.5), 250 mM NaCl and cell lysis was accomplished by sonication with 20 cycles of a 30 s pulse and 2 min rest at 75% power. The lysates were clarified by centrifugation at 31000 g and the supernatants were loaded directly to a Ni-NTA (Qiagen) column equilibrated with lysis buffer plus 20 mM imidazole. The column was washed with 6 volumes of the same buffer, and protein was eluted with 250 mM imidazole. Protein containing fractions were checked by SDS-PAGE and combined prior to dialysis against 50 mM potassium phosphate with 5 mM β-mercaptoethanol and 50% (v/v) glycerol. Dialyzed enzymes were stored at −20°C and were active for at least three months. Yields ranged from 10–150 mg of purified protein per liter of culture depending on the enzyme, but different preparations of each enzyme were generally consistent. (see Supplementary Table 1 and Supplementary Figure 1)
Enzymatic Assays
Enzymatic activity is reported in terms of units (U), where 1 U is the amount of enzyme required to convert 1 μmole/min of substrate into product, and the specific activity is reported as U/mg. Enzymatic activities were determined only for guaB, guaA, purB, purA, purF and purD in Figure 1b, and forall enzymes in Figure 1a, c except folD and glyA by coupling the reaction to consumption of ATP which is monitored by the change in A340 due to the action of lactate dehydrogenase on ADP and NADH (17). To assay inorganic pyrophosphatase, the phosphate detection method of Michelson was used (42). When substrates were not available, the amount of enzyme was estimated empirically, using an assumed specific activity of 1 U/mg. Table 1 shows the amount of each enzyme added in U or mg for each synthesis.
Nucleotide Synthesis
Synthesis of 2,8-13C-ATP (1)
Stoichiometric substrates: 212 mg (1.2 mmoles) glucose, 1 g. (18.8 mmoles) NH4Cl, 687 mg (4.7 mmoles) glutamine, 500 mg (4.7 mmoles) 13C3-serine, 235 mg (2.35 mmoles) KHCO3. Fuel reagents: 2.12 g (9.4 mmoles) α-KG, 7.7 g. (23.5 mmoles) creatine phosphate. Catalytic cofactors: 6.43 mg (0.012 mmoles) ATP, 6.6 mg (0.012 mmoles) GTP, 8.8 mg (0.012 mmoles) NADP+, 68 mg (0.12 mmoles) THF, 38 mg (0.24 mmoles) fumarate. Substrates and cofactors were combined in 50 mM KH2PO4/K2HPO4 pH 7.6, 20 mM MgCl2, 20 mM DTT, 100 μg/mL ampicillin, 50 μg/mL kanamycin. Enzymes were added according to Table 1(1), giving a final volume of 120 mL. HPLC chromatograms of the reaction timecourse are included in Supplementary Figure 2.
Synthesis of U-15N-GTP (2)
Stoichiometric substrates: 212 mg (1.2 mmoles) glucose, 1.27 g. (23.5 mmoles) 15NH4Cl, 1.0 g. (7 mmoles) 15Namide-glutamine, 500 mg (4.7 mmoles) 15N-serine, 235 mg (2.35 mmoles) KHCO3. Fuel reagents: 2.7 g. (11.8 mmoles) α-KG, 9.6 g. (29 mmoles) creatine phosphate. Catalytic cofactors: 6.43 mg (0.012 mmoles) ATP, 64.3 mg (0.12 mmoles) dATP, 8.8 mg (0.012 mmoles) NADP+, 8.4 mg (0.012 mmoles) NAD+, 68 mg (0.12 mmoles) THF, 19 mg (0.12 mmoles) fumarate. Substrates and cofactors were added to 50 mM KH2PO4/K2HPO4 pH 7.6, 20 mM MgCl2, 20 mM DTT, 100 μg/mL ampicillin, 50 μg/mL kanamycin. Enzymes were added according to Table 1(2) giving a final volume of 120 mL.
Synthesis of U-13C,15N-GTP (3)
Stoichiometric substrates 93 mg (0.5 mmoles) 13C6-glucose, 605 mg (1.1 mmoles) 15NH4Cl, 165 mg (1.5 mmoles) 13C,15N- serine, 85 mg (1 mmole) NaH13CO3. Fuel Reagents: 1.13 g (5 mmoles) α-KG, 5.25 g (16 mmoles) creatine phosphate. Catalytic cofactors: 2.68 mg (0.005 mmoles) ATP, 26.8 mg (0.050 mmoles) dATP, 3.7 mg (0.005 mmoles) NADP+, 3.5 mg (0.005 mmoles) NAD+, 5 mg (0.0085 mmoles) THF, 8 mg (0.05 mmoles) fumarate. Substrates and cofactors were added to 25 mM KH2PO4/K2HPO4 pH 7.6, 10 mM MgCl2, 10 mM DTT, 50 μg/mL ampicillin, 25 μg/mL kanamycin and flushed thoroughly with argon. Enzymes were added according to Table 1(3) giving a final volume of 100 mL. Argon was gently bubbled again and the glass flask was sealed. An equal amount of additional creatine phosphate was added during this reaction due to increased consumption of (d)ATP by glnA.
Synthesis of U-13C-GTP (4)
Stoichiometric substrates: 93 mg (0.5 mmoles) 13C6-glucose, 594 mg (1.1 mmoles) NH4Cl, 441 mg (3 mmoles) glutamine, 163.5 mg (1.5 mmoles) 13C,- serine, 85 mg (1 mmole) NaH13CO3. Fuel Reagents: 1.13 g (5 mmoles) α-KG, (14 mmoles) 4.6 g creatine phosphate. Catalytic cofactors: 2.68 mg (0.005 mmoles) ATP, 26.8 mg (0.050 mmoles) dATP, 3.7 mg (0.005 mmoles) NADP+, 3.5 mg (0.005 mmoles) NAD+, 5mg (0.0085 mmoles) THF, 8 mg (0.05 mmoles) fumarate. Substrates and cofactors were added to 25 mM KH2PO4/K2HPO4 pH 7.6, 10 mM MgCl2, 10 mM DTT, 50 μg/mL ampicillin, 25 μg/mL kanamycin and flushed thoroughly with argon. Enzymes were added according to Table 1(4) giving a final volume of 100 mL. Argon was gently bubbled again and the glass flask was sealed.
ATP/GTP formation was monitored by HPLC using a Vydac Nucleotide Ion Exchange Column (250 mm × 4.6 mm), using a linear gradient of Buffer A (25 mM Na2HPO4:NaH2PO4 (1:1) adjusted to pH 2.8 with acetic acid) and Bufffer B (125 mM Na2HPO4:NaH2PO4 (1:1) adjusted to pH 2.9 with acetic acid) over 30 minutes at a flow rate of 2 ml/min. ATP/GTP formation was monitored at 260 nm.
Nucleotide Purification
The reaction was sterilized by filtration through a 0.2 μm filter, and ammonium bicarbonate was added to a final concentration of 0.5 M, and the pH was adjusted to 9.65 with ammonium hydroxide. The solution was filtered again and loaded to a 15 g column of Affigel 601 (Biorad) boronate affinity resin equilibrated with 0.5 M ammonium bicarbonate pH 9.65 at room temperature. The column was washed with the same buffer, and nucleotides were eluted with water acidified with CO2. The products were verified by HPLC, NMR and mass spectrometry. Final HPLC and mass spectrometry data for nucleotides 1–4 is given in the Supplementary Material.
RNA Synthesis
HIV-2 TAR RNA (5′ GGCCAGAUUGAGCCUGGGAGCUCUCUGGCC3′) was synthesized by in vitro transcription with T7 RNA polymerase using a mixture of unlabeled UTP and CTP from Sigma and 13C2,8-ATP and U-15N-GTP. RNA was synthesized in a 20 mL reaction under optimized conditions: 21 mM total NTP’s (5.25 mM each), 40 mM Tris HCl (pH 8.1), 0.1 mM spermidine, 10 mM DTT, 28 mM MgCl2, 0.001% Triton X-100, 80 mg/mL polyethylene glycol (8000 MW), 300 nM each DNA strand (Invitrogen), and 0.65 mg/mL T7 RNA polymerase, incubated at 37°C for 4 hours. The RNA was purified on denaturing 20% polyacrylamide gels, electro-eluted and desalted, lyophilized and diluted in 10 mM sodium phosphate (pH 6.5), 150 mM NaCl, 10% D2O for recording NMR spectra.
NMR Experiments
The spectral benefits of specifically labeling the adenosine and guanosine residues with 13C and 15N isotopes was assessed with two-dimensional correlation spectroscopy. One and two bond coupled 1H, 15N-HSQC correlation experiments were collected using INEPT transfer delay times (2Δ) of 5 and 42 μs, respectively.(43) Spectra were recorded on a 500 MHz Bruker Avance spectrometer equipped with a 5mm TXI-HCN triple resonance probe with a z-axis gradient. Homonuclear and isotope-edited NOESY experiments (Tmix = 250 ms) were collected on a 900 MHz Bruker Avance spectrometer equipped with a triple resonance 5mm TXI-HCN probe with triple-axis gradients. Water suppression was achieved through the use of flip-back pulses and the Watergate scheme.(44, 45) 1H, 13C-HSQC spectra were collected using echo/antiecho gradient selection. Spectra were collected at 10°C, referenced to 2,2-dimethyl-2-silapentane-5-sulfonate, processed with NMRPipe and viewed in NMRDraw. (46)
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM-74330). We thank Fabio Agnelli for collection of 13C-NMR spectra in the Supporting Information and for preparations of glucokinase and glucose-6-phosphate dehydrogenase used in the syntheses.
Footnotes
Supporting Information Available: This material is free of charge via the Internet at http://pub.acs.org.
References
- 1.Melnick JS, Sprinz KI, Reddick JJ, Kinsland C, Begley TP. An efficient enzymatic synthesis of thiamin pyrophosphate. Bioorganic & Medicinal Chemistry Letters. 2003;13:4139–4141. doi: 10.1016/j.bmcl.2003.07.026. [DOI] [PubMed] [Google Scholar]
- 2.Rocha-Uribe A, Hernandez E. Solvent-free enzymatic synthesis of structured lipids containing CLA from coconut oil and tricaprylin. Journal of the American Oil Chemists Society. 2004;81:685–689. [Google Scholar]
- 3.Zhu DM, Mukherjee C, Hua L. ‘Green’ synthesis of important pharmaceutical building blocks: enzymatic access to enantiomerically pure alpha-chloroalcohols. Tetrahedron-Asymmetry. 2005;16:3275–3278. [Google Scholar]
- 4.van den Heuvel RHH, Fraaije MW, Laane C, van Berkel WJH. Enzymatic synthesis of vanillin. Journal of Agricultural and Food Chemistry. 2001;49:2954–2958. doi: 10.1021/jf010093j. [DOI] [PubMed] [Google Scholar]
- 5.Ohdan K, Fujii K, Yanase M, Takaha T, Kuriki T. Phosphorylase coupling as a tool to convert cellobiose into amylose. J Biotechnol. 2007;127:496–502. doi: 10.1016/j.jbiotec.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Koreishi M, Tani K, Ise Y, Imanaka H, Imamura K, Nakanishi K. Enzymatic synthesis of beta-lactam antibiotics and N-fatty-acylated amino compounds by the acyl-transfer reaction catalyzed by penicillin V acylase from Streptomyces mobaraensis. Bioscience Biotechnology and Biochemistry. 2007;71:1582–1586. doi: 10.1271/bbb.70052. [DOI] [PubMed] [Google Scholar]
- 7.Huang KT, Wu BC, Lin CC, Luo SC, Chen C, Wong CH, Lin CC. Multi-enzyme one-pot strategy for the synthesis of sialyl Lewis X-containing PSGL-1 glycopeptide. Carbohydr Res. 2006;341:2151–5. doi: 10.1016/j.carres.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 8.Sugiyama M, Hong Z, Liang PH, Dean SM, Whalen LJ, Greenberg WA, Wong CH. D-Fructose-6-phosphate aldolase-catalyzed one-pot synthesis of iminocyclitols. J Am Chem Soc. 2007;129:14811–7. doi: 10.1021/ja073911i. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi S, Komeda H, Asano Y. New enzymatic method of chiral amino acid synthesis by dynamic kinetic resolution of amino acid amides: Use of stereoselective amino acid amidases in the presence of alpha-amino-epsilon-caprolactam racemase. Applied and Environmental Microbiology. 2007;73:5370–5373. doi: 10.1128/AEM.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roessner CA, Spencer JB, Stolowich NJ, Wang J, Nayar GP, Santander PJ, Pichon C, Min C, Holderman MT, Scott AI. Genetically engineered synthesis of precorrin-6x and the complete corrinoid, hydrogenobyrinic acid, an advanced precursor of vitamin B12. Chem Biol. 1994;1:119–24. doi: 10.1016/1074-5521(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 11.Kajiwara Y, Santander PJ, Roessner CA, Perez LM, Scott AI. Genetically engineered synthesis and structural characterization of cobalt-precorrin 5A and -5B, two new intermediates on the anaerobic pathway to vitamin B-12: Definition of the roles of the CbiF and CbiG enzymes. Journal of the American Chemical Society. 2006;128:9971–9978. doi: 10.1021/ja062940a. [DOI] [PubMed] [Google Scholar]
- 12.Scott AI. Genetically-Engineered Synthesis of Natural-Products. Journal of Natural Products. 1994;57:557–573. doi: 10.1021/np50107a001. [DOI] [PubMed] [Google Scholar]
- 13.Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat Biotechnol. 2003;21:1343–6. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Enzymatic total synthesis of enterocin polyketides. Nat Chem Biol. 2007;3:557–8. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 15.Tolbert TJ, Williamson JR. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. Journal of the American Chemical Society. 1996;118:7929–7940. [Google Scholar]
- 16.Tolbert TJ, Williamson JR. Preparation of specifically deuterated and C-13-labeled RNA for NMR studies using enzymatic synthesis. Journal of the American Chemical Society. 1997;119:12100–12108. [Google Scholar]
- 17.Scott LG, Tolbert TJ, Williamson JR. Preparation of specifically H-2- and C-13-labeled ribonucleotides. Rna-Ligand Interactions Pt A. 2000;317:18–38. doi: 10.1016/s0076-6879(00)17004-1. [DOI] [PubMed] [Google Scholar]
- 18.Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of Isotopically Labeled Ribonucleotides for Multidimensional NMR-Spectroscopy of RNA. Nucleic Acids Research. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A. Preparation of C-13 and N-15 Labeled RNAs for Heteronuclear Multidimensional Nmr-Studies. Nucleic Acids Research. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–22. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- 21.Cromsigt J, Schleucher J, Gustafsson T, Kihlberg J, Wijmenga S. Preparation of partially H-2/C-13-labelled RNA for NMR studies. Stereo-specific deuteration of the H5″ in nucleotides. Nucleic Acids Research. 2002;30:1639–1645. doi: 10.1093/nar/30.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaFrancois CJ, Fujimoto J, Sowers LC. Synthesis and utilization of C-13(8)-enriched purines. Nucleosides Nucleotides & Nucleic Acids. 1999;18:23–37. doi: 10.1080/07328319908045591. [DOI] [PubMed] [Google Scholar]
- 23.SantaLucia J, Shen LX, Cai ZP, Lewis H, Tinoco I. Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Research. 1995;23:4913–4921. doi: 10.1093/nar/23.23.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–82. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 25.Jiang F, Patel DJ, Zhang X, Zhao H, Jones RA. Specific labeling approaches to guanine and adenine imino and amino proton assignments in the AMP-RNA aptamer complex. J Biomol NMR. 1997;9:55–62. doi: 10.1023/a:1018623601946. [DOI] [PubMed] [Google Scholar]
- 26.Gmeiner WHaP, Dale C. An Efficient synthesis of [8–13C]Adenine. J Org Chem. 1988;53:1322–1323. [Google Scholar]
- 27.Barrio MD, Scopes DI, Holtwick JB, Leonard NJ. Syntheses of all singly labeled [N]adenines: Mass spectral fragmentation of adenine. Proc Natl Acad Sci U S A. 1981;78:3986–3988. doi: 10.1073/pnas.78.7.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abad JL, Gaffney BL, Jones RA. 15N-Multilabeled Adenine and Guanine Nucleosides. Syntheses of [1,3, NH(2)-(15)N(3)]- and [2-(13)C-1,3, NH(2)-(15)N(3)]-Labeled Adenosine, Guanosine, 2′-Deoxyadenosine, and 2′-Deoxyguanosine. J Org Chem. 1999;64:6575–6582. doi: 10.1021/jo982372k. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Pagano AR, Weimin W, Shallop A, Gaffney BL, Jones RA. Use of a 13C Atom to Differentiate Two 15N-Labeled Nucleosides. Synthesis of [15NH2–15N2]- and [2–13C-1, NH2–15N2]-Guanosine, and [1,7, NH2–15N3]- and [2–13C-1,7, NH2–15N3]-2′-Deoxyguanosine. J Org Chem. 1997;62:7832–7835. [Google Scholar]
- 30.Bouhss A, Sakamoto H, Palibroda N, Chiriac M, Sarfati R, Smith JM, Craescu CT, Barzu O. Enzymatic-Synthesis of Guanine-Nucleotides Labeled with N-15 at the 2-Amino Group of the Purine Ring. Analytical Biochemistry. 1995;225:18–23. doi: 10.1006/abio.1995.1101. [DOI] [PubMed] [Google Scholar]
- 31.Lagoja IM, Herdewijn P. Chemical synthesis of C-13 and N-15 labeled nucleosides. Synthesis-Stuttgart. 2002:301–314. [Google Scholar]
- 32.Crans DC, Kazlauskas RJ, Hirschbein BL, Wong CH, Abril O, Whitesides GM. Enzymatic regeneration of adenosine 5′-triphosphate: acetyl phosphate, phosphoenolpyruvate, methoxycarbonyl phosphate, dihydroxyacetone phosphate, 5-phospho-alpha-D-ribosyl pyrophosphate, uridine-5′-diphosphoglucose. Methods Enzymol. 1987;136:263–80. doi: 10.1016/s0076-6879(87)36027-6. [DOI] [PubMed] [Google Scholar]
- 33.Chenault HK, Whitesides GM. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl Biochem Biotechnol. 1987;14:147–97. doi: 10.1007/BF02798431. [DOI] [PubMed] [Google Scholar]
- 34.Zhao HM, van der Donk WA. Regeneration of cofactors for use in biocatallysis. Current Opinion in Biotechnology. 2003;14:583–589. doi: 10.1016/j.copbio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Chenault HK, Simon ES, Whitesides GM. Cofactor regeneration for enzyme-catalysed synthesis. Biotechnol Genet Eng Rev. 1988;6:221–70. doi: 10.1080/02648725.1988.10647849. [DOI] [PubMed] [Google Scholar]
- 36.Hennig M, Scott LG, Sperling E, Bermel W, Williamson JR. Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J Am Chem Soc. 2007;129:14911–21. doi: 10.1021/ja073825i. [DOI] [PubMed] [Google Scholar]
- 37.Scott LG, Geierstanger BH, Williamson JR, Hennig M. Enzymatic synthesis and 19F NMR studies of 2-fluoroadenine-substituted RNA. J Am Chem Soc. 2004;126:11776–7. doi: 10.1021/ja047556x. [DOI] [PubMed] [Google Scholar]
- 38.Kappock TJ, Ealick SE, Stubbe J. Modular evolution of the purine biosynthetic pathway. Current Opinion in Chemical Biology. 2000;4:567–572. doi: 10.1016/s1367-5931(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 39.Gross A, Abril O, Lewis JM, Geresh S, Whitesides GM. Practical Synthesis of 5-Phospho-D-Ribosyl Alpha-1-Pyrophosphate (Prpp) - Enzymatic Routes from Ribose 5-Phosphate or Ribose. Journal of the American Chemical Society. 1983;105:7428–7435. [Google Scholar]
- 40.Chen LH, Babbitt PC, Vasquez JR, West BL, Kenyon GL. Cloning and expression of functional rabbit muscle creatine kinase in Escherichia coli. Addressing the problem of microheterogeneity. J Biol Chem. 1991;266:12053–7. [PubMed] [Google Scholar]
- 41.Luo S, Kim G, Levine RL. Mutation of the adenylylated tyrosine of glutamine synthetase alters its catalytic properties. Biochemistry. 2005;44:9441–6. doi: 10.1021/bi050554k. [DOI] [PubMed] [Google Scholar]
- 42.MIchelson OB. Photometric Determination of Phosphorous as Molybdovanadophosphoric Acid. Analytical Chemistry. 1957;29:60–62. [Google Scholar]
- 43.Sklenar V, Peterson RD, Rejante MR, Feigon J. Correlation of nucleotide base and sugar protons in a N-15-labeled HIV-1 RNA oligonulceotide by H-1-N-15 HSQC experiments. Journal of Biomolecular Nmr. 1994;4:117–122. doi: 10.1007/BF00178339. [DOI] [PubMed] [Google Scholar]
- 44.Grzesiek S, Bax A. The Importance of Not Saturating H2o in Protein Nmr -Application to Sensitivity Enhancement and Noe Measurements. Journal of the American Chemical Society. 1993;115:12593–12594. [Google Scholar]
- 45.Piotto M, Saudek V, Sklenar V. Gradient-Tailored Excitation for Single-Quantum Nmr-Spectroscopy of Aqueous-Solutions. Journal of Biomolecular Nmr. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 46.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 47.Mueller EJ, Meyer E, Rudolph J, Davisson VJ, Stubbe J. N-5-Carboxyaminoimidazole Ribonucleotide - Evidence for a New Intermediate and 2 New Enzymatic-Activities in the De-Novo Purine Biosynthetic-Pathway of Escherichia-Coli. Biochemistry. 1994;33:2269–2278. doi: 10.1021/bi00174a038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.