SUMMARY
Obesity and nutrient homeostasis are linked by mechanisms that are not fully elucidated. Here we describe a secreted protein, adropin, encoded by a gene, Energy Homeostasis Associated (Enho), expressed in liver and brain. Liver Enho expression is regulated by nutrition: lean C57BL/6J mice fed high-fat diet (HFD) exhibited a rapid increase, while fasting reduced expression compared to controls. However, liver Enho expression declines with diet-induced obesity (DIO) associated with 3 months of HFD or with genetically induced obesity, suggesting an association with metabolic disorders in the obese state. In DIO mice, transgenic overexpression or systemic adropin treatment attenuated hepatosteatosis and insulin resistance independently of effects on adiposity or food intake. Adropin regulated expression of hepatic lipogenic genes and adipose tissue peroxisome proliferator-activated receptor gamma, a major regulator of lipogenesis. Adropin may therefore be a factor governing glucose and lipid homeostasis, which protects against hepatosteatosis and hyperinsulinemia associated with obesity.
INTRODUCTION
The obesity “epidemic” has been associated with increased prevalence of metabolic diseases associated with excess adiposity (Cowie et al., 2006; Perlemuter et al., 2007). Obesity results from a failure to adapt to abundant calories and reduced physical activity (Hill, 2006). Obesity is frequently associated with insulin resistance, which proceeds to type 2 diabetes when pancreatic β cells cannot compensate by increasing insulin production (Biddinger and Kahn, 2006). Several mechanisms linking obesity with insulin resistance have been proposed, including inflammation and impaired function of hypothalamic centers involved in energy homeostasis (Flier, 2004; Hotamisligil, 2006; Morino et al., 2006). An association between disorders of lipid metabolism and insulin resistance is also evident. Excess energy is normally stored in the triacylglycerol (TG) pool in white adipose tissue (WAT). However, the ectopic accumulation of fatty acids (steatosis) in liver, skeletal muscle, and the pancreas has been linked to insulin resistance and type 2 diabetes (Morino et al., 2006). While steatosis could be a factor causing insulin resistance, hyperinsulinism exacerbates steatosis by further stimulating lipogenesis (Morino et al., 2006; Perlemuter et al., 2007; Petersen et al., 2007).
Peptides secreted from peripheral organs regulate lipid metabolism in key insulin-target tissues and are important for energy homeostasis and maintaining insulin sensitivity (Reitman, 2007; Scherer, 2006; Sharma and Staels, 2007). Much attention has been given to adipokines secreted by adipocytes. Adipokines act as paracrine factors within WAT, and as endocrine hormones acting on the liver, muscles, and central nervous system (CNS) to regulate energy homeostasis (Scherer, 2006). For example, adiponectin promotes insulin sensitivity and stimulates fat oxidation in liver and muscle (Scherer, 2006; Sharma and Staels, 2007) while promoting the storage of TG preferentially in WAT (Kim et al., 2007). Leptin acts on hypothalamic neurons that act as relays to sites regulating ingestive behavior, peripheral lipid metabolism, and insulin sensitivity (Morton et al., 2006). Reduced function of adiponectin and leptin contribute to insulin resistance associated with obesity. However, obesity is also associated with secretion of factors from WAT promoting insulin resistance, including resistin and proinflammatory cytokines (Scherer, 2006; Sharma and Staels, 2007).
While receiving less attention, liver-secreted factors are also critical for energy homeostasis (Reitman, 2007). These act as paracrine and endocrine factors that regulate carbohydrate, lipid, and amino acid metabolism during periods of altered nutritional status (Reitman, 2007; LeRoith and Yakar, 2007). Here we describe a novel secreted peptide initially identified during microarray analysis of liver gene expression in mouse models of obesity. Adropin (derived from the Latin roots “aduro” [to set fire to] and “pinquis” [fats or oils]) is encoded by the Energy Homeostasis Associated gene (gene symbol: Enho). Liver Enho expression is regulated by energy status and dietary nutrient content, and is altered with obesity. Transgenic overexpression or systemic adropin treatment improves diet-induced obesity (DIO), insulin resistance, and glucose tolerance. Adropin also attenuates components of the metabolic distress associated with obesity independently of effects on body weight or weight loss. Collectively, our data suggest that adropin is a secreted factor involved in energy homeostasis that could provide a promising new lead for developing therapies against the metabolic disorders associated with obesity.
RESULTS
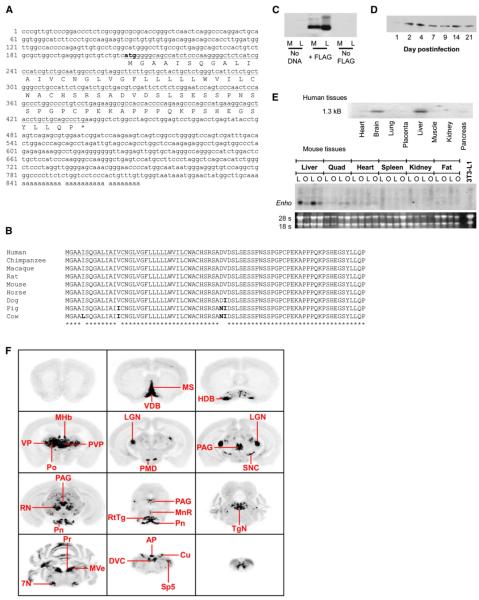
Hypothalamic obesity is associated with insulin resistance and hepatic steatosis (Sutton et al., 2006). To further investigate hypothalamic regulation of liver metabolism, we performed a global analysis of gene expression in C57BL/6J (B6) melanocortin-3 receptor-deficient (Mc3r-/-) mice. During validation of microarray data, we identified an unknown liver transcript downregulated with obesity. RIKEN cDNA 2310040A07 encodes a short (<1 kb) mRNA, encoding a highly conserved 76 aa open reading frame (ORF) (Figures 1A and 1B). Bioinformatics analysis using SignalP 3.0 (Emanuelsson et al., 2007) suggested that the peptide is likely (87% probability) to be secreted, with the most probable cleavage site between residues 33 and 34. Secretion of peptide was confirmed in HEK293 cells and mice by using carboxy-terminal FLAG-tagged expression vectors. FLAG immunoreactivity (IR) was observed in conditioned media of HEK293 cells transfected with pCMV-Enho:FLAG (Figure 1C). FLAG-IR was also observed in sera from B6 mice injected with 108-109 plaque-forming units (pfu) of recombinant adenovirus that expresses the FLAG-labeled adropin into the tail vein (Figure 1D).
Figure 1. Enho mRNA Encodes a Highly Conserved Secreted Peptide.
(A) Enho mRNA sequence with the conserved ORF underlined.
(B) Conservation of adropin sequence in mammals.
(C) Presence of FLAG-IR in cell lysate (L) and conditioned media (M) of HEK293 cells transfected with vectors expressing adropin with or without a C-terminal FLAG.
(D) Western blot of FLAG-IR in sera collected from B6 mice 1-21 days after injection with 108 pfu of adenovirus expressing FLAG-tagged adropin.
(E) Northern blots showing distribution of Enho mRNA in human (upper panel) and mouse tissues (lower panel). Tissues were obtained from lean B6 (L) or obese leptin-deficient (O) mice.
(F) A series of autoradiographs summarizing Enho mRNA expression sites across the B6 mouse brain: 7N, facial nucleus; AP, area postrema; Cu, cuneate nucleus; DVC, dorsal vagal complex; HDB, nucleus of the horizontal limb of the diagonal band; LGN, lateral geniculate nucleus; MHb, medial habenula; MnR, median raphe; MS, medial septum; MVe, medial vestibular nucleus; PAG, periaqueductal gray; PMD, dorsal premamillary nucleus; Pn, pontine nuclei; Po, posterior thalamic nuclear group; Pr, prepositus nucleus; PVP, paraventricular thalamic nucleus; RN, red nucleus; RtTg, reticulotegmental nucleus of the pons; SNC, substantia nigra pars compacta; Sp5, spinal trigeminal nucleus; TgN, tegmental nucleus; VDB, nucleus of the vertical limb of the diagonal band; VP, ventral posterolateral thalamic nucleus.
Analysis of tissue distribution by northern blot revealed a transcript of ∼1.3 kB abundant in liver and whole brain of mice and humans (Figure 1E). As nothing was known about expression of the transcript in the brain, we further analyzed its distribution in the mouse by in situ hybridization histochemistry using three separate riboprobes. Dense Enho expression was observed in areas of the thalamus, midbrain, and hindbrain (Figure 1F). Abundant Enho mRNA was observed in the medial habenula, the dorsal premamillary nucleus of the hypothalamus, the medial septal complex (medial septum and the nucleus of the diagonal band), and the thalamus (the posterior thalamic nuclear group, the lateral and medial geniculate nuclei, and the paraventricular, anterodorsal, ventral posterolateral, and ventral posteromedial thalamic nuclei). In the midbrain, high levels of Enho mRNA were evident in the substantia nigra pars compacta, interpeduncular nucleus, red nucleus, periaqueductal gray, and median raphe. Enho mRNA was observed in the pontine nuclei and the reticulotegmental nucleus of the pons. Dense Enho mRNA was also identified in regions of the medulla, including the medial vestibular nucleus, prepositus nucleus, facial nucleus, inferior olive, lateral reticular nucleus, dorsal vagal complex, area postrema, and cuneate nucleus. Sense probes produced no detectable hybridization (data not shown). Together, these observations suggest that the transcript is abundant in areas involved in the control of complex behaviors, including circadian rhythms and stress response, and in areas that regulate peripheral metabolism through the autonomic nervous system.
Regulation of Liver Enho Expression
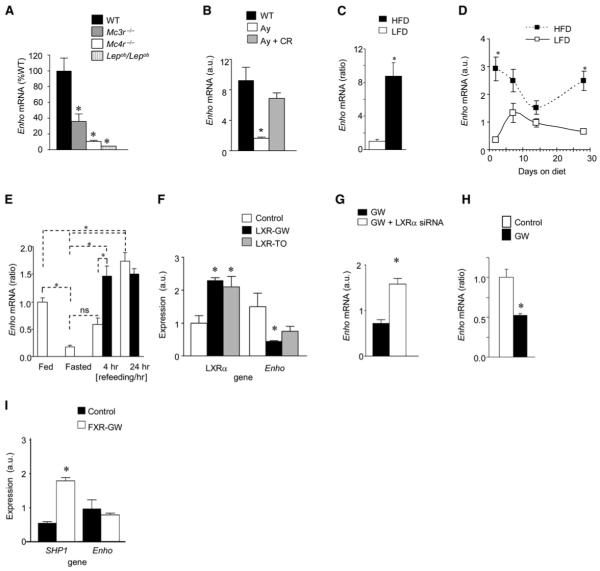
As the transcript was initially identified by exhibiting reduced expression in liver of Mc3r-/- mice, its expression in lean and obese states was further investigated by northern and quantitative RT-PCR. Obesity due to deficiency in melanocortin receptor signaling (lethal yellow Ay/a, Mc3r-/-, and Mc4r-/-) or leptin (Lepob/Lepob) is associated with marked reductions in liver Enho mRNA compared to lean wild-type (WT) controls (Figures 1E and 2A). We next determined whether altered Enho expression is related to obesity or is a specific consequence of disrupted melanocortin signaling. Prevention of obesity in Ay/a mice by calorie restriction (CR) (body weight [BW] in g, 20.7 ± 1.4 g) normalized liver Enho mRNA compared to obese ad libitumfed Ay/a mice (BW of 48.2 ± 1.4 g) (Figure 2B). Reduced hepatic Enho mRNA in Ay/a mice is thus a secondary effect of obesity.
Figure 2. Regulation of Liver Enho by Obesity and Nutrition.
(A) Liver Enho expression in 6-month-old male Mc3r-/-, Mc4r-/-, and Lepob/Lepob mice compared to lean controls. The Mc3r-/- and Mc4r-/- mice used for this analysis were backcrossed >10 generations onto the B6 background. * p < 0.05 compared to WT, n = 5-6 per group.
(B) Liver Enho mRNA is also reduced in Ay/a mice compared to lean B6 controls, which develop obesity due to hyperphagia associated with expression of an Mc4r antagonist. The reduced expression of Enho in liver is not observed in Ay/a mice subject to calorie restriction (CR), suggesting that the decline is secondary to obesity and not a specific defect due to reduced melanocortin receptor signaling (n = 4 per group). * p < 0.05 versus obese Ay/a mice and lean controls.
(C) Feeding lean male B6 mice HFD (60% kJ/fat) for 2 days is associated with increase liver Enho expression compared to mice fed LFD (10% kJ/fat) * p < 0.01.
(D) Differential effects of purified LFD or HFD on the expression of liver Enho mRNA in male B6 mice after 2, 7, 14, or 28 days. * p < 0.05 compared to LFD within time point (n = 4 per group).
(E) Fasting is associated with reduced liver Enho expression compared to controls that had been fed a purified LFD (10% kJ/fat) for 10 days. The rebound in expression is affected by dietary fat content, with mice refed LFD (open bars) exhibiting a delayed response compared to mice refed with HFD (black bars) (n = 5-6 per group). * p < 0.05, ns, no significant difference. Note that the mice used for the studies shown in (C) and (D) were 3 months old, and had been weaned onto standard rodent chow.
(F) Reduced Enho mRNA in HepG2 cells after 48 hr treatment with LXR agonists (GW3965 [GW]; TO9 [TO]; both 1 μM for 24 hr).
(G) An siRNA targeting LXRα increases Enho mRNA in HepG2 cells treated with GW-3965.
(H) Liver Enho mRNA in 12-week-old B6 mice is significantly reduced 4 hr after intravenous injection with 10 mg/kg GW3965 (n = 4, *p < 0.05).
(I) The FXR agonist GW-4064 (1 μM for 24 hr) increased expression of Shp1 but did not effect Enho expression in HepG2 cells. Values shown in each panel are mean ± SEM.
To investigate whether nutritional status affects liver Enho mRNA, we examined regulating by fasting and short- to longterm exposure to high-fat diet (HFD: 60% kJ from fat, 20% kJ/ carbohydrate, 20% kJ/protein). DIO in B6 mice induced by 3 months of HFD was associated with reduced liver Enho mRNA compared to lean controls (expression as a ratio of control: 1.00 ± 0.18 versus 0.53 ± 0.19, n = 4 for DIO, n = 5 for control, p = 0.06). However, while chronic exposure to DIO reduced expression, the results from studies investigating acute regulation by diet showed a dynamic response. A 9-fold increase in liver Enho mRNA was observed in mice fed HFD for 2 days compared to mice fed low-fat diet (LFD: 10% kJ from fat, 70% kJ/carbohydrate, 20% kJ/protein) (Figure 2C). Further analysis of expression in mice subject to 2 days, 7 days, 14 days, or 28 days of either HFD or LFD (n = 3-4 per group) showed a complex biphasic pattern of regulation, with significantly increased expression in the HFD-fed relative to LFD-fed mice on day 2 or day 28 (Figure 2D). The decline in liver Enho mRNA with DIO is thus presumed to occur after a prolonged (>1 month) exposure to HFD and may represent malfunction of normal regulation by macronutrients in the obese state.
Liver Enho mRNA in lean B6 mice was also regulated by fasting, which reduced expression 80% relative to chow-fed animals (Figure 2E). Consistent with results shown in Figures 2C and 2D, demonstrating stimulation by HFD, the rebound in expression during refeeding was affected by macronutrient content of the diet. Mice refed with LFD exhibited a delayed response relative to mice refed with HFD (Figure 2E).
Together, these data indicate that hepatic Enho expression is influenced by fasting and the macronutrient composition of the diet, with high expression levels observed during short-term intake of diets high in fat content. However, chronic exposure to HFD is associated with reduced expression, perhaps indicating deregulation of liver Enho expression with obesity.
The rapid regulation of liver Enho mRNA by macronutrient content suggested an involvement of intracellular lipid sensors. Peroxisome proliferator activated receptor alpha (Ppara) is activated during the metabolic adaptation to fasting (Reitman, 2007) and therefore appeared an unlikely candidate for stimulating Enho expression. The liver X receptors (LXRα and LXRβ, also known as Nr1h2 and Nr1h3) and farnesoid X receptor (FXR, also known as Nr1h4) are nuclear receptors that are regulated by sterols and bile acids and are involved in carbohydrate and lipid homeostasis (Kalaany and Mangelsdorf, 2006). Treatment of HepG2 cells with the LXR agonists GW3965 and TO9 increased expression of LXRα mRNA (Figure 2F). This is the primary LXR isoform in HepG2 cells, and its expression is stimulated by LXR agonists (Laffitte et al., 2003). In these cells, presence of the LXR agonist reduced Enho expression (Figure 2F), a response blocked by an antisense RNA targeting LXRα (Figure 2G) and previously shown to reduce LXRα mRNA and protein (Stayrook et al., 2008). To determine whether activation of LXRα would regulate Enho mRNA in mouse liver, GW3965 (10 mg/kg) was administered intravenously to lean, chow-fed, male B6 mice. Mice treated with GW3965 exhibited a significant reduction in liver Enho mRNA 4 hr after injection (Figure 2H). Treatment of HepG2 cells with the FXR agonist GW4064 significantly increased the expression of Shp1, as previously reported (Goodwin et al., 2000), but did not affect Enho expression (Figure 2I).
Together, these results indicate that liver Enho mRNA expression is regulated by LXRα, a nuclear receptor involved in cholesterol and triglyceride metabolism (Kalaany and Mangelsdorf, 2006). While we predicted that lipid sensors would stimulate Enho expression, the opposite effect was observed. The link between the response observed with activation of LXRα and the stimulation of hepatic Enho expression by high fat intake thus remains unclear. However, the rapid regulation by LXRα agonists observed in cultured cells and in whole animals is further evidence linking the Enho gene with carbohydrate and lipid homeostasis.
Improved Glucose Homeostasis in Adropin-Transgenic Mice
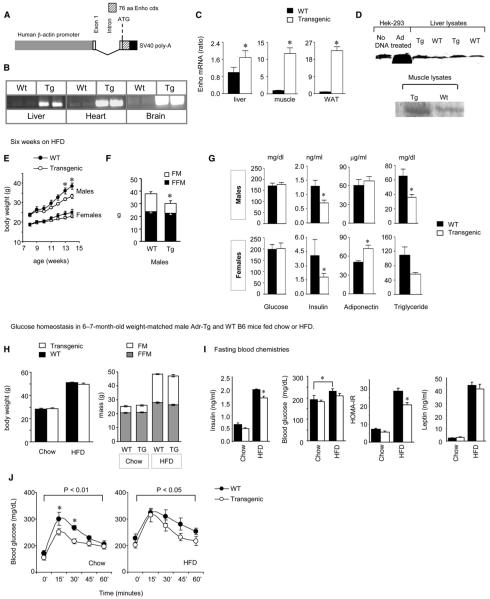
Thus far, the data obtained from the analysis of expression in whole animals and HepG2 cells suggested that the Enho gene is associated with carbohydrate and lipid metabolism, and that severe obesity is associated with loss of normal regulation. To further investigate the association of altered Enho expression with energy status, we generated transgenic B6 mice overexpressing adropin (Adr-Tg) (Figure 3A). Widespread expression of the transgene was confirmed by RT-PCR using transgene-selective primers (Figure 3B) and quantifying expression using primers simultaneously detecting the endogenous and transgene cDNA (Figure 3C). Polyclonal antibodies raised against the C terminus of the putative secreted domain revealed increased adropin IR in tissue lysates where endogenous protein was detected (Figure 3D). Adr-Tg mice appear grossly normal, with no evidence of significant increases in mortality associated with the transgene.
Figure 3. Adr-Tg Mice Exhibit Attenuated Metabolic Distress Associated with Chronic Exposure to HFD.
(A) Schematic showing the transgene construct with the adropin ORF ligated into a vector containing the human β-actin promoter.
(B) Representative RT-PCR analysis using transgene-specific primers; expression was observed in liver, heart, and whole brain.
To investigate how the adropin transgene would affect metabolism in situations of obesity, we challenged Adr-Tg and wildtype B6 mice to HFD (60% kJ/fat). At 8 weeks of age, chowfed Adr-Tg had normal BW; however, examination of Adr-Tg mice and WT controls fed HFD from 8 to 14 weeks of age suggested protection from DIO (Figure 3E). Males (Adr-Tg, n = 3; WT, n = 4) and females (Adr-Tg, n = 7; WT, n = 8) were fed an HFD for 6 weeks. As predicted, weight gain was significantly greater in males compared to females (least square means [LSM] for BW gain in g for males, 12.2 ± 1.0; for females, 5.2 ± 0.7; p < 0.01). Weight gain was significantly attenuated in Adr-Tg mice (LSM for BW gain in g: for WT, 10.4 ± 0.8; for Adr-Tg, 7.0 ± 0.9; effect of genotype, p = 0.01), due exclusively to reduced fat mass (FM) (Figure 3F). Similar results were observed in a second group of male mice fed HFD for 6 weeks (BW gain in g for WT, 13.2 ± 1.8; for Adr-Tg, 6.8 ± 1.5; p < 0.05; FM gain for WT, 12.3 ± 1.7; for Adr-Tg, 5.6 ± 2.0; p < 0.05; FFM gain for WT, 1.5 ± 0.4; for Adr-Tg, 1.5 ± 0.8, p = 0.78, N = 3 for Adr-Tg, N = 7 for WT). For this group, we investigated the effects of a longer duration of HFD. These results suggested that while DIO in Adr-Tg mice is delayed, it is not prevented. BW parameters of WT and Adr-Tg converged after 3 months on HFD (data not shown).
To investigate whether reduced weight gain would improve the metabolic distress associated with obesity, glucose homeostasis was assessed in Adr-Tg mice with reduced FM (Figures 3E and 3F). To determine whether adropin affects glucose homeostasis independently of BW, we also examined Adr-Tg mice matched with controls for BW and adiposity. Glucose homeostasis examined in mice fed HFD for 6 weeks indicated improvements consistent with reduced obesity (Figure 3G). Under fasting conditions, serum insulin levels were lower in Adr-Tg mice (LSM in ng/ml: WT 2.7 ± 0.5, Adr-Tg 1.3 ± 0.5, p = 0.05); blood glucose was not significantly different (LSM in mg/dL: Adr-Tg 189 ± 11; WT 185 ± 10, p = 0.77). Female Adr-Tg mice exhibited an increase in total serum adiponectin, while serum TG was also reduced in Adr-Tg (Figure 3G). These observations suggested that reduced adiposity due to the overexpression of adropin is also associated with improvement in glucose homeostasis. However, these studies did not determine whether adropin affects carbohydrate and lipid metabolism independently of reducing weight.
Secreted factors that regulate energy homeostasis can also affect glucose homeostasis independently of reducing food intake or causing weight loss (Asilmaz et al., 2004; Obici et al., 2001; Zhou et al., 2007). We therefore assessed glucose homeostasis in male 6- to 7-month-old Adr-Tg and WT mice (n = 7-8/group) fed standard rodent chow or HFD for 3 months, which were matched for BW and adiposity (Figure 3H). Adr-Tg mice again exhibited improved glucose homeostasis, indicated by reduced fasting insulin, protection from HFD-induced hyperglycemia, lower homeostasis model assessment of insulin resistance (HOMA-IR) values (Figure 3I), and significantly enhanced glucose clearance in Adr-Tg mice (Figure 3J). Improved glucose tolerance was also observed in Adr-Tg mice fed chow (Figure 3J). Significantly, the improvements in glucose homeostasis of the HFD-fed group were not due to significant differences in energy expenditure (VO2 in ml/gBW/hr for WT: 2296 ± 38; Adr-Tg: 2337 ± 55; p = 0.55), substrate metabolism as indicated by the respiratory quotient (RQ) (WT: 0.765 ± 0.003; Adr-Tg: 0.764 ± 0.003; p = 0.88), or physical activity (X-movements in beam breaks/hr for WT: 417 ± 28; Adr-Tg: 397 ± 20, p = 0.55). Together, these results strongly suggest that adropin regulation of glucose homeostasis is independent of changes in BW, food intake, and whole body energy expenditure.
Delayed DIO in Adr-Tg Mice Is Due to Altered Metabolism
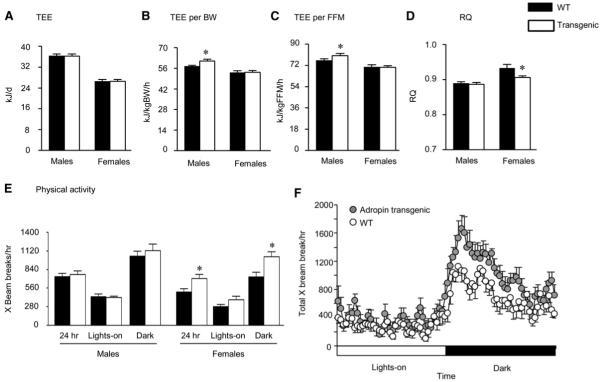
To investigate how the transgene delays DIO, chow-fed male and female Adr-Tg and WT controls (n = 7-8, age 8-10 weeks) were housed in metabolic chambers measuring energy expenditure, whole body substrate oxidation indicated by the RQ, and physical activity. Adr-Tg and WT mice had similar BW (male Adr-Tg: 25.1 ± 0.7 g, male WT: 26.4 ± 0.6 g, p = 0.18; female Adr-Tg: 20.4 ± 0.3 g, female WT: 20.8 ± 0.4 g, p = 0.45). Gender-specific effects of the transgene were observed on energy metabolism. Resting energy expenditure (REE) and total energy expenditure (TEE) expressed per mouse were not significantly different in Adr-Tg and WT mice of either sex (Figure 4A) (REE in kJ/day for male WT: 26.6 ± 0.7, male Adr-Tg, 27.2 ± 0.8, p = 0.55; for female WT: 17.7 ± 0.5, female Adr-Tg: 17.0 ± 0.2, p = 0.22). However, TEE expressed either per gram of BW or fat-free mass (FFM) was modestly but significantly increased in male Adr-Tg mice compared to controls (+3.7 kJ/kgBW/hr, p < 0.05, Figure 4B; +4.1 kJ/kgFFM/hr, p < 0.05, Figure 4C). Similar differences were evident when activity data was used to calculate REE, suggesting an increase in basal metabolic rate in male Adr-Tg mice (in kJ/gBW/hr for WT, 42.0 ± 0.9; Adr-Tg, 45.3 ± 1.3; p < 0.05; in kJ/kgFFM/hr for WT, 55.8 ± 1.3; Adr-Tg, 59.7 ± 1.9; p < 0.05). A difference of ∼4 kJ/day amounts to 180 kJ or approximately 5 g of fat over 6 weeks, which is comparable to the actual difference in weight gain observed in Adr-Tg mice relative to controls.
Figure 4. Analysis of Whole Body Energy Metabolism in Chow-Fed Adr-Tg and WT Mice.
(A) Total 24 hr energy expenditure (TEE) of male and female Adr-Tg mice compared to controls.
(B and C) TEE expressed per g body weight (B) or FFM (C) (*p < 0.05).
(D) RQ of male and female Adr-Tg and WT mice (*p < 0.05).
(E and F) Spontaneous locomotory activity of female Adr-Tg and WT mice (*p < 0.05). In (E), the data are presented as mean X beam breaks over 24 hr, during the lights-on period (0600-1800 hr; L or white bar) and during the dark period (1800-0600 hr; D or black bar), by gender. All data are mean ± SEM and are the mean from 3 days of recordings; n = 7-8 each genotype and sex.
(C) Quantitative analysis of Enho mRNA in liver, muscle, and retroperitoneal WAT of WT and Adr-Tg mice (n = 3 per group, * p < 0.05).
(D) Western blot analysis of adropin IR in control HEK293 cells (no DNA), cells infected with adenovirus vector expressing adropin (Ad treated), and liver and muscle tissue lysates from Adr-Tg (Tg) or WT mice.
(E) Body weight of WT and Adr-Tg mice fed HFD for 6 weeks (N = 3-8 for each genotype and sex). * p < 0.05 versus WT (within gender).
(F) Reduced FM and normal FFM for male mice shown in (E) after 6 weeks on HFD. *p < 0.05 for FM (n = 3 for WT, 8 for Adr-Tg).
(G) Blood chemistries for Adr-Tg mice fed HFD for 6 weeks. Transgenics exhibited reduced insulin, normal blood glucose, increased serum adiponectin (in females), and reduced serum TG compared to WT mice.
(H-J) Improvements in glucose homeostasis in Adr-Tg mice are independent of reduced weight gain. Body weight and composition data from male Adr-Tg and WT mice aged 6-7 months and maintained on chow or HFD are shown (60% kJ/fat) (H). Adr-Tg mice exhibited protection from hyperinsulinemia and hyperglycemia associated with obesity; reduced HOMA-IR values also suggested improved insulin sensitivity. Serum leptin levels were affected by diet (p < 0.01), correlating with increased adiposity, but not by genotype (I). Glucose clearance was also significantly improved in Adr-Tg mice relative to controls, irrespective of diet (J). All data are mean ± SEM.
In contrast, female Adr-Tg mice had normal energy expenditure (Figures 4A, 4B, and 4C) (REE in kJ/gBW/hr for WT, 35.5 ± 0.7; Adr-Tg, 34.8 ± 0.8; REE in kJ/kgFFM/hr for WT, 47.2 ± 1.1; Adr-Tg, 46.0 ± 0.8). However, a significantly lower RQ during the dark period in female Adr-Tg mice suggests altered substrate metabolism (Figure 4D). Female Adr-Tg mice also exhibit increased spontaneous locomotor activity during the dark period (Figures 4E and 4F).
Food intake was not significantly different in chow-fed Adr-Tg and WT mice, irrespective of gender (intake in kJ/day for male WT, 60.1 ± 3.2; Adr-Tg 56.6 ± 1.5; for female WT 51.4 ± 4.6; Adr-Tg 47.2 ± 1.8). Food intake of Adr-Tg was also not significantly different when first exposed to HFD (intake in kJ/day for male WT, 75.9 ± 2.5; Adr-Tg 74.2 ± 2.5; for female WT, 51.4 ± 4.6; Adr-Tg 47.2 ±1.8). Together, these observations indicate that delayed DIO in Adr-Tg mice is primarily due to altered metabolism, and does not involve significant suppression of food intake.
The hypothalamus regulates energy expenditure and glucose homeostasis through the autonomic nervous system (Morton et al., 2006). To investigate the impact of adropin on the hypothalamus, we measured serum leptin and brown adipose tissue (BAT) uncoupling protein-1 (Ucp1) expression to determine whether neuroendocrine function is altered in Adr-Tg. Serum leptin levels in 6-month-old mice fed LFD or HFD exhibited the predicted increase associated with DIO (p < 0.01) with no effect of genotype (Figure 3I). The expression of Ucp1 in BAT was also normal under HFD-fed or chow-fed conditions (expression in AU in HFD-fed WT: 1.15 ± 0.08, HFD-fed Adr-Tg: 1.13 ± 0.13, p = 0.85; chow-fed WT: 1.11 ± 0.18, chow-fed Adr-Tg: 0.91 ± 0.10, p = 0.33). Ucp1 expression is regulated by the sympathetic nervous system and leptin (Cannon and Nedergaard, 2004). These observations suggest that autonomic activity and leptin sensitivity are unlikely to be altered in Adr-Tg mice.
Attenuated Hepatic Steatosis in Adr-Tg Mice
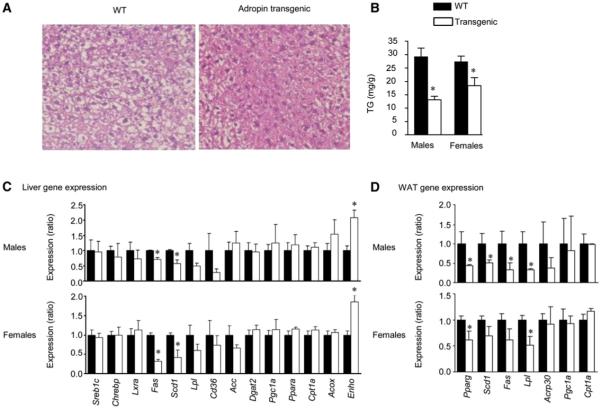
Hepatic steatosis is frequently observed with obesity and insulin resistance (Perlemuter et al., 2007). After 6 weeks on HFD, liver TG content was significantly reduced in Adr-Tg mice compared to control littermates (Figures 5A and 5B) associated with reduced expression of key enzymes involved in lipogenesis and TG synthesis, stearoyl CoA desaturase 1 (Scd1), and fatty acid synthase (Fas) (Figure 5C), with normal expression of genes involved in fat oxidation (Figure 5C). Attenuated hepatic steatosis of Adr-Tg mice may therefore result at least partially from reduced lipogenesis.
Figure 5. Attenuated Hepatic Steatosis and Altered Expression of Genes Involved in Lipid Metabolism in Adr-Tg Mice.
(A) Hematoxylin- and eosin-stained liver sections showing less severe macrovesicular fatty change in livers of Adr-Tg mice compared to WT.
(B) Liver TG content is lower in Adr-Tg mice (n = 3-8 each genotype and sex, *p < 0.05 versus WT).
(C) Expression of stearoyl-CoA desaturase-1 (Scd1), fatty acid synthase (Fas), and lipoprotein lipase (lpl) mRNA is reduced in liver of male and female Adr-Tg mice compared to WT (n = 3-8 each genotype and sex, *p < 0.05 versus WT). Liver data shown in (A)-(C) was from mice fed HFD for 3 months.
(D) Expression of genes involved in fatty acid metabolism in WAT from male and female Adr-Tg and WT mice. * p < 0.05. Values shown are mean ± SEM from mice fed HFD for 6 weeks.
Adipose Tissue Lipogenic Gene Expression in Adr-Tg
As adipocytes also have a critical role in lipid homeostasis, we examined expression of lipogenic genes in retroperitoneal WAT. Expression of the Pparg gene, encoding a nuclear receptor that regulates expression of genes involved in lipid synthesis (Sharma and Staels, 2007), and a subset of Pparg-regulated genes (Scd1, Fas, lipoprotein lipase [Lpl]) was significantly reduced in Adr-Tg mice compared to controls (Figure 5D). As observed for liver, the expression of genes involved in fat oxidation was not significantly different in WAT of Adr-Tg (Figure 5D). Overexpression of adropin therefore affects the expression of genes involved in lipogenesis in both liver and adipose tissue.
Adropin(34-76) Improves Glucose Homeostasis
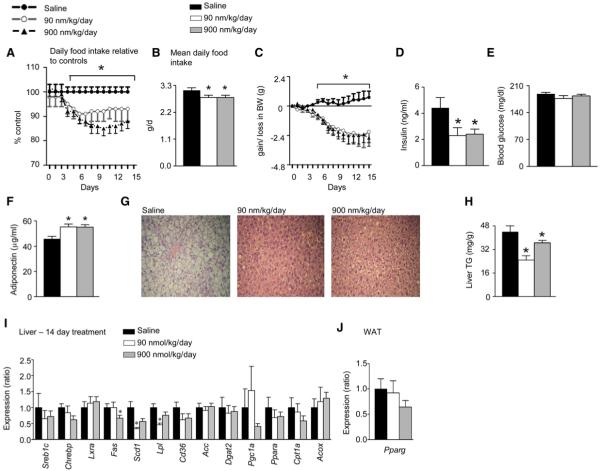
Analysis of Adr-Tg mice suggests delayed obesity and a prevention of the deterioration of glucose homeostasis associated with weight gain. However, these studies did not determine whether delivering adropin to obese, insulin-resistant mice would rapidly reverse the deterioration of glucose homeostasis. We therefore sought to establish whether treatment with a synthetic form of adropin could improve glucose homeostasis in DIO mice, using a synthetic peptide corresponding to the secreted domain predicted by SignalP 3.0. Groups of individually housed 6-month-old male DIO mice were administered adropin(34-76) at 0, 90, or 900 nmol/kg/day (n = 12 per dose) by i.p. injection at 0900 hr and 1800 hr for 14 days. The groups were matched for BW (41.5 ± 1.5, 41.8 ± 1.1, and 42.6 ± 1.0 g, respectively). After 3 days of treatment, a reduction in food intake was observed in mice treated with 90 or 900 nmol/kg/day of adropin(34-76) (Figure 6A), causing a modest but significant reduction of cumulative food intake over the treatment period (Figure 6B). Weight loss coinciding with the reduction in food intake after the third day of treatment was also evident, with BW stabilizing at 2.1-2.7 g (∼6%) below starting weight after 9 days (Figure 6C). Adropin(34-76) treatment reduced hyperinsulinemia without affecting blood glucose (Figures 6D and 6E), increased serum adiponectin (Figure 6F), reversed hepatic steatosis (Figures 6G and 6H), and decreased expression of hepatic expression of genes involved in TG production and uptake (Figure 6I). However, while appearing to be lower in the 900 nmol/kg/day group, the expression of Pparg in WAT was not significantly different from controls (Figure 6J).
Figure 6. Adropin(34-76) Therapy Affects Energy Metabolism in DIO B6 Mice and Cultured Adipocytes.
(A-J) Administration of adropin(34-76) by i.p. injection at 0900 and 1800 hr for 14 days is associated with reduced food intake (A and B) and weight loss (C). Adropin treatment reduced fasting insulin (D) without affecting blood glucose (E), and increased serum adiponectin (F), suggesting improved insulin sensitivity. Adropin treatment improves hepatic steatosis (G) and significantly reduces liver TG accumulation (H), associated with reduced expression of genes involved in lipid synthesis in liver (I). Pparg expression in WAT of mice treated with adropin(34-76) was not significantly different from controls treated with saline (J). The values shown are mean ± SEM with N = 12/treatment group.
The significant results observed with 2 weeks of treatment demonstrate that a synthetic form of adropin is effective at reversing insulin resistance associated with obesity and chronic intake of a HFD. Moreover, these results were observed with the lowest dose (90 nmol/kg) of a synthetic peptide not subject to modifications that enhance the pharmokinetic profile.
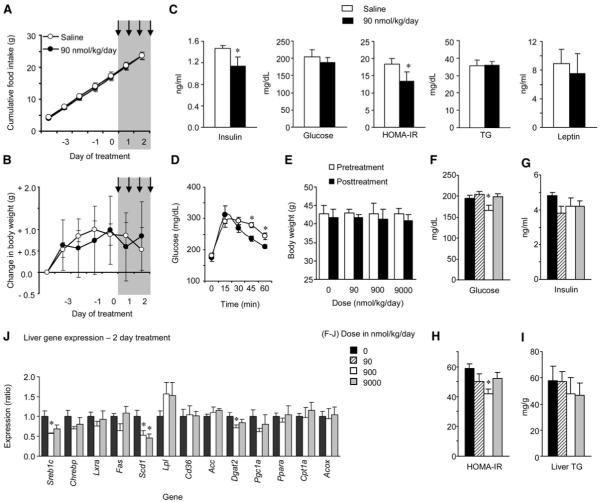
Improvements in glucose homeostasis and hepatic steatosis associated with 2 weeks of therapy could result from weight loss. We therefore determined whether evidence for similar effects of adropin(34-76) on glucose homeostasis in DIO mice would be observed with shorter treatment duration prior to significant effects on BW. The dose of 90 nmol/kg/day required several days to affect food intake and BW (Figures 6A, 6B, and 6C). We therefore repeated a study using this dose with 2 days of bidaily injections (n = 8/group). As previously observed, 2 days of treatment did not significantly affect cumulative food intake or BW (Figures 7A and 7B). However, there were significantly lower fasting insulin and HOMA-IR values in the treatment group, suggesting improved insulin sensitivity (Figure 7C). There was no significant effect on fasting serum TG or leptin (Figure 7C), suggesting that peptide treatment had not markedly affected circulating lipids or leptin sensitivity. However, DIO mice treated with adropin(34-76) for 2 days did have significantly better glucose tolerance relative to controls (Figure 7D).
Figure 7. Short-Term Treatment with Synthetic Adropin(34-76) Improves Glucose Homeostasis in DIO B6 Mice Independently of Weight Loss or Reduced Food Intake.
(A-D) Treatment of male DIO mice with 90 nmol/kg/day adropin(34-76) administered as two i.p. injections/day over 2 days did not significantly affect food intake (A) or weight gain (B) (n = 8/group). Treatment followed a 5 day lead-in period, with the injection period marked by the gray area. Following treatment, there were significant improvements in glucose homeostasis, indicated by reductions in fasting insulin and HOMA-IR values (C) and improved glucose tolerance (D). * p < 0.05 compared to saline. However, there were no changes observed in serum TG (C).
(E-J) A second study assessed a dose-response to 2 days of injections on glucose homeostasis (F-H) and liver metabolism (I, J) (n = 6/group). There was no significant effect of treatment on body weight (E, the white and solid bars are body weight pre- and posttreatment). There was evidence for improved glucose homeostasis (F-H), with significant reductions in fasting glucose (F) and HOMA-IR (H). While liver TG content was not significantly affected by treatment (I), there were changes in the expression of genes involved in fatty acid metabolism, which was statistically significant for Scd1 and diacylglycerol O-transferase 2 (Dgat2) (J). * p < 0.05 versus saline. The values shown are mean ± SEM.
A second study examined the effects of a wider dose range (0-9000 nmol/kg/day, n = 6/group) administered for 2 days on liver metabolism. Again, no significant difference in BW pre- or postinjection was observed, even with the highest dose of 9000 nmol/kg/day (Figure 7E). Evidence for improvements in glucose homeostasis was again observed, with a significant reduction in glucose and HOMA-IR observed with the 900 nmol/kg dose (Figures 7F and 7H). While serum insulin levels were lower, the effect did not achieve statistical significance in this study (Figure 7G). Liver TG content was also not affected after 2 days of treatment (Figure 7I). However, the expression of genes involved in fatty acid metabolism was significantly different in the treatment groups, with reductions in the expression of Sreb1c, Scd1, and Dgat2 mRNA (Figure 7J).
Together, these observations show that improvements in glucose homeostasis and hepatic lipid metabolism associated with with adropin(34-76) therapy occur rapidly and are independent of reduced BW or altered food intake.
DISCUSSION
Our studies have identified a gene encoding a secreted peptide involved in energy homeostasis. Hepatic expression of the Enho transcript is altered with obesity and changes in energy status and in response to dietary macronutrients, which initially suggested that it was associated with energy homeostasis. The Enho gene encoding adropin is also expressed in areas of the brain involved in metabolic regulation. We demonstrated the effects of adropin on energy homeostasis using transgenic overexpression of the ORF and by systemic treatment using a synthetic peptide. These therapies significantly affected weight gain and improved glucose homeostasis. Adropin also affected glucose homeostasis and lipid metabolism independently of weight loss. Adropin thus appears to have a significant role in energy homeostasis, and may be involved in the metabolic adaptation to fasting and dietary macronutrients. Adropin may thus provide an important new lead for the development of therapies to improve glucose homeostasis.
Impaired function of peptide factors that regulate energy homeostasis contributes to a deterioration of glucose and lipid homeostasis associated with obesity, with leptin and adiponectin secreted by adipocytes being well-investigated examples (Flier, 2004; Scherer, 2006). Our results suggest that deregulation of adropin production may also be a factor linking obesity with steatosis and insulin resistance. The effects of adropin replacement therapy observed in DIO mice are consistent with a significant role for this peptide in maintaining glucose homeostasis. However, confirmation that a decline in adropin is a major factor in the deterioration of glucose and lipid metabolism in the obese state will require investigating adropin deficiency due to targeted gene deletion.
The observation that hepatic Enho expression correlates with dietary fat intake and regulation by LXR agonists suggests a role in adapting substrate metabolism to macronutrient intake. The efficient coordination of substrate oxidation with fat and carbohydrate content of the diet is an important feature of energy homeostasis. Individuals with a family history of type 2 diabetes exhibit impaired metabolic flexibility and do not efficiently match fat oxidation with consumption (Heilbronn et al., 2007). Impaired coordination of substrate oxidation with dietary macronutrient content is also a factor in obesity of mice and humans with MC4R mutations (Butler, 2006; Nogueiras et al., 2007). The observation that hepatic Enho expression is regulated by dietary fats and LXRα suggests a role in adapting substrate metabolism to macronutrient intake. If the hypothesis that adropin acts as a metabolic signal to adapt substrate metabolism with fat intake is correct, then a decline in adropin synthesis with obesity may be a factor in the impaired ability to match lipid metabolism with dietary fat intake, promoting steatosis, dyslipidemia, and glucose intolerance.
In mice, transition from low-to high-fat diets is associated with a rapid shift in substrate metabolism to increase fat oxidation and reduce lipogenesis (Butler, 2006; Sampath and Ntambi, 2005). This response involves nuclear receptors that act as lipid and glucose sensors and that subsequently act to regulate the expression of clusters of genes expressing key metabolic enzymes (Kalaany and Mangelsdorf, 2006; Sampath and Ntambi, 2005; Uyeda and Repa, 2006). The regulation of Enho transcription by macronutrients observed in the current study could involve intracellular sensors of carbohydrates and fatty acids. In the current study, we observed that LXRα agonists rapidly reduced the expression of Enho mRNA in cultured HepG2 cells and in liver of live mice. LXRs stimulate lipogenesis in liver and promote storage of energy as TG in adipose tissue (Kalaany and Mangelsdorf, 2006). As adropin therapy is associated with suppression of lipogenic gene expression and reduces adiposity, suppression of adropin synthesis by LXRα may facilitate the effects of LXRs to promote synthesis and storage of TG.
Both LXRα and LXRβ have recently been suggested to act as glucose sensors (Mitro et al., 2007), although these data are controversial (Denechaud et al., 2008). We have shown that the stimulation of LXRα suppresses hepatic Enho expression. Therefore, stimulation of LXR by diets with a high carbohydrate intake may provide a hypothesis for explaining the regulation of liver Enho expression by dietary macronutrients. However, at this time, the mechanisms by which the macronutrient content of the diet regulates liver Enho expression remain unclear. A simple model involving stimulation of liver Enho transcription by the other known regulators of lipid metabolism in liver such as Ppara, Srebp1, and ChREBP is unlikely. The activity of these transcription factors is stimulated when Enho expression is low, such as after fasting (Ppara) (Reitman, 2007) and consumption of high-carbohydrate, low-fat diets (Srebp1, ChREBP) (Horton et al., 2002; Uyeda and Repa, 2006).
Several observations suggest that adropin may have a role in the regulation of lipogenesis. Adropin therapy rapidly reduces the expression of genes involved in lipogenesis in liver. Transgenic mice overexpressing adropin (Adr-Tg) mice exhibit lower expression of genes involved in lipogenesis in liver and adipose tissue. A lower RQ observed in female Adr-Tg mice also suggests altered substrate metabolism. While these effects on liver gene expression could be due to weight loss and improvements in insulin sensitivity, suppression of liver lipogenic genes was also observed prior to significant effects on food intake or weight loss. In the fed state, liver Enho expression also correlates negatively with lipogenic activity, with lower expression observed during refeeding with high-carbohydrate diets and increased expression on diets with high fat and low carbohydrate content. However, liver Enho expression also declines with fasting, which is a state of minimal lipogenesis. In this situation, the loss of an inhibitory signal may not dominate other endocrine and neural inputs that regulate carbohydrate and lipid metabolism (Reitman, 2007). However, it is also possible that a decline in adropin production may provide a regulatory signal involved in the homeostatic metabolic adaptation to fasting, analogous to the instigation of metabolic adaptations to fasting that are associated with a decline in serum leptin (Morton et al., 2006).
Adr-Tg mice exhibited normal leptin and expression of Ucp1 in BAT, suggesting that adropin does not affect energy metabolism through altered leptin sensitivity or BAT thermogenesis. It is likely that adropin regulates energy homeostasis through other mechanisms that include suppressing lipogenesis. Female Adr-Tg mice also exhibited a significant increase in physical activity, which may be a mechanism for regulating whole body substrate metabolism. While activity of male Adr-Tg mice was not significantly different, there was evidence for a modest increase in energy expenditure when adjusted for body mass. While speculative, the increase in serum adiponectin associated with adropin treatment may also stimulate oxidative metabolism (Scherer, 2006).
While adropin(34-76) treatment reduced food intake in DIO B6 mice, this response was only evident after 3 days of treatment. This may suggest an indirect mechanism secondary to improvements in lipid metabolism and insulin sensitivity. However, Enho is expressed in the brain, including the area postrema and dorsal vagal complex, which are involved in regulating ingestive behaviors. It is also worth noting that Enho mRNA is expressed in other areas of the central nervous system involved in relaying sensory information (brainstem), regulating complex behaviors (thalamus), temperature regulation (periaqueductal gray), and circadian rhythms (lateral geniculate nucleus and medial habenula). Whether adropin functions as a neuropeptide in the central nervous system to affect homeostatic regulation of metabolism remains to be investigated.
In summary, adropin is a newly discovered secreted peptide that is involved in energy homeostasis and lipid metabolism. Expression of Enho and secretion of adropin by the liver is proposed to be involved in peripheral lipid homeostasis in response to variable macronutrient consumption. This could involve both paracrine effects on hepatocyes or actions as an endocrine signal of macronutrient consumption and energy status. Adropin may form the basis for the development of new therapeutic targets for treating metabolic disorders associated with obesity.
EXPERIMENTAL PROCEDURES
Animal procedures complied with NIH guidelines and were approved by the Pennington Biomedical Research Center or Columbia University Institutional Animal Care and Use Committee.
Animals
Melanocortin receptor knockout mice have been previously described (Sutton et al., 2006). Lean and DIO B6 (60% kJ HFD from 6 through 26 weeks), Lepob/Lepob, and Ay/a mice were purchased from the Jackson Laboratory (Bar Harbor, MN). Mice were housed in a 12 hr light/dark period (lights on: 0600-1800 hr). Low-fat (10% kJ/fat, D12450B) or high-fat (60% kJ/fat, D12492) diets were purchased from Research Diets, Inc. (New Brunswick, NJ). For CR, 8-week-old Ay/a mice were fed 70% of their prior measured daily ad libitum intake for 16 weeks. FM and FFM were estimated using nuclear magnetic resonance (Bruker Mice Minispec NMR Analyzer; Bruker Optics, Inc.; Billerica, MA) and validated against dual X-ray absorptiometry and standard chemical composition analysis (Tinsley et al., 2004). For i.v. administration of the LXRα agonist, GW3965 (10 mg/kg) was administered into the tail vein of B6 mice (aged 12 weeks, BW of 25 g); liver was collected for RNA extraction 4 hr after the injection.
Expression Vectors
The adropin ORF was amplified from mouse liver cDNA using primers with Not1 (5′-ggggcggccgcaccatgggggcagccatctcccaa-3′) and Xho1 (5′-gggctcgagggccagagcccttcagggctgcag-3′) restriction enzyme sites and ligated into p3 × FLAG-CMV-14 Expression Vector (Sigma-Aldrich; St. Louis, MO). Constructs expressing the native form, or with a C-terminal FLAG epitope, were verified by sequencing. Expression of the native and FLAG-tagged protein was confirmed in transiently transfected HEK293 cells (Lipofectamine 2000; Invitrogen) using both an anti-FLAG antibody and a polyclonal antibody raised against adropin (Sigma-Genosys; The Woodlands, TX). The DNA encoding the native or FLAG-tagged forms was ligated into the BstXI and Xba1 sites of pAd5-Blue and recombinant adenovirus generated using previously described methods (Moraes et al., 2001). Viral particles were purified using a Vivapure AdenoPACK 100 (Vivascience AG; Hannover, Germany). Mice were administered 100 μl of 5 × 108-9 pfu of virus into the tail vein.
Adropin-Transgenic Mice
The adropin ORF was subcloned into a human β-actin promoter vector. After excision from vector sequences, transgenic animals were generated by pronuclear injection into B6 fertilized eggs.
Energy Expenditure
Physical activity and energy expenditure were measured using a 16-chamber Comprehensive Laboratory Animal Monitoring System (Columbus Instruments; Columbus, OH) (Butler, 2006). Spontaneous physical activity was measured using an OPTO-M3 sensor system. REE was calculated using measurement made when X movements averaged < 0.5 beam breaks/hr.
Adropin(34-76) Treatment Study
Synthetic adropin(34-76) was provided by Biomeasure, Inc./IPSEN (Milford, MA). Peptide was synthesized using standard Fluorenylmethyloxycarbonyl (Fmoc) chemistry on PEG resin. In each deprotection step, the Fmoc group was removed by treatment with 20% piperidine in N,N-dimethylformamide (DMF) for 30 min. The resin was washed with DMF. In each coupling step, five equivalents of Fmoc amino acid, five equivalents of 2-(1-H-benzotriazole-1-yl)-1,1,2,3-tetramethyluronium hexafluorophosphate (HBTU), and one equivalent of diisopropylethylamine (DIEA) in DMF were used. The coupling time was ∼1 hr. At the end of the peptide chain assembly, the peptide was cleaved off from the resin using trifluoroacetic acid (TFA) containing 5% phenol, 2% ethanedithiol, and 2% anisol. The crude linear peptide was precipitated with ether, cyclized in aqueous solution containing 10% dimethyl sulfoxide (DMSO, pH 8.5), and purified by reverse-phase high-pressure liquid chromatography (HPLC). The final product was analyzed by amino acid analysis, analytical HPLC, and mass spectrometry.
For treatment, adropin(34-76) suspended in 0.9% saline with 0.1% bovine serum albumin was administered by i.p. injection at 0800 and 1800 hr. The doses used were based on prior experience testing novel secreted peptides. DIO B6 mice were acclimated for 10 days to stainless steel wire-bottomed cages; food intake was measured by weighing food in the hopper and correcting for spillage. After the final injection, mice were fasted overnight and euthanized at 1100 hr. Trunk blood was collected, and tissue samples (liver and retroperitoneal WAT) were frozen in liquid N2. A portion of the liver from three animals per group was fixed with formalin and processed by Pathology Laboratory, LSU School of Veterinary Medicine.
Microarray Analysis
Microarrays were performed by the Genomics Core Facility of the Pennington Biomedical Research Center using total RNA isolated from lean and obese Mc3rKO and age-matched littermates as described previously (Koza et al., 2006). Validation of gene expression involved was performed in either 96- or 384-well plates with TaqMan Universal PCR Master Mix or SYBR Green PCR Master Mix (Applied Biosystems; Foster City, CA) and an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems), as previously described (Koza et al., 2006).
Quantitative RT-PCR and Northern Blot
Total RNA was extracted using either TRI reagent or TRIzol (Invitrogen; Carlsbad, CA) and reverse transcribed using Superscript III Reverse Transcription System (Invitrogen; Carlsbad, CA). Analysis of gene expression with cyclophilin B as reference was performed in 384-well plates, using TaqMan Universal PCR Master Mix or SYBR Green PCR Master Mix and an ABI PRISM 7900HT Sequence Detection System. Sequences of oligonucleotide primers and probe (Integrated DNA Technologies; Coralville, IA) for Enho were designed using Primer Express 2.0 (Applied Biosystems): forward 5′-atggcctcgtaggcttcttg-3′, reverse 5′-ggcaggcccagcagaga-3′, probe 5′-FAM-tgctactgctctgggtc-BHQ-3′. Sequences of primer-probe combinations for other genes were designed and validated as reported previously (Sutton et al., 2006).
For northern analysis, a 210 bp DNA probe corresponding to Enho coding sequence was labeled with 32P-dATP (>3000 Ci/mmol) using Prime-It II Random Primer Labeling Kit (Stratagene; La Jolla, CA). (1 × 106)-(1 × 107) cpm of denatured probe was hybridized with a Multiple Tissue Northern Human RNA Blot (BD Biosciences Clontech; Palo Alto, CA). For analysis of gene expression in mice, total RNA was extracted from tissues or differentiated 3T3-L1 adipocytes as previously described (Floyd and Stephens, 2002). Blots were exposed to a phosphor screen and images captured using a Molecular Dynamics Phosphorimager (Sunnyvale, CA).
Generation of Enho cRNA Probe and In Situ Hybridization Histochemistry
cDNA obtained from mouse brain was used as a template to amplify three sequences by PCR: a 401 bp sequence corresponding to nt 98-498 of the Enho transcript (primers: 5′-CTGAATTCGACAGGACAGCCACCTTGGAT-3′ and 5′-AGAAGCTTCGATTCCACGCTCTGACTCAG-3′); a 524 bp sequence corresponding to nt 38-561 (primers: 5′-CTGAATTCTCAACTCAGGCCCAGGACTG-3′ and 5′-AGAAGCTTAATCTAGGCTGCTGGGTCCA-3′); and a 234 bp sequence corresponding to nt 205-438, which contains coding sequence only (primers: 5′-CTGAATTCATGGGGGCAGCCATCTCCCA-3′ and 5′-AGAAGCTTTCAGGGCTGCAGCAGGTAGC-3′). The respective amplification products were ligated into the pcDNA3 vector (Invitrogen; Paisley, UK); inserts were verified by DNA sequencing. Plasmids linearized by digestion with EcoRI were used to generate antisense 35S-labeled cRNA using T7 RNA polymerase (Ambion). Sense probes were also generated by linearizing plasmids with HindIII and using SP6 RNA polymerase (Ambion).
Eight-week-old male B6 mice were anesthetized with pentobarbital (50 mg/kg i.p.) and then perfused transcardially with diethylpyrocarbonate (DEPC)-treated phosphate buffered saline (PBS) followed by 10% neutral buffered formalin. After postfixing in the same fixative for 4 hr at 4°C, brains were then immersed in 20% sucrose in DEPC-treated PBS at 4°C overnight and cut coronally at 25 μm on a freezing microtome. Tissue sections were mounted onto SuperFrost slides (Fisher Scientific; Loughborough, UK) and air dried. Sections were fixed in 4% formaldehyde in DEPC-treated PBS for 20 min at 4°C, dehydrated in ascending concentrations of ethanol, cleared in xylene for 15 min, rehydrated in descending concentrations of ethanol, and placed in prewarmed sodium citrate buffer (95-100°C, pH 6.0). Slides were placed in an LG Intellowave commercial microwave oven (Slough, UK) for 10 min at 95-100°C, dehydrated in ascending concentrations of ethanol, and air dried.
In situ hybridization was performed as previously described (Heisler et al., 2006). Briefly, 35S-labeled Enho cRNA probes diluted to 106 cpm/ml in hybridization solution and coverslips were applied to mounted sections, which were incubated for 16 hr at 57°C. Sections were incubated in 0.002% RNase A (QIAGEN; Crawley, UK) for 30 min and then sequentially washed in decreasing concentrations of SSC. Sections were dehydrated in graded ethanol containing 0.3 M NH4OAc followed by 100% ethanol. Slides were air dried and placed in X-ray film cassettes with BMR-2 film (Kodak; Rochester, NY) for 3 days.
Cell Culture
Culture and treatment of HepG2 cells with LXR and FXR agonists, and inhibition of LXRα using siRNA, were described previously (Stayrook et al., 2008; Wang et al., 2008). LXR and FXR agonists are from Sigma-Aldrich (St. Louis, MO).
Western Blot
Lysates or conditioned media were homogenized on ice in lysis buffer (Cell Signaling; Beverly, MA) and centrifuged, and aliquots diluted with 2× electrophoresis buffer (50 mM Tris HCl, pH 6.7, 4% w/v glycerol, 4% SDS, 1% 2-mercptoethanol, and 0.02 mg/ml bromophenol blue). Ten micrograms protein was separated on a 4%-20% SDS-polyacrylamide gradient gel (BioRad; Hercules, CA), transferred to Immobilon-P polyvinylidene diflouride membranes in Towbin-transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol, and 0.01% SDS), and then incubated with antibodies. Membranes were washed five times in PBST and incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. Membranes were then washed as above and the antigen-antibody-peroxidase complex detected by enhanced chemiluminescence (Amersham Biosciences; Piscataway, NJ).
A polyclonal antibody was generated using 14 amino acid antigen corresponding to amino acids 37-50 of adropin, which was used to elicit an antisera response in two New Zealand white rabbits (Sigma-Genosys). The third production bleed from one rabbit was affinity purified (Sigma-Genosys) and used for western blot analysis.
Serum Measurements, Glucose Tolerance Tests, and Determination of Hepatic Lipid Content
Blood glucose was measured using a Glucometer Elite XL (Bayer Corporation; Elkhart, IN). Plasma lipids were measured on a Beckman Synchron CX7 for terminal experiments or commercially available kits (Wako Diagnostics; Richmond, VA). For glucose tolerance tests, mice were fasted overnight and blood samples taken at time 0 (baseline) or 15-60 min after an i.p. injection of glucose (1 mg/kg). Hepatic TG content was measured as previously described (Sutton et al., 2006). Serum insulin was measured using a commercially available kit (Crystal Chem; Downer’s Grove, IL). HOMA-IR was calculated as follows: (fasting insulin [μU/ml] × fasting glucose [mmol/l]) / 22.5.
Statistics
All data presented are mean ± SEM. JMP IN 5.1 (SAS Institute, Inc.; Cary, NC) was used for statistical analysis; two or multiple groups of data used either Student’s t test or one-way and two-way ANOVA. Significance was assumed for P values < 0.05.
ACKNOWLEDGMENTS
This work was supported by grants from the Pennington Biomedical Research Foundation and the American Diabetes Association (1-04-JF09) and by a Sponsored Research Agreement with Biomeasure, Inc./IPSEN; Milford, MA to A.A.B. A.A.B. and R.L.M. are supported in part by CNRU Center Grant #1P30 DK072476, entitled “Nutritional Programming: Environmental and Molecular Interactions” and sponsored by the NIDDK. The UAB Transgenic Mouse Facility is supported by NIH grants P30 CA13148 and P30 AR48311. L.K.H. is supported by NIDDK R01DK065171 and the Wellcome Trust. H.A.H., J.Z.D., and M.D.C. are employees of Biomeasure, Inc./IPSEN; A.A.B. has been a paid consultant for Biomeasure, Inc.
The authors thank the staff of the PBRC Genomics Core Facility, Diana Albarado, and Emily Meyer for technical assistance. We also thank Drs. Les Kozak, Heike Munzberg, and Christopher Morrison for providing comments during manuscript preparation.
REFERENCES
- Asilmaz E, Cohen P, Miyazaki M, Dobrzyn P, Ueki K, Fayzikhodjaeva G, Soukas AA, Kahn CR, Ntambi JM, Socci ND, et al. Site and mechanism of leptin action in a rodent form of congenital lipodystrophy. J. Clin. Invest. 2004;113:414–424. doi: 10.1172/JCI19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddinger SB, Kahn CR. From mice to men: insights into the insulin resistance syndromes. Annu. Rev. Physiol. 2006;68:123–158. doi: 10.1146/annurev.physiol.68.040104.124723. [DOI] [PubMed] [Google Scholar]
- Butler AA. The melanocortin system and energy balance. Peptides. 2006;27:281–290. doi: 10.1016/j.peptides.2005.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cowie CC, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, Saydah SH, Williams DE, Geiss LS, Gregg EW. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999-2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- Denechaud PD, Bossard P, Lobaccaro JM, Millatt L, Staels B, Girard J, Postic C. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J. Clin. Invest. 2008;118:956–964. doi: 10.1172/JCI34314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Flier JS. Obesity wars. Molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- Floyd ZE, Stephens JM. Interferon-gamma-mediated activation and ubiquitin-proteasome-dependent degradation of PPARgamma in adipocytes. J. Biol. Chem. 2002;277:4062–4068. doi: 10.1074/jbc.M108473200. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Heilbronn LK, Gregersen S, Shirkhedkar D, Hu D, Campbell LV. Impaired fat oxidation after a single high-fat meal in insulin-sensitive nondiabetic individuals with a family history of type 2 diabetes. Diabetes. 2007;56:2046–2053. doi: 10.2337/db06-1687. [DOI] [PubMed] [Google Scholar]
- Heisler LK, Jobst EE, Sutton GM, Zhou L, Borok E, Thornton-Jones Z, Liu HY, Zigman JM, Balthasar N, Kishi T, et al. Serotonin reciprocally regulates melanocortin neurons to modulate food intake. Neuron. 2006;51:239–249. doi: 10.1016/j.neuron.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hill JO. Understanding and addressing the epidemic of obesity: an energy balance perspective. Endocr. Rev. 2006;27:750–761. doi: 10.1210/er.2006-0032. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, Skaf J, Kozak LP. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Joseph SB, Chen M, Castrillo A, Repa J, Wilpitz D, Mangelsdorf D, Tontonoz P. The phospholipid transfer protein gene is a liver X receptor target expressed by macrophages in atherosclerotic lesions. Mol. Cell. Biol. 2003;23:2182–2191. doi: 10.1128/MCB.23.6.2182-2191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoith D, Yakar S. Mechanisms of disease: metabolic effects of growth hormone and insulin-like growth factor 1. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:302–310. doi: 10.1038/ncpendmet0427. [DOI] [PubMed] [Google Scholar]
- Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V, Kreusch A, Saez E. The nuclear receptor LXR is a glucose sensor. Nature. 2007;445:219–223. doi: 10.1038/nature05449. [DOI] [PubMed] [Google Scholar]
- Moraes MP, Mayr GA, Grubman MJ. pAd5-Blue: direct ligation system for engineering recombinant adenovirus constructs. Biotechniques. 2001;31:1050, 1052, 1054–1056. doi: 10.2144/01315st05. [DOI] [PubMed] [Google Scholar]
- Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55(Suppl 2):S9–S15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- Nogueiras R, Wiedmer P, Perez-Tilve D, Veyrat-Durebex C, Keogh JM, Sutton GM, Pfluger PT, Castaneda TR, Neschen S, Hofmann SM, et al. The central melanocortin system directly controls peripheral lipid metabolism. J. Clin. Invest. 2007;117:3475–3488. doi: 10.1172/JCI31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obici S, Feng Z, Tan J, Liu L, Karkanias G, Rossetti L. Central melanocortin receptors regulate insulin action. J. Clin. Invest. 2001;108:1079–1085. doi: 10.1172/JCI12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlemuter G, Bigorgne A, Cassard-Doulcier AM, Naveau S. Nonalcoholic fatty liver disease: from pathogenesis to patient care. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:458–469. doi: 10.1038/ncpendmet0505. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Savage DB, Bilz S, Solomon G, Yonemitsu S, Cline GW, Befroy D, Zemany L, Kahn BB, et al. The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proc. Natl. Acad. Sci. USA. 2007;104:12587–12594. doi: 10.1073/pnas.0705408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ML. FGF21: a missing link in the biology of fasting. Cell Metab. 2007;5:405–407. doi: 10.1016/j.cmet.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 2005;25:317–340. doi: 10.1146/annurev.nutr.25.051804.101917. [DOI] [PubMed] [Google Scholar]
- Scherer PE. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes. 2006;55:1537–1545. doi: 10.2337/db06-0263. [DOI] [PubMed] [Google Scholar]
- Sharma AM, Staels B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue-understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- Stayrook KR, Rogers PM, Savkur RS, Wang Y, Su C, Varga G, Bu X, Wei T, Nagpal S, Liu XS, et al. Regulation of human 3 alphahydroxysteroid dehydrogenase (AKR1C4) expression by the liver X receptor alpha. Mol. Pharmacol. 2008;73:607–612. doi: 10.1124/mol.107.039099. [DOI] [PubMed] [Google Scholar]
- Sutton GM, Trevaskis JL, Hulver MW, McMillan RP, Markward NJ, Babin MJ, Meyer EA, Butler AA. Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or -4 receptors. Endocrinology. 2006;147:2183–2196. doi: 10.1210/en.2005-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley FC, Taicher GZ, Heiman ML. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obes. Res. 2004;12:150–160. doi: 10.1038/oby.2004.20. [DOI] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rogers PM, Su C, Varga G, Stayrook KR, Burris TP. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J. Biol. Chem. 2008;283:26332–26339. doi: 10.1074/jbc.M804808200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sutton GM, Rochford JJ, Semple RK, Lam DD, Oksanen LJ, Thornton-Jones ZD, Clifton PG, Yueh CY, Evans ML, et al. Serotonin 2C receptor agonists improve type 2 diabetes via melanocortin-4 receptor signaling pathways. Cell Metab. 2007;6:398–405. doi: 10.1016/j.cmet.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]