Abstract
We designed phosphorothioate-modified DNA probes linked to superparamagnetic iron oxide nanoparticles (SPION) for in vivo magnetic resonance imaging (MRI) of fosB and ΔfosB mRNA after amphetamine (AMPH) exposure in mice. Specificity of both the fosB and ΔfosB probes was verified by in vitro reverse transcriptase-PCR amplification to a single fragment of total cDNA obtained from acutely AMPH-exposed mouse brains. We confirmed time-dependent uptake and retention profiles of both probes in neurons of GAD67-green fluorescent protein knock-in mice. MRI signal of SPION-labeled fosB probe delivered via intracerebroventricular route was elevated in both acutely and chronically AMPH-exposed mice; the signal was suppressed by dopaminergic receptor antagonist pretreatment. SPION-labeled ΔfosB probe signal elevation occurred only in chronically AMPH-exposed mice. The in vivo target specificity of these probes permits reliable MRI visualization of AMPH-induced differential elevations of fosB and ΔfosB mRNA in living brains.
Introduction
Neuroscience research on drug addiction has focused primarily on the morphological and molecular adaptations of neurons in response to chronic drug exposure. Results from animal models of drug abuse have suggested a correlation between altered gene expression and drug exposure that leads to addiction. Specifically, the expression of inducible transcription factors including the Fos family (cFos, FosB, Fra-1, and Fra-2) occurs rapidly but transiently after acute exposure to drugs, resolving within 4–12 h (Hope et al., 1992, 1994; Nguyen et al., 1992; Hiroi et al., 1997; Hope, 1998; Thiriet et al., 2001; Uslaner et al., 2001; Novoselov et al., 2002; Ostrander et al., 2003; Parelkar and Wang, 2004; Okabe et al., 2005). The Fos and Jun proteins combine to form a heteroduplex dimer of activator protein-1 (AP-1), which binds alone or in concert with other activator proteins to the promoter regions of ∼1000 genes to regulate their transcription. Although drugs of abuse are chemically diverse, their actions in the brain converge to produce some common effects (Nestler, 2001), and can activate similar brain regions in the mesolimbic dopaminergic neurons, including the nucleus accumbens, striatum, and medial prefrontal cortex in the forebrain. Current gene assays such as in situ hybridization and in situ reverse transcriptase-PCR used to investigate gene transcript modification associated with chronic exposure rely on postmortem brain samples or biopsy. A technique such we have developed for live brain real-time assessment of transiently altered neural mRNA levels after stimulation may provide first-hand information on neural activation.
The pivotal role of a long-lasting fosB protein (ΔFosB), which is a truncated form of FosB, is well established in sensitivity to drug addiction (Nestler, 2001), food intake (Teegarden et al., 2008), and stress (Perrotti et al., 2004). Although immunohistochemistry can robustly differentiate the expression of these two protein variants, there is currently no direct gene assay technique that is established to differentiate the induction profiles of fosB and ΔfosB mRNA in vivo (Mandelzys et al., 1997). To visualize in live brains the differential fosB gene expression profile after brain stimulation, we developed MR probes that link T2* contrast agent [superparamagnetic iron oxide nanoparticles (SPION)] with a oligodeoxynucleotide (ODN) sequence complementary to fosB or ΔfosB mRNA to visualize endogenous mRNA targets via in vivo hybridization (Liu et al., 2007a,b,c, 2008). The presence of this SPION-sODN probe in brain cells results in localized signal reduction in T2*-weighted MR images, in which that rate of signal reduction (R2*) reflects the regional iron concentration at different stages of amphetamine (AMPH) exposure in live mouse brains (Boxerman et al., 1995).
Materials and Methods
Probe design.
SPION (a T2 susceptibility contrast agent) was homemade (Liu et al., 2007a) or purchased commercially (Molday Ion, BioPhysics Assay Laboratory). We generated two phosphorothioate-modified ODN (sODN): sODN-fosB [5′-CCTTAG CGGATGTTGACCCTGG-3′, complementary to sequence No. 1925–1946 of fosB mRNA of the mouse (mmFosB, Accession No. X14897)] and sODN-ΔfosB (5′-ACTTGAACTTCACTCGGCCAGCGG-3′ on 1901–1924 of ΔfosB mRNA). The sODN was labeled with Rhodamine (Rhd) or fluorescein isothiocyanate (FITC) on the 5′end and biotinylation on the 3′-end. Biotinylated sODN was conjugated to SPION-NeutrAvidin (NA) via Avidin-biotin linkage (Liu et al., 2007a).
In vitro PCR.
Combination of sODN-fosB to a common upstream primer in sense sequence (USP, 5′-GATCGCCGAGCTGCAAAAAG-3′) was used in the PCR (Liu et al., 2007b). The product was separated by agarose gel (1%) electrophoresis. The cDNA (a generous gift from Dr. Y. Nakabeppu, Kyushu University, Maidashi Higashi-ku, Fukuoka, Japan) was a fragment cut by BamH1 and PstI. Total striatal cDNA was reverse transcribed from d(T)18 primed mRNA (Cui et al., 1999). Gel image was acquired by FluorChem Q System (Alpha Innotech).
Amphetamine treatments.
All procedures and animal care practices adhered strictly to Association for Assessment and Accreditation of Laboratory Animal Care, Society for Neuroscience, and institutional guidelines for experimental animal health, safety, and comfort. For acute AMPH (A1), amphetamine-naive, male C57black6 mice (24 ± 3 g, Taconic Farm) received a single dose of AMPH (4 mg/kg, i.p.). For chronic AMPH (A7/W/A or A7/W/S), age-matched male mice received intraperitoneal injection of AMPH (4 mg/kg) or saline (vehicle, 10 ml/kg) in their home cages every other day for a total of seven injections (Paulson et al., 1991; Xue et al., 1996; Johnston et al., 2004; Giordano et al., 2006). The injection period was followed by a 2 week period of no drug (abstinence). AMPH-induced locomotion activity was measured (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Delivery of contrast conjugates for magnetic resonance imaging.
On the day of magnetic resonance imaging (MRI) acquisition, we anesthetized the mice with pure O2 plus 2% halothane at a flow rate of 800 ml/min for probe delivery. MRI contrast probes (SPION-fosB or SPION-ΔfosB, at 120 pmol SPION-sODN per kg) were delivered using an intracerebroventricular (ICV) delivery route (Liu et al., 2007a). The animals were allowed to wake up in their home cages to minimize stress and anesthesia-induced gene expression and therefore facilitate probe distribution, uptake, retention, and exclusion under physiological conditions. AMPH (4 mg/kg, i.p.) or saline was given (intraperitoneally) 3 h later; again the animals were returned to the same cage. MRI was conducted 4 h after drug administration (Liu et al., 2007a). Three hours after ICV delivery using BBB bypass has been predetermined as the minimal time required for sODN-RNA hybridization in rodent brains (Liu et al., 1994).
Delivery of sODN-fosB probe for uptake and retention.
Rhd-sODN was mixed with lipofectin (Invitrogen) at 1 nmol/μg lipofectin and was delivered using an ICV delivery route (8 nmol/kg) to a total of six mice [four C56black6 and two glutamate decarboxylase (GAD67)-green fluorescent protein (GFP) knock-in mice which express GFP in gamma-aminobutyric acid (GABA)-ergic neurons] (Tamamaki et al., 2003).
In vivo MRI acquisition.
All in vivo MRI acquisitions were performed in a 9.4 tesla MRI scanner (Bruker-Avance System). Animals were again anesthetized with 2% halothane in oxygen (800 ml/min flow rate) during scanning. Protocols for MRI acquisition, subtraction R2* maps construction, and data analysis are described (Liu et al., 2009). To obtain maximal homogeneity field strength inside the gradient for signal sensitivity and reproducibility, automatic or manual shimming was performed before each MRI scan series.
Postmortem tissue preparation.
For probe uptake study (Cui et al., 1999; Liu et al., 2007a, 2008), mice were decapitated and brains were quickly removed and frozen in OTC place on the top of liquid nitrogen. Brains were sectioned in 20 or 50 μm thickness and pretreated with 4% paraformaldehyde on glass slide for 15 min followed by gentile de-ionized H2O rinse before cover slides were placed. These steps were important to maintain probe integrity for 20 μm tissue sections. We were not able detect sODN-fosB signal if the sample were treated with ethanol or organic solvents, or from retrograde perfusion preparation with paraformaldehyde. For probe localization in animals treated with amphetamine, brain were sectioned in 50 μm using a Vibratome and counterstained with counter-staining with Cy3-IgG against neuronal nuclear (NeuN) antigen (Millipore Bioscience Research Reagents, clone A60, 1:200 dilution). Probe uptake was acquired using with a mercury light source and filters for fluorescein (495 nm broad spectrum wavelengths), Cy3, or rhodamine (560 nm broad spectrum wavelengths), and an Olympus Digital Imaging System with Hamamatsu ORCA High Resolution Monochrome Cooled CCD Camera, Extended Range Digital Camera (Deep Cooled), IEEE 1394 Interface, Microsuite 5 Biological Suite Software Ki, 25 mm Smartshutter; digitized data were stored in computer and external hard drive.
Histological and immunohistochemical staining.
Elevation of ΔFosB antigen was determined using alkaline phosphatase-labeled rabbit polyclonal IgG antibodies against FosB (H-75) (Santa Cruz Biotech) and enhanced with 3,3′-diaminobenzidine tetrahydrochloride (Sigma), as described previously (Liu et al., 2007a); this antibody detects one 35KD epitope of Fos B fusion protein but not the 40–50KD FosB (Nestler et al., 2001).
Statistical analysis (MRI).
The region of interest (ROI) analysis included averaging R2* values from the brain regions and computing the mean and SEM from the averaged values in each group of animals for statistical comparisons. Once we obtained the first set of MRI data, we calculated the number of animals required in each group to achieve 95% power for a p value of 0.01. We made these calculations using an in-house software package (Liu et al., 2007a), and compared the statistical significance of these values using a t test (one tail, type II, or equal variant, GraphPad Prism IV, GraphPad Software). A p value of <0.05 was statistically significant.
Results
Probe design, binding, and specificity
The relative location of sODN-fosB or sODN-ΔfosB in their mRNA is shown in Figure 1, A or B. Initial assessment in the binding specificity of fosB probe to the fosB cDNA showed that the sODN-fosB probe together with a USP-amplified 146 bp fragment using PCR from the fosB cDNA; this probe, however, did not amplify any fragment from the ΔfosB cDNA, a cDNA clone that shares homology with most of fosB cDNA (Fig. 1C). We further evaluated the probe binding stringency against total cDNA obtained from the striatum of C57black6 mice, as the probe should exhibit selectivity against thousands of gene transcripts. We would expect to observe amplification of a smear or several fragments if it was not specific and bound to several cDNA of different sizes. Figure 1D demonstrates that these primers did in fact amplify only one fragment from the total striatal cDNA (n = 4). The same was observed with sODN-ΔfosB and the USP in reverse-transcription PCR, but in this case one 123 bp fragment was amplified, as predicted from the mRNA sequence in Figure 1B. The result supported specific antisense interaction between sODN-fosB (the ligand) and its mRNA target (the receptor). The specificity of these two probes is shown by an ability to distinguish fosB and ΔfosB mRNA in vivo, as described below.
Figure 1.
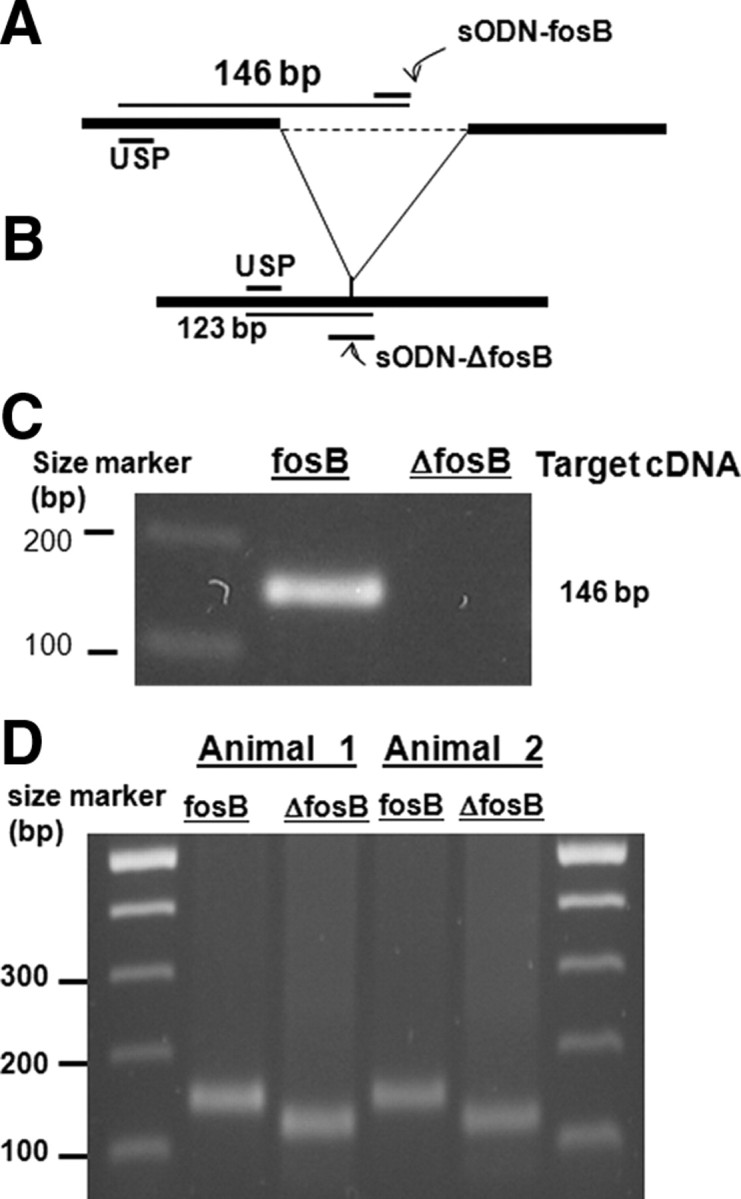
Probe design and specificity. A shows a relative location of sODN-fosB within the splice site of the fosB mRNA. B shows the location of sODN-ΔfosB flanking the spliced site of ΔfosB mRNA. C shows a combination of sODN-fosB to a common USP ODN in sense sequence allows amplification of 146 bp from fosB cDNA, but not ΔfosB cDNA by PCR. D shows one cDNA fragment was amplified from total cDNA of mouse striatum (C57black6 mice) when the sODN-fosB or sODN-ΔfosB was used (two of four mice tested were shown) (Cui et al., 1999). A combination of sODN-ΔfosB and USP-ODN allows amplification of 123-bp ΔfosB cDNA. Because of partial homology in this 123 bp fragment to FosB cDNA, we did not perform diagnostic sequencing, but the specificity of sODN-ΔfosB was tested by in vivo hybridization and MRI (see Fig. 3D).
Neuronal uptake of sODN after ICV delivery
We next examined the uptake and retention dynamics of sODN probe in vivo after ICV delivery of rhodamine (Rhd)-labeled sODN-fosB (8 nmol/kg) (Fig. 2A). GAD67-GFP knock-in mice expressing GFP in GABAergic neurons were used for this study. Figure 2B shows that Rhd-sODN-fosB (red) uptake occurs as an apparent wave propagating away from the lateral ventricles into the parenchyma. Under higher magnification, we found that the Rhd-sODN-fosB signal was distributed homogenously within GFP-positive, presumptive GABAnergic neurons (green) as well as GFP-negative cells (Fig. 2C, closed arrows) (n = 4), or as punctuate at peri-nuclear location (Fig. 2C, asterisk). The homogeneous labeling was seen in cells immediately adjacent to the lateral ventricles while the punctate formations were seen in cells away from the lateral ventricles.
Figure 2.
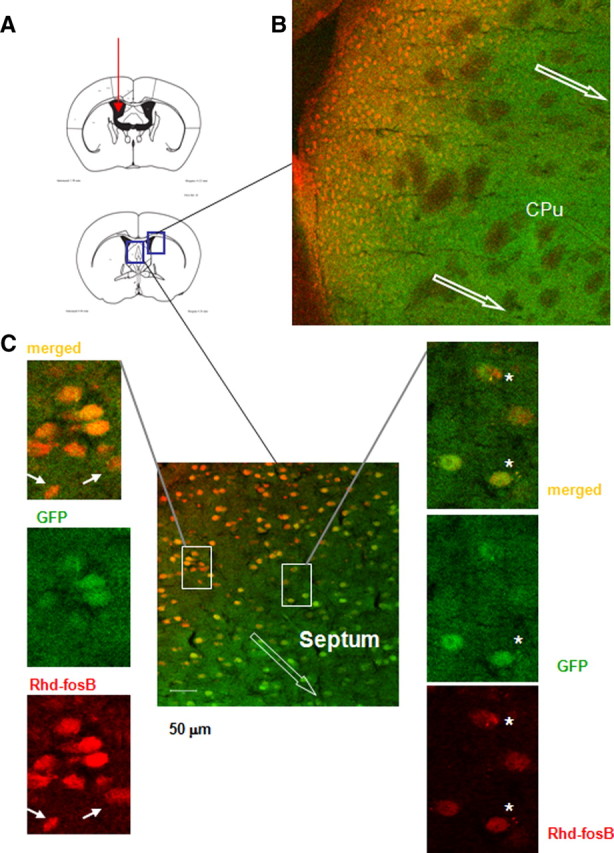
Delivery and uptake of rhodamine-sODN-FosB by confocal microscopy to normal resting mice. A shows the location of ICV and the relative location of tissue sample (box) in B. The direction of migration is indicated by open arrows in a migration front. C shows two types of sODN distribution: Rhd-sODN in green GABAnergic neurons can be seen as overlapping of red (Rhd-sODN-FosB) and GFP in neurons (behind the migration line) and Rhd-sODN in punctate formations (asterisk) ahead of the migration front.
Probe dynamics in neural cells after ICV delivery
We delivered the sODN probe labeled with a strong MRI contrast agent (SPION) and acquired whole-brain MRI of anesthetized mice at four time points referenced to probe delivery (SPION-fosB, ICV, 120 pmol/kg). We calculated R2* maps to assess distribution of the SPION-containing probe (Fig. 3A). While there is endogenous but relatively low R2* signals in the pre-ICV map, we observed elevated R2* signal in and around the infusion site (thick open arrow) and bilaterally within the ventricles (dotted arrows) at 30 min. At this early time point, the spread of R2* signal away from the ventricles may be attributed to localized distortion of the magnetic field because of high concentration of iron-containing probes, which did not cause R2* enhancement in the contralateral cortices (thin arrow). Animals were allowed to wake up after MRI in their home cages to recover with minimal disturbance. R2* maps acquired at the 3 h time point in the animals under anesthesia showed widespread enhancement in the striatum, septum, and cortex (thin arrows) of the cortex contralateral to infusion hemispheres, as previously elevated R2* signal in the infusion track and ventricles diminished. To mimic the experimental paradigm to study AMPH-induced fosB gene expression profiles, we injected saline (10 ml/kg, i.p.) to the mice after the MRI scans at this time point. Again, these animals were returned to their home cages and R2* maps were acquired at 7 h, under anesthesia; we observed a decline in these maps similar to the pre-ICV map (Fig. 3A, fourth row). In conclusion, MRI R2* maps acquired at different time points after ICV delivery of SPION-sODN indicated that global availability of the probe distant to the infusion site had been achieved at 3 h (Liu et al., 2007a). For subsequent statistical analysis, we determined the mean R2* values in several brain regions (Fig. 3B) of each animal and calculated group averages. ROIs in the contralateral hemisphere of the brain, covering −2.7 to 2.10 mm to the bregma based on the mouse brain atlas (Paxinos and Franklin, 2001), included the medial prefrontal cortex, nucleus accumbens, caudate putamen, hippocampus, somatorsensory, and motor cortices.
Figure 3.
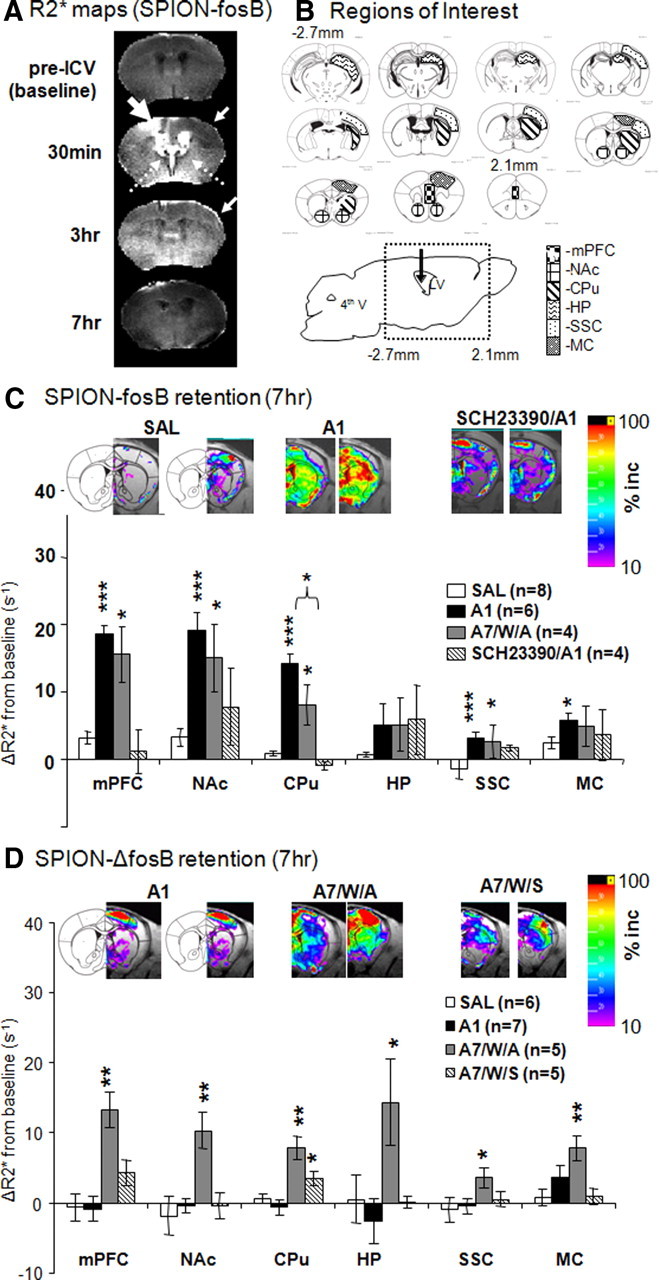
Distribution and retention of SPION-fosB and SPION-ΔfosB probes in vivo. A compares the R2* maps (caudal view) obtained from live animals at four time points referenced to ICV infusion of SPION-fosB probe. A thick arrow points to the site of ICV infusion. B depicts outlines of ROI for statistical analyses (Paxinos and Franklin, 2001). These regions include the mPFC, NAc, and CPU, HP, somatosensory (SSC), and MC. C shows quantitative analyses of fosB expression in ROI from R2* maps of acute and chronic AMPH in this study. Representative SPION-fosB signal maps are presented above the bar graph based on the following equation and signal cutoffs are 10–100%: ΔR2* = (R2* − R2*baseline)/R2*baseline× 100%. We observed significantly reduced fosB mRNA induction in the CPU after AMPH challenge dose after chronic AMPH exposure and a period of no drug (p = 0.03). D shows quantitative analyses of ΔfosB expression in ROI of SAL, acute AMPH exposure (A1), or chronic AMPH with challenge (A7/W/A) and without challenge (A7/W/S). Significant level (compared with the group received saline); *p < 0.05, **p < 0.01, ***p < 0.001.
SPION-fosB signal elevation after AMPH has regional specificity and is suppressed by SCH23390 pretreatment
We examined the feasibility of detecting AMPH-induced fosB gene transcription using SPION-fosB probe and MRI. Four groups of animals were used: (1) no AMPH (saline control or SAL, n = 8), (2) acute AMPH (A1, n = 6), (3) repeated AMPH followed by a period of drug abstinence and a subsequent AMPH challenge dose (4 mg/kg, i.p., A7/W/A, n = 4), and (4) acute AMPH with SCH23390 (0.1 mg/kg, s.c.) pretreatment 40 min before AMPH (SCH23390/A1, n = 4). SCH23390 is an effective D1/D5 receptor antagonist that depresses AMPH-induced neuronal activity within the dopaminergic pathway. The MRI experimental paradigm was similar in all animal groups (i.e., AMPH or SAL at 3 h and MRI at 7 h after probe delivery). Representative averaged maps of percentage R2* signal increase (10–100%) from baseline of SAL, A1, and SCH23390/A1 groups are displayed above the bar graph in Figure 3C. We observed sporadic SPION-fosB signal in the SAL group and robust signal elevation in localized brain regions in the A1 group, most notably in the stratum. The elevation was substantially depressed in SCH23390/A1. We observed significant R2* elevations in A1 and A7/W/A in all ROIs except the hippocampus (HP) and motor cortex (MC). We also observed significant reductions in AMPH-induced SPION-fosB signals in the striatum (p = 0.03) of the A7/W/A group compared with the A1 group (Fig. 3C). This result suggests that SPION-fosB reports changes in gene transcription as a result of neuronal activation by AMPH exposure in vivo.
MRI detects SPION-ΔfosB signal elevation only in chronicallyAMPH-exposed animal brains
We examined AMPH-induced ΔfosB gene transcription using SPION-ΔfosB probe and MRI after the protocol established above. Here, we included animals groups similar to the previous SPION-fosB study: (1) SAL (n = 6), (2) A1 (n = 7), and (3) A7/W/A (n = 5). We also included an additional group of animals that received chronic exposure to AMPH followed by a period of abstinence; this sensitized group was given SAL (as a placebo) instead of AMPH challenge on the day of MRI acquisition (denoted as A7/W/S, n = 5). Representative averaged maps of percentage R2* signal increase (10–100%) from baseline of A1, A7/W/A and A7/W/S groups are displayed above the bar graph in Figure 3D. We observed robust elevation of the SPION-ΔfosB signal only in the A7/W/A group and in some restricted brain regions such as the striatum and cortex of the A7/W/S group (Fig. 3D). ROI analysis showed significant R2* elevation in A7/W/A group compared with the SAL group in all brain regions under investigation including the HP. Furthermore, we observed a slight but significant R2* elevation in the striatum [caudate–putamen (CPU)] of A7/W/S group (p = 0.04).
Histological confirmation of MRI observations
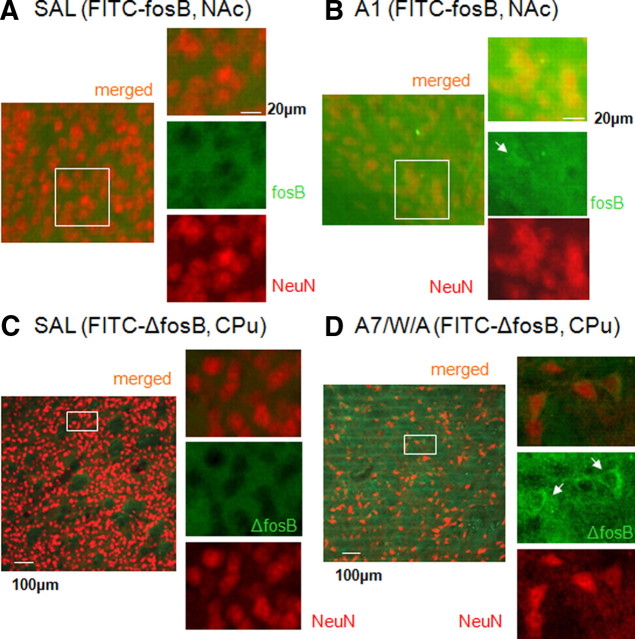
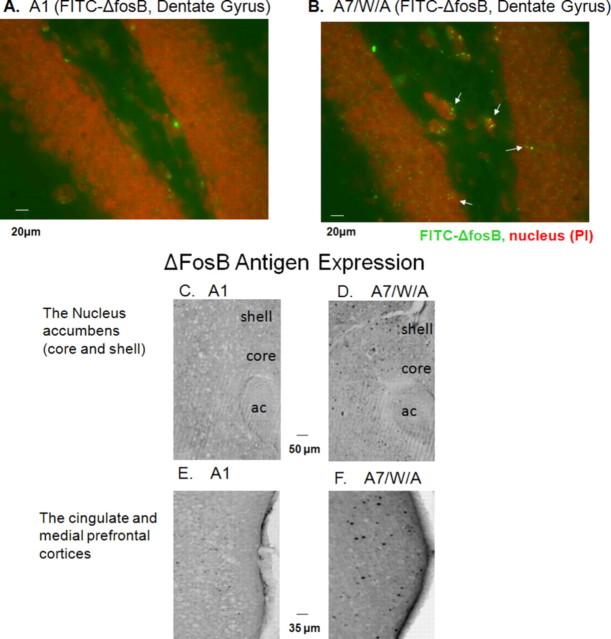
To validate our MRI observations, we examined the uptake of FITC-labeled sODN-fosB or FITC-labeled sODN-ΔfosB (120 pmol/kg, ICV) in SAL, A1, or A7/W/A groups as in the MRI studies. We observed FITC-sODN-fosB signal in the cytoplasm in the nucleus accumbens (NAc) of A1 brains, with overlapping of FITC and anti-NeuN-Cy3 in the peri-nuclear regions (Fig. 4B) (n = 4); however, we observed little FITC signal, most likely of background nature, and little overlap between FITC and anti-NeuN-Cy3 in the SAL group received sOND-fosB (Fig. 4A) (n = 4) or sODN-ΔfosB (Fig. 4C) (n = 4). Similarly, we observed much enhanced FITC-sODN-ΔfosB signal in the cytoplasm of the A7/W/A group compared with the SAL control group. In another groups of 8 mice, we delivered FITC-sODN-ΔfosB (8 nmol/kg, ICV), then AMPH at 3 h, but collected brain samples 1 h later. Consistent with the result in Figure 3D, we observed elevated retention of FITC signal, as punctate formations surrounding the nucleus [counterstained by propidium iodide (PI)] in the neuronal formation of the hippocampus of A7/W/A group compared with A1 group (Fig. 5 A, B).
Figure 4.
Distribution of FITC-sODN-fosB or FITC-sODN-ΔfosB in C57black mice. A and B show FITC-sODN-fosB in SAL or AI group. Overlapping of green (FITC-sODN) and red (antibodies to neuronal nuclear protein) indicate the distribution of sODN in the neurons. C, D show FITC-sODN-ΔfosB in SAL or A7/W/A group. Samples were obtained 4 h after AMPH (7 h after ICV infusion according to MRI protocol).
Figure 5.
A and B show elevated retention of FITC signal as punctate formations surrounding the nucleus (counterstained by PI) in the neuronal formation of the hippocampus of A7/W/A group (n = 4) compared with A1 group (n = 4). Samples were obtained 1 h after AMPH. Elevation of ΔFosB-positive cells is increased in A7/W/A mice (no ICV infusion of any probe), predominantly in the shell of the NAc (C) and the mPFC (E). Very few ΔFosB positive cells are present in A1 samples (D and F). N = 6 each group.
We further performed immunohistochemistry using antibodies against ΔfosB to validate our combination assay on mRNA elevation. We observed very little ΔFosB expression in the A1 group [A1, n = 6, left panels, Fig. 5C,E for the NAc and medial prefrontal cortex (mPFC), respectively], but we did detect elevated ΔFosB expression in some of the cells in the A7/W/A group (n = 6, right panels, Fig. 5D,F within the same ROI). This observation was consistent with MRI detection of different ΔfosB mRNA induction profiles for the A1 and A7/W/A groups. Based on the different R2* maps of SPION-ΔfosB in vivo, we conclude that SPION-ΔfosB and MRI detected a different gene expression profile for ΔfosB mRNA in chronically AMPH-exposed animal brains compared with acutely AMPH-exposed brains. Because sODN-fosB or sODN-ΔfosB binds to one mRNA (Fig. 1C), we further conclude that the in vivo target specificity of our SPION-sODN probes allows MRI visualization of differential inductions of fosB and ΔfosB mRNA in living brains under AMPH influence.
Discussion
We have examined potential gene interactions at the transcription level after chronic AMPH exposure, using a common rodent model of AMPH sensitization and a novel MRI contrast-enhanced technique (Giordano et al., 2006; Liu et al., 2007c), While we detected a positive correlation between molecular MRI and gene activation assay of fosB mRNA, we have demonstrated unique characteristics AMPH in vivo that are not realized from traditional molecular assays involving postmortem samples: a lack of ΔfosB mRNA induction in the acute exposure group, and after a 2 week abstinence period in chronic exposure paradigms. We observed significant elevation of ΔfosB mRNA expression only when a challenge dose of AMPH was given to sensitized mice; this induction occurred at the same time we observed desensitization of AMPH-induced fosB mRNA in the AMPH-sensitized animals. Our in vivo MRI results show that the SPION-fosB and SPION-ΔfosB probes provided sufficient in vivo target specificity and image sensitivity to report regional changes of fosB and ΔforB mRNA, respectively. Moreover, the data suggest that an active brain exhibits stringent controls on nonspecific binding of SPION-ΔfosB to closely related fosB mRNA. We will discuss the implications of these observations below.
We have considered suggestions using null signal in FosB knock-out mice to demonstrate the in vivo target specificity of sODN-fosB probe. However, the FosB knock-out strain generally has a mutant fosB gene that transcribes mutant mRNA although the wild-type FosB antigen is absent (Brown et al., 1996). Unless there is a deletion mutation at the fosB locus, the mutant mRNA would be a target of our probe. To obtain information from a good control for SPION-fosB, we tested the specificity of SCH23390, known to reduce the expression of amphetamine-induced FosB expression.
Correlations between molecular MR imaging and gene activation assay
Conventional understanding of probe design suggest that our sODN-ΔfosB probe, based on its size, could not be stringent enough for mRNA reporting, especially sODN-ΔfosB has partial sequence complementary to the flanking region of the splice site in the fosB mRNA. Therefore, we formulated a hypothesis that SPION-ΔfosB would report elevation of mRNA of the Fos families after acute exposure to AMPH. We selected this testing system in live mice because cells grown in culture conditions are known to indiscriminately take up short DNA (Overhoff and Sczakiel, 2005) or iron oxide-based contrast agent without sODN (Bulte et al., 1999) and we have no way, therefore, of determining the retention of SPION or SPION-sODN in these cells. If SPION-ΔfosB probe did not have adequate stringency in live brains, we would expect to observe SPION retention similar to that of SPION-fosB, at least in the A1 paradigm. However, SPION-ΔfosB did not report fosB mRNA elevation in the A1 paradigm. We concluded that SPION-ΔfosB exhibited sufficient in vivo specificity for our in vivo MRI detection of gene activities after AMPH exposure.
The same conclusion goes for the specificity of the fosB probe as it did not amplify the ΔfosB cDNA clone in vitro (Fig. 1C). Four evidences presented here support that the observed SPION-fosB signal elevation in MRI after AMPH is consistent with reports in the literature of fosB mRNA induction by AMPH. First, our R2* maps derived from images acquired immediately and 3 h after probe delivery to the CSF in live brains showed evidence of global distribution of the SPION-fosB probe. The biodistribution of Rhd-sODN-fosB in neurons within 3 h of delivery consisted in two forms as hybrids in whole cells near the ventricle and as punctate formations that migrate from the lumen of the ventricle to the parenchyma. The punctate formations in the migration front is consistent with the notion that cellular transfer of sODN or iron oxide nanoparticles is mediated by endocytosis, and that uptake is in the form of endosomes (Liang et al., 1996; Bulte et al., 1999). Once the endosome is internalized within the cell bodies, in our studies shown here, it releases the cargo (Rhd-sODN-fosB) to the cells, and the cargo is observed as blending with GFP in GABAnergic neurons as well as non-GABAnergic neurons. The blending is observed mostly behind the migration front or in the regions that have long periods of exposure to internalized Rhd-sODN-fosB. In contrast, the punctate formations in front of the migratory Rhd-sODN-fosB may indicate an alternative explanation. Matured mRNA is transported from the nucleus to the cytoplasm in the punctate formations (Vargas et al., 2005), the presence of punctate formations after sODN infusion may suggest, although need further investigation, a hybridization to, or targeting of cellular mRNA by sODN. Second, we have shown that PCR amplifies only one cDNA fragment from the total cDNA of mouse striatum by using either sODN-fosB or sODN-ΔfosB probe. The absence of a smear supports the notion of binding of sODN-fosB or sODN-ΔfosB probe to only one target cDNA. The inability of PCR product to reveal specific targeting of fosB or ΔfosB cDNA is compensated by a lack of in vivo hybridization in normal live brain using SPION-ΔfosB and MRI in A1 paradigm. Third, AMPH induced robust mRNA transcription of the Fos families, as well as SPION-fosB retention in the striatum in both acute and chronic AMPH exposure groups. Can a change to an intracellular mRNA transcript be reflected in the hybrid it forms with SPION-fosB? We tested this mechanism using SCH23390, a dopaminergic receptor antagonist that has been shown to suppress AMPH-induced mRNA transcription of the immediate early genes (IEG); we detected significantly lower fosB mRNA induction in the dopaminergic brain regions in vivo (e.g., mPFC, NAc, and CPU). Fourth, our MRI technique detected significant desensitization of fosB mRNA in the striatum of animals in the chronic AMPH exposure group (A7/W/A) compared with those in the acute AMPH exposure group. Our observations are consistent with other reports that have shown chronic AMPH exposure desensitizes c-fos mRNA induction in the striatum to a subsequent drug dose (Renthal et al., 2008). Therefore, our novel MRI method for detection of AMPH-induced gene activities in live mouse brains not only demonstrates specific interaction between probes and intracellular mRNA that is consistent with those of advanced molecular biology approaches, but also provides results not reported previously in real time without the trauma of tissue sample collection to the mouse and without the complications that can accompany mRNA purification.
The resulting R2* enhancement subsided by 7 h in normal brains, most likely because of clearance by active exclusion in live brain cells and relatively low fosB mRNA expression. This clearance time is different from our previous observation of 24 h when the mice were anesthetized using ketamine and xylazine (Liu et al., 2007b). Our current data provide strong evidence that, unless AMPH is administered to increase the transcription of fosB genes, normal and resting live brains actively clear unbound SPION probes. Although the mechanism that governs clearance of free probes from live brains remains unknown, active clearance provides a window for MRI detection of the profiles of bound SPION-fosB probe in mice treated with or exposed to psycho-stimulants.
Implication of null SPION-ΔfosB signal elevation in the acute paradigm
Unlike the baseline measures of c-fos and fosB mRNA detected with SPION-cfos and SPION-fosB in normal mice, we detected no endogenous ΔfosB mRNA in normal resting mice or in the A1 group with the SPION-ΔfosB probe and MRI. Null retention was also reported using an MR probe (SPION-Ran) that has no target mRNA transcripts (Liu et al., 2007a). Therefore, we can conclude that null retention of SPION-ΔfosB reflects low or no expression of ΔfosB mRNA in the brains of normal resting mice. This result is consistent with other reports that normal mice exhibit little basal ΔfosB mRNA or ΔFosB protein expression (Alibhai et al., 2007; Conversi et al., 2008). Our finding of null retention of SPION-ΔfosB probe in the A1 group suggests that SPION-ΔfosB exhibits discriminatory properties of closely related mRNA such as fosB mRNA. Therefore SPION-ΔfosB shows stringency for reporting gene activities. In addition, our data on the retention of SPION-ΔfosB differ from previous studies using in vitro and postmortem assays (Kelz et al., 1999; Giordano et al., 2006; Alibhai et al., 2007; Conversi et al., 2008). The ability of sODN-ΔfosB to separate different targets of the same families of genes in vivo supports antigen detection. Because most mRNA transcripts have a short half-life and our MRI requires no mRNA purification, detection of gene transcripts using SPION-ΔfosB represents most recent gene transcription in live brains with no disturbance from autopsy trauma. These brain probes directly report real-time gene activation of living subjects.
Implication for chronic drug exposure
We have observed different expression profiles for ΔfosB mRNA in acute and chronic AMPH exposure groups after AMPH challenge; these differences were observed in all of the brain regions we investigated, including the hippocampus. Alternative splicing of fosB mRNA predicts that ΔfosB mRNA expression would accompany the elevation of fosB mRNA activation (Nakabeppu and Nathans, 1991, Marinescu et al., 2007). This mechanism does not appear to account for our observations in the A1 group or in the striatum of the sensitization (A7/W/S) group because there was no significant elevation in ΔfosB mRNA in both treatment groups. Rather, our observation is consistent with the notion that the product of ΔfosB mRNA (ΔFosB antigen) serves as one of the epigenetic modulators of certain immediate early genes (Renthal et al., 2008), (Kumar et al., 2005). In addition, this induction of hippocampal ΔfosB mRNA in the A7/W/A group not only provides evidence for the sensitivity of our probe for detecting the genetic basis of neural activities by MRI, but also suggests additional studies for ΔfosB mRNA as a biomarker for memory and learning processes (Fleischmann et al., 2003; Gass et al., 2004).
Footnotes
This project was supported by grants from American Heart Association (09GRNT2060416), National Institutes of Health (R01EB002066 to B.R.R., R03EB008134 to J.B.M., R21DA024235 and R01DA026108 to C.H.L., R01NS045845 and R21NS057556 to P.K.L., and R01DA020796 and P30NS045776 to P.G.B.), and National Center for Research Resources (P41RR014075 to B.R.R.). We thank Dr. Yusaku Nakabeppu (Kyushu University, Japan) for cloned cDNA of fosB and ΔfosB, and Dr. Helen D'Arceuil and Ms. N. Eusemann for suggestions and excellent editing. Two patent applications have been submitted on this technology: (1) Imaging nucleic acids (US 60-603907) and (2) Targeting brain cells via ophthalmic delivery which included SPION-ΔfosB probe (US 60-962499).
The authors declare no competing financial interests.
References
- Alibhai IN, Green TA, Potashkin JA, Nestler EJ. Regulation of fosB and DeltafosB mRNA expression: in vivo and in vitro studies. Brain Res. 2007;1143:22–33. doi: 10.1016/j.brainres.2007.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxerman JL, Hamberg LM, Rosen BR, Weisskoff RM. MR contrast due to intravascular magnetic susceptibility perturbations. Magn Reson Med. 1995;34:555–566. doi: 10.1002/mrm.1910340412. [DOI] [PubMed] [Google Scholar]
- Brown JR, Ye H, Bronson RT, Dikkes P, Greenberg ME. A defect in nurturing in mice lacking the immediate early gene fosB. Cell. 1996;86:297–309. doi: 10.1016/s0092-8674(00)80101-4. [DOI] [PubMed] [Google Scholar]
- Bulte JW, Zhang S, van Gelderen P, Herynek V, Jordan EK, Duncan ID, Frank JA. Neurotransplantation of magnetically labeled oligodendrocyte progenitors: magnetic resonance tracking of cell migration and myelination. Proc Natl Acad Sci U S A. 1999;96:15256–15261. doi: 10.1073/pnas.96.26.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conversi D, Bonito-Oliva A, Orsini C, Colelli V, Cabib S. DeltaFosB accumulation in ventro-medial caudate underlies the induction but not the expression of behavioral sensitization by both repeated amphetamine and stress. Eur J Neurosci. 2008;27:191–201. doi: 10.1111/j.1460-9568.2007.06003.x. [DOI] [PubMed] [Google Scholar]
- Cui JK, Hsu CY, Liu PK. Suppression of postischemic hippocampal nerve growth factor expression by a c-fos antisense oligodeoxynucleotide. J Neurosci. 1999;19:1335–1344. doi: 10.1523/JNEUROSCI.19-04-01335.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, Wagner EF, Gass P. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass P, Fleischmann A, Hvalby O, Jensen V, Zacher C, Strekalova T, Kvello A, Wagner EF, Sprengel R. Mice with a fra-1 knock-in into the c-fos locus show impaired spatial but regular contextual learning and normal LTP. Brain Res Mol Brain Res. 2004;130:16–22. doi: 10.1016/j.molbrainres.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Giordano TP, 3rd, Satpute SS, Striessnig J, Kosofsky BE, Rajadhyaksha AM. Up-regulation of dopamine D(2)L mRNA levels in the ventral tegmental area and dorsal striatum of amphetamine-sensitized C57BL/6 mice: role of Ca(v)1.3 L-type Ca(2+) channels. J Neurochem. 2006;99:1197–1206. doi: 10.1111/j.1471-4159.2006.04186.x. [DOI] [PubMed] [Google Scholar]
- Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine's psychomotor and rewarding effects. Proc Natl Acad Sci U S A. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci U S A. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT. Cocaine and the AP-1 transcription factor complex. Ann N Y Acad Sci. 1998;844:1–6. [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–1244. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Johnston M, Zakharov A, Papaiconomou C, Salmasi G, Armstrong D. Evidence of connections between cerebrospinal fluid and nasal lymphatic vessels in humans, non-human primates and other mammalian species. Cerebrospinal Fluid Res. 2004;1:2. doi: 10.1186/1743-8454-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Jr, Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang YJ, Marotti L, Self DW, Tkatch T, Baranauskas G, Surmeier DJ, Neve RL, Duman RS, Picciotto MR, Nestler EJ. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Liang WW, Shi X, Deshpande D, Malanga CJ, Rojanasakul Y. Oligonucleotide targeting to alveolar macrophages by mannose receptor-mediated endocytosis. Biochim Biophys Acta. 1996;1279:227–234. doi: 10.1016/0005-2736(95)00237-5. [DOI] [PubMed] [Google Scholar]
- Liu CH, Kim YR, Ren JQ, Eichler F, Rosen BR, Liu PK. Imaging cerebral gene transcripts in live animals. J Neurosci. 2007a;27:713–722. doi: 10.1523/JNEUROSCI.4660-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Huang S, Kim YR, Rosen BR, Liu PK. Forebrain ischemia-reperfusion simulating cardiac arrest in mice induces edema and DNA fragmentation in the brain. Mol Imaging. 2007b;6:156–170. [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Huang S, Cui J, Kim YR, Farrar CT, Moskowitz MA, Rosen BR, Liu PK. MR contrast probes that trace gene transcripts for cerebral ischemia in live animals. FASEB J. 2007c;21:3004–3015. doi: 10.1096/fj.07-8203com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, You Z, Ren J, Kim YR, Eikermann-Haerter K, Liu PK. Noninvasive delivery of gene targeting probes to live brains for transcription MRI. FASEB J. 2008;22:1193–1203. doi: 10.1096/fj.07-9557com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, You Z, Liu CM, Kim YR, Whalen MJ, Rosen BR, Liu PK. Diffusion-weighted magnetic resonance imaging reversal by gene knockdown of matrix metalloproteinase-9 activities in live animal brains. J Neurosci. 2009;29:3508–3517. doi: 10.1523/JNEUROSCI.5332-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PK, Salminen A, He YY, Jiang MH, Xue JJ, Liu JS, Hsu CY. Suppression of ischemia-induced fos expression and AP-1 activity by an antisense oligodeoxynucleotide to c-fos mRNA. Ann Neurol. 1994;36:566–576. doi: 10.1002/ana.410360405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelzys A, Gruda MA, Bravo R, Morgan JI. Absence of a persistently elevated 37 kDa fos-related antigen and AP-1-like DNA-binding activity in the brains of kainic acid-treated fosB null mice. J Neurosci. 1997;17:5407–5415. doi: 10.1523/JNEUROSCI.17-14-05407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu V, Loomis PA, Ehmann S, Beales M, Potashkin JA. Regulation of retention of FosB intron 4 by PTB. PLoS ONE. 2007;2:e828. doi: 10.1371/journal.pone.0000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular neurobiology of addiction. Am J Addict. 2001;10:201–217. doi: 10.1080/105504901750532094. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. DeltaFosB: a sustained molecular switch for addiction. Proc Natl Acad Sci U S A. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Kosofsky BE, Birnbaum R, Cohen BM, Hyman SE. Differential expression of c-fos and zif268 in rat striatum after haloperidol, clozapine, and amphetamine. Proc Natl Acad Sci U S A. 1992;89:4270–4274. doi: 10.1073/pnas.89.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov IA, Cherepov AB, Raevskiı̆ KS, Anokhin KV. [Locomotor activity and expression of c-Fos protein in the brain of C57BL and Balb/c mice: effects of D-amphetamine and sydnocarb] Eksp Klin Farmakol. 2002;65:18–21. [PubMed] [Google Scholar]
- Okabe C, Takeshima H, Murphy NP. Methamphetamine sensitization in nociceptin receptor knockout mice: locomotor and c-fos expression. Eur J Pharmacol. 2005;507:57–67. doi: 10.1016/j.ejphar.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Ostrander MM, Badiani A, Day HE, Norton CS, Watson SJ, Akil H, Robinson TE. Environmental context and drug history modulate amphetamine-induced c-fos mRNA expression in the basal ganglia, central extended amygdala, and associated limbic forebrain. Neuroscience. 2003;120:551–571. doi: 10.1016/s0306-4522(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Overhoff M, Sczakiel G. Phosphorothioate-stimulated uptake of short interfering RNA by human cells. EMBO Rep. 2005;6:1176–1181. doi: 10.1038/sj.embor.7400535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelkar NK, Wang JQ. mGluR5-dependent increases in immediate early gene expression in the rat striatum following acute administration of amphetamine. Brain Res Mol Brain Res. 2004;122:151–157. doi: 10.1016/j.molbrainres.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Paulson PE, Camp DM, Robinson TE. Time course of transient behavioral depression and persistent behavioral sensitization in relation to regional brain monoamine concentrations during amphetamine withdrawal in rats. Psychopharmacology. 1991;103:480–492. doi: 10.1007/BF02244248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. London: Academic; 2001. [Google Scholar]
- Perrotti LI, Hadeishi Y, Ulery PG, Barrot M, Monteggia L, Duman RS, Nestler EJ. Induction of ΔFosB in reward-related brain structures after chronic stress. J Neurosci. 2004;24:10594–10602. doi: 10.1523/JNEUROSCI.2542-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ. ΔFosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Teegarden SL, Nestler EJ, Bale TL. Delta FosB-mediated alterations in dopamine signaling are normalized by a palatable high-fat diet. Biol Psychiatry. 2008;64:941–950. doi: 10.1016/j.biopsych.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet N, Zwiller J, Ali SF. Induction of the immediate early genes egr-1 and c-fos by methamphetamine in mouse brain. Brain Res. 2001;919:31–40. doi: 10.1016/s0006-8993(01)02991-2. [DOI] [PubMed] [Google Scholar]
- Uslaner J, Badiani A, Norton CS, Day HE, Watson SJ, Akil H, Robinson TE. Amphetamine and cocaine induce different patterns of c-fos mRNA expression in the striatum and subthalamic nucleus depending on environmental context. Eur J Neurosci. 2001;13:1977–1983. doi: 10.1046/j.0953-816x.2001.01574.x. [DOI] [PubMed] [Google Scholar]
- Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci U S A. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue CJ, Ng JP, Li Y, Wolf ME. Acute and repeated systemic amphetamine administration: effects on extracellular glutamate, aspartate, and serine levels in rat ventral tegmental area and nucleus accumbens. J Neurochem. 1996;67:352–363. doi: 10.1046/j.1471-4159.1996.67010352.x. [DOI] [PubMed] [Google Scholar]