Abstract
In early Drosophila embryos, the transcription factor Dorsal regulates patterns of gene expression and cell fate specification along the dorsal-ventral axis. How gene expression is produced within the broad lateral domain of the presumptive neurogenic ectoderm is not understood. To investigate transcriptional control during neurogenic ectoderm specification, we examined divergence and function of an embryonic cis-regulatory element controlling the gene short gastrulation (sog). While transcription factor binding sites are not completely conserved, we demonstrate that these sequences are bona fide regulatory elements, despite variable regulatory architecture. Mutational analysis of conserved putative transcription factor binding sites revealed that sites for Dorsal and Zelda, a ubiquitous maternal transcription factor, are required for proper sog expression. When Zelda and Dorsal sites are paired in a synthetic regulatory element, broad lateral expression results. However, synthetic regulatory elements that contain Dorsal and an additional activator, also drive expression throughout the neurogenic ectoderm. Our results suggest that interaction between Dorsal and Zelda drives expression within the presumptive neurogenic ectoderm, but they also demonstrate that regulatory architecture directing expression in this domain is flexible. We propose a model for neurogenic ectoderm specification in which gene regulation occurs at the intersection of temporal and spatial transcription factor inputs.
INTRODUCTION
Patterned specification of cell fate results from differential gene expression. Differential control of gene expression is accomplished by site-specific transcription factors, which bind DNA to regulate expression over developmental space and time and are themselves regulated at the level of expression or activity. Cis-regulatory regions determine how individual genes respond to varying levels and combinations of transcription factors found in different cells during development. However, cell type is discrete and developmental pattern is precise. Therefore, patterning depends on the function of cis-regulatory regions to integrate information from transcription factors to produce differential gene expression states in the developing embryo. The architecture of these regulatory regions is complex and the logic behind the organization needs to be determined empirically (e.g. Brown et al., 2007; Davidson, 2001; Deplancke et al., 2006; Ochoa-Espinosa and Small, 2006; Zinzen et al., 2006).
Dorsal-ventral axis patterning during Drosophila embryogenesis is a well-studied system that is poised for understanding cis-regulatory mechanisms driving development. Over 25 cis-regulatory sequences have been identified for over 60 genes known to control different aspects of dorsal-ventral patterning (Stathopoulos and Levine, 2005). Three presumptive germ layers form along the dorsal-ventral axis in the developing Drosophila embryo: mesoderm in ventral regions, neurogenic ectoderm in lateral regions and ectoderm and amnioserosa in dorsal regions. The specification of these germ layers is dependent upon the NFκB-like transcription factor, Dorsal, which localizes to the nucleus in a gradient with highest amounts in ventral regions and lowest amounts in dorsal regions (rev. in Moussian and Roth, 2005). Although Dorsal has been studied extensively, questions remain about how this analog gradient of nuclear Dorsal can direct discrete target gene expression outputs.
Combinatorial interactions between Dorsal and other transcription factors surely contribute to the distinct outputs of gene expression. In ventral and ventral-lateral regions, a synergistic relationship between the bHLH transcription factor, Twist, and Dorsal, has been demonstrated to establish the mesodermal and ventral-neurogenic cell fates (Ip et al., 1992; Jiang and Levine, 1993; Markstein et al., 2004). Furthermore, in dorsal regions of the embryo, the ectoderm and amnisoserosa form as a result of repression by Dorsal and activation by ubiquitous transcription factors to regulate the expression of genes such as decapentaplegic (dpp) (Liang et al., 2008; Rusch and Levine, 1997). However, the regulatory architecture required to support expression in a broad lateral domain, encompassing the entire presumptive neurogenic ectoderm region of the early embryo, has not been clearly defined (rev. in Stathopoulos and Levine, 2004).
Only two regulatory elements that direct expression in a broad lateral domain within the early embryo have been identified, those controlling expression of the genes short-gastrulation (sog) and thisbe (ths). These regulatory elements were found by searching for clusters of high-affinity Dorsal binding sites in the genome and have been validated (Markstein and Levine, 2002; Stathopoulos and Levine, 2002). These regulatory elements have similar binding site composition: both contain multiple Dorsal binding sites, sites for the ventral repressor, Snail, and the presence of an overrepresented sequence, TTCCAGC, also called GCTGGAA, which we will refer to as the T motif (Stathopoulos et al., 2002). These cis-regulatory elements also contain the CAGGTAG motif and other similar heptamers, collectively referred to as TAGteam sites (De Renzis et al., 2007; ten Bosch et al., 2006). The maternal transcription factor, Zelda, also known as vielfaltig (Staudt et al., 2006), binds specifically to these heptamers and is a critical player in zygotic genome activation (Liang et al., 2008). However, the requirement of all these putative binding sites (i.e. Dorsal, Snail, T motif, Zelda) to direct sog and ths early embryonic expression has not been rigorously tested. One reason that more neurogenic ectoderm regulatory elements have not been found could be that variable combinations of cis and trans factors are capable of directing expression in the presumptive neurogenic ectoderm.
It has been demonstrated that flexibility can occur in regulatory element structure with little to no effect on transcriptional output. Regulatory regions with variable binding site composition are capable of generating expression in the same tissue in Caenorhabditis elegans (Guhathakurta et al., 2002; Hunt-Newbury et al., 2007). Studies in sea urchin have found that flexibility in both cis and trans- regulators can exist while still producing conserved expression of the Endo16 gene (Romano and Wray, 2003). More recently, a study comparing even-skipped gene regulatory elements in Drosophilids and Sepsids showed that although there is minimal sequence conservation, functional conservation of regulatory elements remains (Hare et al., 2008). Additionally, an extensive study of co-expressed genes in Ciona demonstrates that different motif architectures are tolerated to generate co-regulation of genes (Brown et al., 2007). Such flexibility in the organization and composition of binding sites within cis-regulatory sequences might provide a method for “buffering” during development, allowing organisms to develop reproducibly even when the regulatory regions of DNA are altered throughout the course of evolution.
In this analysis, we explore the transcriptional architecture required to pattern the neurogenic ectoderm in D. melanogaster embryos. Specifically, our goal was to define the transcription factor binding sites necessary and sufficient to direct expression within the broad lateral domain of early embryos. We define the underlying logic within the minimal cis-regulatory element, which supports expression of sog in Drosophila early embryos, using both evolutionary comparisons and synthetic reporter constructs. Collectively, our results support the view that flexible regulatory element structures are capable of producing similar transcriptional outputs.
EXPERIMENTAL PROCEDURES
Regulatory element alignments and annotations
Cartwheel (http://cartwheel.caltech.edu/) and JASPAR (http://jaspar.genereg.net/) were used to generate Position Weight Matrices (PWMs) from in vitro binding data (Brown et al., 2005; Sandelin et al., 2004). These matrices were used to scan putative regulatory regions for motifs of interest. For a complete list of motifs, Cartwheel-generated consensus sequences, threshold values and probabilities of these matrices occurring randomly in a one kilobase (Kb) sequence, see Table 1.
| Name | Consensus | Probability (p) | Threshold | Reference |
|---|---|---|---|---|
| TAGTeam | YAGGYAD | 3.7E-04 | 14.7 | ten Bosch et al., 2006 |
| Snail | CAGGTG | 9.8E-04 | 27.5 | Mauhin et al., 1993 |
| Snail | DCADRDNN | 9.2E-04 | 21 | Papatsenko personal comm. |
| Snail | CACCT | 9.8E-04 | match 4/4 | Markstein et al., 2002b |
| Snail | MMRCAWGT | 2.4E-04 | match 8/8 | Stathopoulos and Levine, 2005 |
| Hb | GCATAAAAAA | < 1 hit/kb | 29.21 | Stanojević et al., 1989 |
| Schnurri | GRCGMCWVWBHGTCTG | < 1 hit/kb | 25 | Pyrowolakis et al., 2004 |
| D-STAT | TTTCCCGGAAA | < 1 hit/kb | 42.94 | Yan et al 1996 |
| Twist | ACATATG | 8.5E-04 | 40.16 | Lee et al 1997 |
| Daughterless | CACCTGC | 6.1E-04 | 40.73 | Senger personal comm. |
| bHLH | CANNTG | 3.9E-03 | match 6/6 | Murre et al 1994 |
| Dorsal | GGGAATTCC | 8.4E-04 | 49 | Senger personal comm. |
| NFKappaB | GGGAATTTCC | < 1 hit/kb | 39 | Vlieghe et al., 2006 |
| Dorsal | GGGWDWWWCCM | < 1 hit/kb | match 11/1 | Markstein et al., 2002b |
| TTCCAGC | TTCCAGC | 6.1E-05 | match 7/7 | Stathopoulos et al 2002 |
| Pointed | SNGGAWRY | 9.0E-04 | 14.3 | Xu et al |
| Su(H) | BTGTGGGAAMCGAGAT | < 1 hit/kb | 30 | Bailey et al 1995 |
p*2*1000 = per site probability of finding a motif with these parameters at random
Homologous sequences were obtained for seven of the twelve sog Drosophilid sequences (Papatsenko and Levine, 2005a). A complete list of all the predicted sog Drosophilid homologous sequences is available (http://flydev.berkeley.edu/cgi-bin/Annotation/enhancers/sog.htm; D. Papatsenko, in preparation). Sequences were loaded onto the Cartwheel site and scanned for binding sites using the previously generated PWMs. Cartwheel generates false positive statistics for each of the matrices (listed in Table 1). We used these statistics to set thresholds which correspond to one or fewer false positive match per kilobase of sequence for all of the putative binding sites. Snail does not have particularly good binding site predictions. To adjust for this, four motifs were used to find putative Snail binding sites. The same methods were used to find binding sites in the thisbe (ths) regulatory elements.
The Neu3 D. melanogaster regulatory element sequence we tested was used to find homologous regulatory elements in each of the twelve sequenced Drosophilids. Briefly, UCSC BLAT search was used to find sequences of high similarity in the other Drosophilid genomes (http://genome.ucsc.edu/cgi-bin/hgBlat). In the case of the identification of D. virilis homologous sequences, the Drosophila genome version “April 2004” must be selected.
Vector construction
All of the even-skipped (eve) promoter LacZ (eve.p-lacZ) fusion elements used a modified pLacZattB vector, with the eve minimal promoter inserted in place of the hsp70 minimal promoter (Bischof et al., 2007; Jiang et al., 1991). cis-regulatory modules were amplified from genomic DNA, cloned into the NotI site of the eve.p-LacZ.attB vector and verified by sequencing. Synthetic cis-regulatory elements were constructed from oligonucleotides and cloned into the pGEMT-easy vector (Promega) or directly into the BglII and NotI sites of the eve.p-LacZ.attB. All constructs were verified by sequencing.
Site-directed mutagenesis
Primers were designed to mutate sites within the sog cis-regulatory element using the QuickChange SiteDirected Mutagenesis Kit from Stratagene (for primer sequences see Supplemental Materials & Methods). Genomic DNA was used as a template for PCR reaction to amplify the sog regulatory element. It was cloned into the pGEMT-easy vector, which was subsequently used as the template for mutagenesis reactions.
Generation of transgenic fly lines
Phi-C31 mediated site-specific integration of cis-regulatory element-reporter fusions was done as described into either ZH-attp51D or attp16 (Bischof et al., 2007; Groth et al., 2004; Markstein et al., 2008). Embryo injections were performed in house and with help from Rainbow Transgenic Flies (Newbury Park, CA) and Genetic Services Inc. (Sudbury, MA).
In situ hybridization
Digoxigenin-UTP-labeled LacZ antisense RNA probes were used to detect LacZ reporter gene expression as described previously with a few modifications (Jiang and Levine, 1993; Tautz and Pfeifle, 1989). Briefly, embryos were collected, aged to be 2–4 hours old, dechorinated in 100% Sodium hypochlorite (Sigma #239305) for 3 minutes, washed and transferred to a scintillation vial with 3mL buffer (1.3XPBS, 67mM EGTA pH 8.0), 4mL heptane, 1 mL 37% formaldehyde solution. Embryos were fixed for 20 minutes and then MeOH was used to remove the vitelline membrane. D. mojavensis and D. pseudobscura embryos were fixed with 1.6mL buffer, 8mL heptane, 0.4mL paraformaldehyde solution (Electron Microscopy Sciences #15713-S).
Fly lines
Drosophila species were obtained from the Drosophila Species Stock Center (https://stockcenter.ucsd.edu/info/welcome.php). Dorsal mutant analysis was performed with dl1 cn1 sca1/CyO, l(2)DTS1001, and Twist mutant analysis was carried out using twi cn bw/CyO; both stocks are available from the Bloomington Stock Center.
RESULTS
Broad lateral expression of sog is conserved among Drosophilids
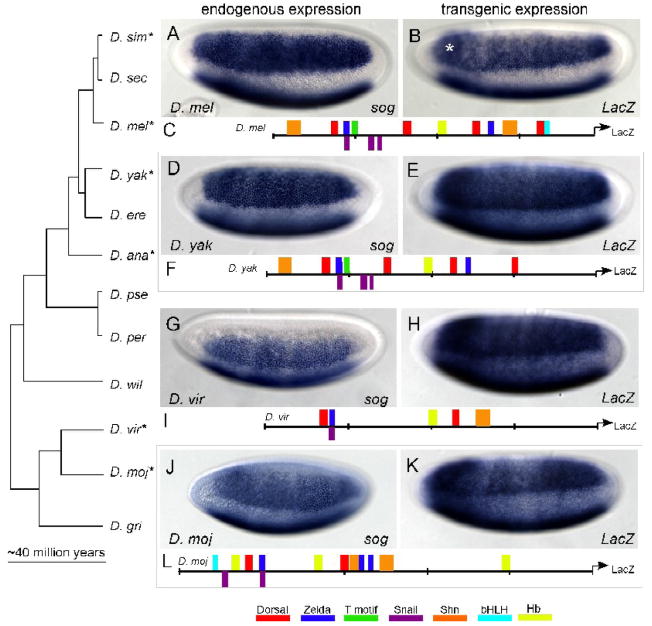
The genomes of twelve Drosophila species have been sequenced, facilitating the analysis of coding and regulatory regions spanning approximately 40 million years of evolution (Clark et al., 2007). In Anopheles, sog expression is different from D. melanogaster in that it is found in ventral regions of the embryo (Goltsev et al., 2007). We decided to determine whether sog expression is conserved or divergent within early embryos from a phylogenetically representative set of seven of the twelve sequenced Drosophilids: D. melanogaster, D. yakuba, D. simulans, D. ananassae, D. pseudobscura, D. mojavensis, and D. virilis. The broad lateral expression pattern of sog in D. melanogaster was conserved when compared with endogenous sog expression in the Drosophilids we examined (Fig. 1D, G, J and Supplemental Figure 1A, C compare with Fig. 1A). We found that expression was maintained in a broad lateral stripe even when the size of the embryos varied. D. yakuba, D. simulans, and D. mojavensis embryos are all slightly smaller than D. melanogaster on average; D. virilis and D. ananassae embryos are longer along the anterior-posterior axis and thinner along the dorsal-ventral axis than D. melanogaster. Nevertheless, sog expression is absent from both ventral and dorsal-most regions of the embryos in these divergent Drosophilids, as observed in D. melanogaster. The sharp ventral border of sog expression due to Snail repression in ventral regions, seen in D. melanogaster, is also apparent in the other Drosophilids we tested.
Fig. 1. Reporter fusions in D. melanogaster reveal conservation of expression and regulatory logic.
Whole mount in situ hybridization using anti-sense probes to mRNA transcripts in various Drosophilid species. In (A, D, G, and J), the expression of sog is depicted. In (B, E, H, K), reporter gene expression is detected by in situ hybridization using a riboprobe recognizing the lacZ gene. sog is expressed broadly in lateral regions of the embryo in (A) D. melanogaster (D. mel), (D) D. yakuba (D. yak), (G) D. virilis (D. vir), and (J) D. mojavensis (D. moj). Expression is absent from ventral and dorsal regions of the embryo and the anterior and posterior poles. Cartoons of putative minimal sog regulatory elements of (C) D. mel, (F) D. yak, (I) D. vir, and (L) D. moj which were fused to a LacZ reporter and integrated into D. melanogaster embryos (B, E, H, and K, respectively). Stronger expression (denoted by the asterisk) present in all transgenic embryos in a band at the anterior is associated with vector sequence and likely due to the lacZ gene, (Jiang et al., 1991). In this figure and all subsequent ones, embryos are oriented with anterior to the left and dorsal side up. The asterisks denotes stronger expression in a band at the anterior is associated with vector sequence, present in all transgenic embryos and most prominent at cellularization (Jiang et al., 1991). The embryos are tilted ventral-laterally in order to show repression in ventral regions; thus both lateral stripes of sog expression are in view, though the domain of expression located at the bottom of the images is only a partial view of the broad lateral expression domain. Zelda refers to all of the TAGteam motifs.
Validation of homologous sog regulatory regions
A sog cis-regulatory module that drives expression in the broad lateral domain of early D. melanogaster embryos was previously identified and verified in a genome wide search for clusters of Dorsal binding sites (Markstein et al., 2002). This minimal cis- regulatory module from D. melanogaster was used to find homologous DNA sequences in the six other Drosophilid species (see Methods). We tested whether these putative cis-regulatory elements were able to support expression of a reporter in the presumptive neurogenic ectoderm of D. melanogaster.
Constructs containing DNA of the presumptive sog cis-regulatory modules isolated from six species were fused to a reporter gene (i.e. LacZ or Cherry) and integrated into the D. melanogaster genome by PhiC31 integration (see Methods). By using site-specific integration methods, we are confident that our comparative analysis of regulatory sequences is not confounded by positional effects, which can result when P-elements are used to generate transgenic lines as a result of random integration of the reporter gene construct into the genome (Levis et al., 1985). All the transgenic constructs direct expression similar to that supported by the minimal sog cis-regulatory module previously identified from D. melanogaster (Fig. 1E, H, K and Supplemental Fig. 1B, D, compare with Fig. 1B), which itself is comparable in expression to the endogenous sog gene at this same stage (Figs. 1A and 2B; and Markstein et al., 2002). These results demonstrate that the homologous sequences are functionally conserved regulatory elements.
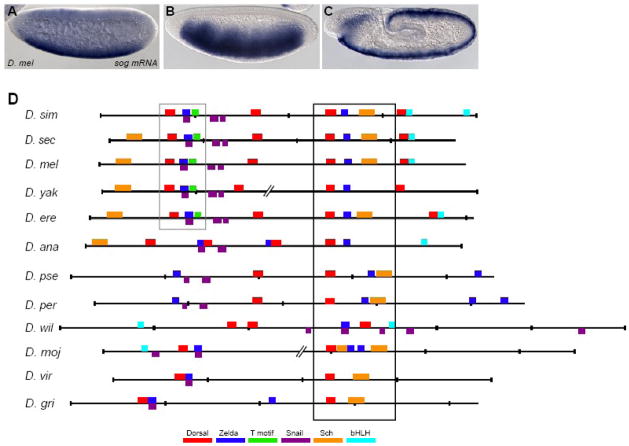
Fig. 2. Alignment of putative sog cis-regulatory elements from other Drosophilids reveals conservation and turnover of binding sites.
(A–C) sog expression is dynamic. Endogenous expression of the sog gene detected by in situ hybridization using a riboprobe within embryos of nuclear cycle ~11 (A), cycle 14/stage 5 (B), and during germ-band elongation after gastrulation (C). (D) Shown are the predicted binding sites for transcription factors and over-represented motifs that are associated with neurogenic ectoderm patterning within the sog cis-regulatory modules identified from 12 Drosophilid species. Position weight matrices (PWMs) were used to find putative transcription factor binding sites using the program Cartwheel (Brown et al., 2005). Alignments were generated on the UCSC genome browser webpage (http://genome.ucsc.edu). Cartoons were generated by Cartwheel and then colored according to the key. Gaps in the alignments, shown as broken lines, were introduced to help visualize conservation. Box domains represents well-conserved region of sog cis-regulatory element. The sites boxed in black are located in the most well-conserved region, and the sites boxed in grey are the second most well-conserved region. Note that closely associated Dorsal and Zelda binding sites are present in all of the alignments, though the location of these sites within the sequence can vary. Full alignment can be viewed in Supplemental Fig. 3.
There are minor differences in the borders of expression for various regulatory element reporters. In particular, there is a slight difference in the ventral borders of both the D. virilis and D. mojavensis sog cis-regulatory element reporters as compared with the other reporter constructs (Fig. 1H and K, compare with 1B). The border appears less sharp than in the D. melanogaster sog regulatory element reporter and when compared to endogenous sog expression. Since the endogenous expression boundary is discrete, we reasoned that there must be changes in cis- or trans-factors that influence the transgenic reporter expression. In fact, there are more putative binding sites for the Snail transcriptional repressor in the D. melanogaster cis-regulatory module (i.e. three sites), than are found in the cis-regulatory modules of D. viliris and D. mojavensis (i.e. one site and two sites, respectively). D. virilis and D. mojavensis may use other transcription factors, which are not functional in the context of the D. melanogaster embryo, to support repression in ventral regions (i.e. cis effects). Alternatively, changes in the Snail protein within these other species (i.e. trans effects) may contribute to changes in binding site preference such that we no longer can predict binding sites using the PWM defined by D. melanogaster data.
Despite subtle differences in the expression patterns supported by these divergent sequences, all the predicted cis-regulatory modules do indeed direct expression of a reporter in a broad lateral domain within early D. melanogaster embryos. We hypothesized that a core set of conserved binding sites and transcription factors bind to all the regulatory elements tested to drive reporter expression in this broad lateral expression domain.
Identification of conserved binding sites within homologous sog regulatory regions
In order to determine the requirements for patterned broad lateral expression, we set out to identify the functional set of transcription factor binding sites within the minimal D. melanogaster sog regulatory element. To date, several predicted transcription factors binding sites and overrepresented motifs, that act to pattern expression during D. melanogaster embryogenesis have been identified (Markstein and Levine, 2002; Markstein et al., 2002; Mauhin et al., 1993; Muller et al., 2003; Ochoa-Espinosa and Small, 2006; Papatsenko and Levine, 2005b; Pyrowolakis et al., 2004; Stanojevic et al., 1989; Stathopoulos and Levine, 2004; Stathopoulos et al., 2002; ten Bosch et al., 2006; Vlieghe et al., 2006; Xu et al., 2000; Yan et al., 1996). We used the results of these in vitro binding studies as well as degenerate binding site predictions (Murre et al., 1994) to construct position weight matrices that describe the binding sites preferences exhibited by Dorsal, Zelda, Smad/Schnurri, D-STAT, Snail, bHLH proteins (including Daughterless and Twist), and Hunchback DNA-binding proteins (see Methods). We also analyzed whether the overrepresented T motif (TTCCGCA) was present, as this motif was found previously to be associated with the broad lateral expression in the early embryo (Stathopoulos et al., 2002).
Using these position weight matrices (PWMs), we scanned for putative transcription factor binding sites in the sog regulatory element from D. melanogaster using the Cartwheel program (http://woodward.caltech.edu/canal/; Brown et al., 2005). We identified the four Dorsal binding sites, two sites for the Snail repressor, one T motif site, and two TAGteam sequences all of which had been previously identified within this cis-regulatory module. We identified several novel sites as well, including two binding sites for Schnurri, the transcriptional co repressor, one degenerate bHLH site, an additional Snail site, and one Hunchback site, an activator which functions along the anterior-posterior axis (Fig. 2D and Supplemental Fig. 2).
We examined the conservation of these binding sites within the homologous regulatory elements from other Drosophilids in an effort to define the essential features of the minimal regulatory element. We searched for the same putative binding sites identified within the D. melanogaster sog regulatory element in the functionally conserved regulatory sequences from D. simulans, D. yakuba, D. annanassae, D. psuedobscura, D. virilis and D. mojavensis (see Fig. 1C, F, I, and L).
Our results reveal conserved clusters of binding sites among otherwise nonconserved sequence (see boxes, Fig. 2D and Supplemental Fig. 3). Using the Cartwheel program, we defined threshold cutoffs for matches to PWMs such that conserved binding sites were found and sites that were likely to appear in the sequence randomly were rejected (see Table 1 and Methods; Brown et al., 2005). These conditions were used for all of the putative binding site sequences except Snail, as the binding data for this factor is not as well defined as for the others (see Methods). Using this program we found only one Dorsal binding site is conserved, in sequence and position, throughout the homologous sog cis-regulatory modules examined. One Zelda site is also conserved, in sequence and position, until the divergence of D. virilis and D. grimshawi; moreover this particular site retains close proximity to the conserved Dorsal site. A previous study of cis-regulatory modules regulated by the Dorsal transcription factor also identified conservation of one Dorsal and one linked TAGteam site within the sog cis-regulatory module (Papatsenko and Levine, 2007). In addition, we identified conserved binding sites for Snail and Schnurri. We found bHLH sites throughout the diverged sequences, however these sites were not conserved in position or exact sequence. Considering approximately 40 million years of evolution between D. melanogaster and D. virilis or D. mojavensis, the fact that specific DNA sequences are conserved suggests they were maintained by selection.
Neurogenic ectoderm specification involves dynamic expression
We examined the expression pattern of sog in embryos and document the dynamic nature of the transcript (Fig. 2A–C). At early stages, approximately nuclear cycle 9/10, sog is expressed ubiquitously throughout the embryo with strongest expression in ventral regions of embryos (Fig. 2A). At cellularization, nuclear cycle 14, sog is expressed in a broad lateral stripe, and later expression refines to encompass the mesectoderm (Fig. 2B and C); these expression patterns have been previously documented (Francois et al., 1994).
We find that expression within all three of these domains (Fig. 2A–C) is controlled by the same sog cis-regulatory module; one regulatory element controls three distinct patterns of gene expression (Fig. 3A, 3B, and data not shown). The sog cis-regulatory module drives reporter gene expression in a ubiquitous domain within embryos at early stages (Fig. 3A), and this early ubiquitous expression as well as the other patterns of expression controlled by the reporter gene are zygotic, as expression is present when the transgene is introduced paternally (data not shown). This observation suggested the hypothesis that an early ubiquitous activator may function together with Dorsal to regulate expression of sog.
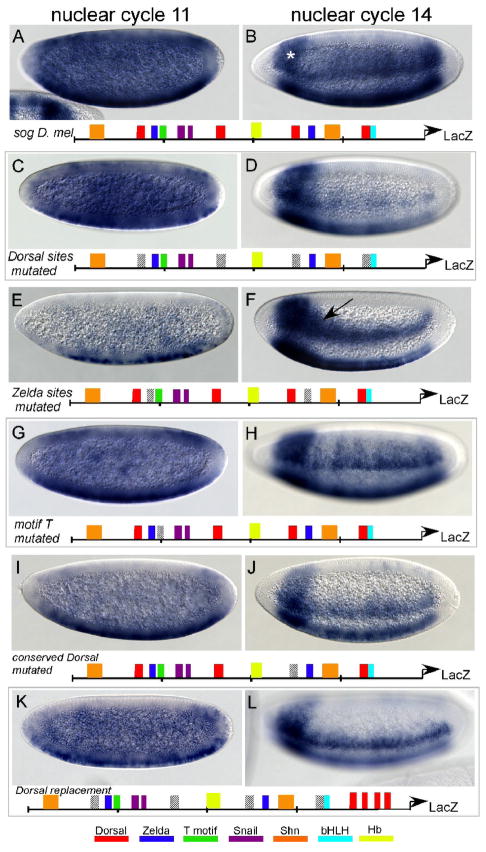
Fig. 3. Mutational analysis of conserved binding sites within the sog cis-regulatory module.
The minimal sog cis-regulatory element, and versions containing mutations introduced by site-directed mutagenesis, were fused to a LacZ reporter and integrated into the D. melanogaster genome at positions 51D and 53C4 (ZH-attp51D and attp16 respectively) by site-directed transgenesis. Embryos at nuclear cycle 9 are depicted in (A,C,E,G,I and K); embryos at nuclear cycle 14/stage 5 are depicted in (B,D,F,H,J and L). The asterisks denotes stronger expression in a band at the anterior is associated with vector sequence, present in all transgenic embryos and most prominent at cellularization (Jiang et al., 1991). Cartoons below the embryo images represent predicted transcription factor sites within the sog cis-regulatory module; the particular sites mutated in each experiment are depicted by diagonal lines.
(A,B) In situ hybridizations reveal expression of the reporter at nuclear cycle 9 (A), which is ubiquitous except for repression in the anterior and the pole cells. Expression at nuclear cycle 14 (B) is supported in lateral regions of the embryo, within a broad domain. (C,D) The four Dorsal binding sites were mutated in the cis-regulatory element. Expression supported by this mutagenized sequenced was unaffected at nuclear cycle 9 (C), but at nuclear cycle 14 (D) expression is restricted to ventral regions of the neurogenic ectoderm.
(E,F) Zelda binding sites were mutated in the cis-regulatory element. Early activation of reporter expression is absent from all but the ventral-most regions of the embryo (E) and at nuclear cycle 14 (H) expression is also diminished compared to wild-type(B). The arrow marks the region of the lateral stripe, closer to the anterior, which shows expanded expression in more dorsal regions, compared with the width of the stripe closer to the posterior.
(G,H) The predicted T motif was mutated in the cis-regulatory element. No effect was identified at nuclear cycle 9 (G), but expression of the reporter by this sequence appeared modulated along the anterior-posterior axis (i.e. “stripy”) at nuclear cycle 14 (H).
(I,J) The well- conserved Dorsal binding site (within the black box in Fig. 2D) was mutated in the cis-regulatory element. Reporter expression was examined at nuclear cycle 9 (I) and nuclear cycle 14 (J); at the later stage (J), expression is restricted to ventral regions of the neurogenic ectoderm.
(K,L) The construct with all the mutagenized Dorsal binding sites (C,D) was amended to contain four Dorsal binding sites proximal to the LacZ reporter. The expression at both stages is similar to the original construct (C,D).
Mutagenesis of sites reveals conservation of function and spatial organization
In order to dissect the core regulatory logic of the sog regulatory element, we took a candidate approach and mutated conserved binding sites within the sog cis-regulatory sequence in order to determine which sites are important for regulation. We mutated sites we thought most likely to promote expression in the neurogenic ectoderm taking into account two criteria: (1) whether the site was conserved in our comparative analysis of orthologous sog cis-regulatory sequences and (2) whether there was evidence to suggest the proteins that recognize these sites function to regulate expression along the dorsal-ventral axis. Predicted sites for Dorsal, Zelda, Schnurri/Smad, and Snail were all conserved, in sequence and relative position, in the comparisons of divergent Drosophilid sequences (Fig. 2D).
However, since our goal was to identify how activation of sog is produced in a broad lateral domain even as the levels of nuclear Dorsal diminish, we limited our analysis to putative activators of sog expression that might function during cellularization. The bHLH protein, Twist, functions with Dorsal to control expression of genes within the presumptive ventral neurogenic ectoderm, in a lateral stripe encompassing 5–7 cells (Jiang and Levine, 1993). In twist mutants embryos, sog expression remains broad in a lateral stripe ~15 cells wide. The only change in expression is that the ventral border extends slightly into ventral regions, presumably due to the fact that lower levels of Snail repressor are present (data not shown). Similarly, when the sog regulatory element is crossed into the twist mutant background, reporter expression remains broad but slightly expanded into ventral regions (data not shown). Considering this information and given that the bHLH site was not conserved in the other Drosophila species, we chose not to investigate whether Twist contributes to sog expression. Schnurri has been documented to function as a transcriptional repressor only after embryos have completed germ-band elongation (Pyrowolakis et al., 2004); therefore we did not expect Schnurri to effect the early embryonic sog expression pattern. Snail protein likely represses sog in ventral regions, because sog expression is expanded in snail mutant embryos (Kosman et al., 1991). For these reasons, we chose to focus our efforts on the requirement of documented transcriptional activators Dorsal and Zelda, as well as on the T motif, since its function was undefined.
Consistent with Dorsal playing a key role in controlling dorsal-ventral patterning, we find that Dorsal sites are required to generate a broad lateral expression pattern. When all four Dorsal binding sites in the sog regulatory element are mutated, early ubiquitous expression of sog is unperturbed, but expression of the reporter at nuclear cycle 14 is restricted to the ventral neurogenic-ectoderm forming a narrow band of expression in 3–5 cells (Fig. 3D, compare with 3B). Furthermore, within the set of sog cis-regulatory modules sequences, we found that only one of the four Dorsal binding sites was conserved, both in sequence and position, in 11 of the 12 species examined (see Fig. 2D; black box). In order to test the significance of this highly conserved Dorsal binding site, we mutated this site to examine the effect on reporter expression. We found that mutating this site alone produced a severe reduction in expression at nuclear cycle 14, which was almost as acute as mutagenesis of all four Dorsal binding sites (Fig. 3J and 3D, respectively, compare with Fig. 3B). These results suggest that Dorsal transcription factor binding to these sites is crucial for broad expanded expression into lateral regions at cellularization.
We also analyzed the requirement for Zelda to direct sog expression. TAGteam sites are recognized by Zelda, a recently described transcription factor that is maternally deposited and thus ubiquitously expressed in the early embryo (Liang et al., 2008). The presence of TAGteam sites in cis-regulatory elements has been associated with ubiquitous expression in the early embryo (De Renzis et al., 2007; ten Bosch et al., 2006). We mutagenized the two TAGteam sites (i.e. Zelda sites) present in the sog cis-regulatory element, and observed that ubiquitous early activation of the reporter is almost completely eliminated (Fig. 3E). At nuclear cycle 14, reporter gene expression is restricted to the ventral neurogenic ectoderm, as observed when Dorsal sites are mutated (Fig. 3F, compare with Fig. 3D). Our mutagenesis results indicate a role for Zelda in directing early ubiquitous expression (Fig. 3E), as well a secondary role for Zelda in controlling expression of sog in a broad lateral domain later (Fig. 3F).
Additionally, we find evidence that an unknown factor, which presumably binds to the T motif, is necessary for proper expression of the reporter in the presumptive neurogenic ectoderm. When the T motif is mutated, reporter expression is still broad, but the expression pattern exhibits modulation along the anterior-posterior axis (see Fig. 3H). This result suggests that there is regulatory input for sog from pair-rule transcription factors. It has been shown that mutations that effect dorsal-ventral patterning also influence anterior-posterior patterning both by altering nuclear movements and through transcriptional changes (Carroll et al., 1987; Keranen et al., 2006). Furthermore, transcription factors that pattern the anterior-posterior axis also bind regions in and around many genes that are dorsal-ventral axis determinants, and the reverse is also true (Li et al., 2008; Zeitlinger et al., 2007). Considering the importance of early patterning events on the ultimate specification of cells, regulatory cross talk between anterior-posterior and dorsal-ventral factors could enable synchronous expression where necessary. Nevertheless, we conclude that the transcription factor that binds to the T motif likely does not contribute to the sog expression domain along the dorsal-ventral axis (i.e. the height of the broad lateral stripe), instead this site facilitates modulation of the sog expression domain along the anterior-posterior axis.
In addition to testing the necessity of these binding sites, we wanted to test whether the presence of these sites alone was sufficient to direct expression or if spacing of sites was important. To address this question, we used the construct with mutated Dorsal sites and replaced four Dorsal binding sites proximal to the reporter. The resulting reporter expression is similar to the transgenic with mutated Dorsal sites (Fig. 3L). This result implies that distance between the Dorsal and Zelda sites (i.e. reletive spacing) is indeed important for creating a broad lateral expression domain.
Analysis of sog expression and reporters in mutant backgrounds
Similar to what we observed in the cis-regulatory construct with mutagenized Zelda cites (Fig. 3F), a refined domain of expression for sog was recently identified in Zelda mutant embryos (Liang et al., 2008) suggesting that we have indeed disrupted Zelda binding to the sog cis-regulatory module sequence. Ubiquitous expression remains in all mutagenized sog cis-regulatory module transgenic embryos (Fig. 3A,C,G, I and K) except those with mutagenized Zelda binding sites, suggesting that Zelda plays a major role in directing early expression of sog.
In dorsal mutant embryos, sog expression is completely eliminated at both early and later stages (data not shown; Francois et al., 1994). Expression of the sog cis-regulatory element reporter gene is also absent at all stages we tested in a dorsal mutant background (i.e. up to stage 6; data not shown). This complete loss of expression is much more severe than when the Dorsal binding sites are mutagenized within the cis-regulatory element (see Fig. 3C,D).
Collectively, our results suggest that Dorsal and Zelda function together to control sog expression within a broad lateral stripe. However, at early stages, the mechanism to generate ubiquitous expression remains unclear. It is conceivable that our mutagenesis experiments, which targeted high-affinity Dorsal binding sites, did not completely eliminate Dorsal binding or, alternatively, Dorsal might fulfill an additional role to indirectly influences the ability of Zelda or another factor to support sog expression (see Discussion). Nevertheless, a role for Dorsal and Zelda proteins in supporting expression is clear.
Constructing synthetic regulatory elements from putative binding sites
The results of our analyses [i.e. the identification of conserved sites (Fig. 1 and 2) as well as sites required for broad lateral expression (Fig. 3)], together, suggested that Dorsal, Zelda, and possibly T motif sites are important for sog expression. Using this newly acquired information, we designed synthetic cis-regulatory elements to attempt to reconstruct the broad pattern found in the presumptive neurogenic ectoderm (Fig. 4).
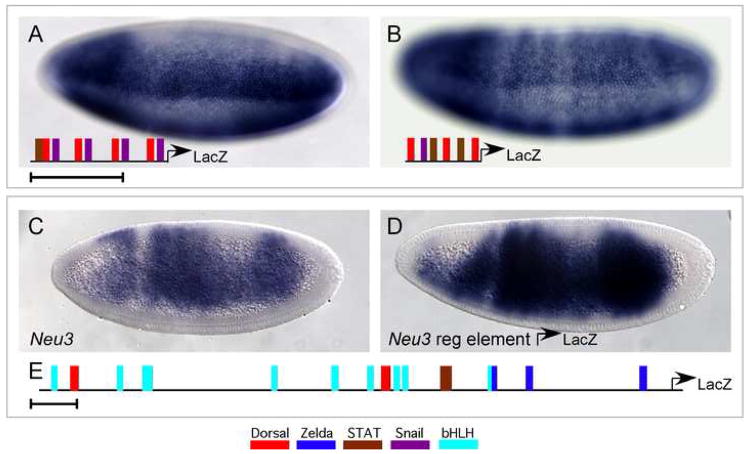
Fig. 4. Synthetic cis-regulatory elements constructed from conserved motifs and binding sites.
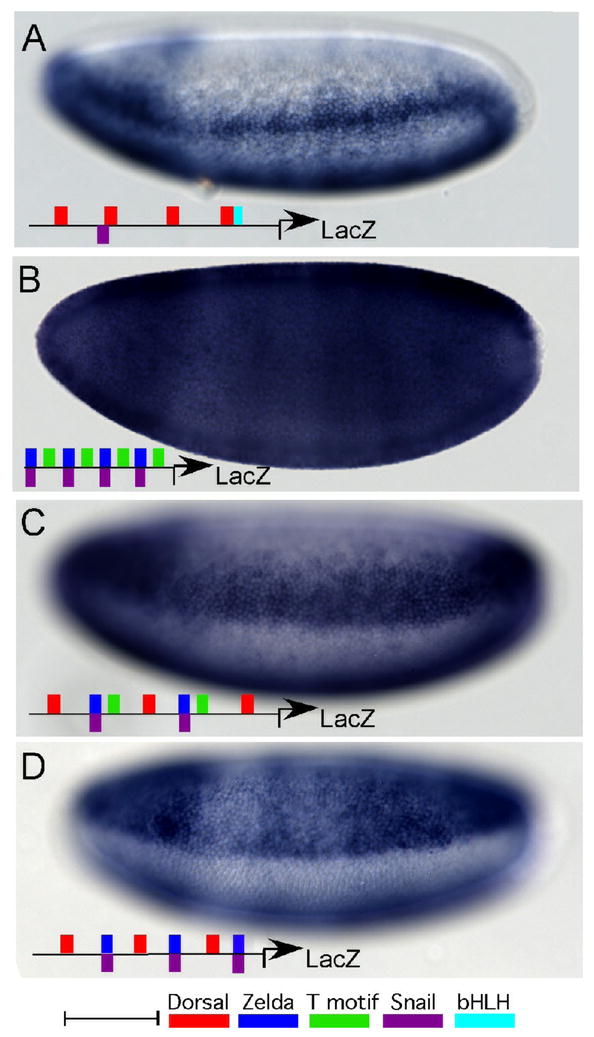
Whole mount in situ hybridizations using a riboprobe to LacZ to analyze expression supported by various synthetic reporter constructs.
(A) Native Dorsal sites taken from the minimal sog cis-regulatory element direct reporter expression in the ventral regions of the neurogenic ectoderm. The exact sequences of the 4 Dorsal binding sites from the sog cis-regulatory module were used including 5 bp of linker sequence upstream and downstream from the predicted binding site. Thus, the sites were separated by 10 bp, or one helical turn of DNA. At least one Dorsal binding site was associated with a Snail binding site, which explains the repression observed ventrally. A bHLH site is associated with another Dorsal site.
(B) Multimerized Zelda and T motif sites direct ubiquitous reporter expression. A ~20bp fragment of the endogenous sog cis-regulatory module in which Zelda and T motif sites are linked (see grey box, Fig. 2D) was multimerized so that four copies were assayed.
(C) Dorsal, Zelda, T motif, and Snail sites direct expression in a broad lateral stripe. A ~30 bp fragment of the endogenous sog cis-regulatory module in which Dorsal, Zelda, and T motif sites are closely associated (see grey box, Fig. 2D) was assayed. Two copies of this element together with one additional Dorsal site was used to construct a synthetic reporter.
(D) Multimerized Dorsal and Zelda sites direct broad lateral reporter expression. A ~31bp fragment of the endogenous sog cis-regulatory module in which Zelda and T motif sites are linked (see grey box, Fig. 2D) was multimerized so that three copies were assayed.
Scale bar represents 50 bp.
Neither Dorsal nor Zelda and T motif alone are able to support expression in a broad lateral domain. When the four native Dorsal sites from the minimal sog cis-regulatory element are used to drive reporter expression, early expression is broad, encompassing the ventral and ventral-lateral but not the dorsal-most region of the embryo (data not shown). This result suggests that Dorsal, possibly functioning with a bHLH transcription factor, is capable of generating broader expression, at least transiently, in the early embryo. Expression of this synthetic regulatory element at nuclear cycle 14 is detected in a ventral-lateral stripe, consisting of ~5 cells (Fig. 4A). This is similar to what was observed previously when the proximal element for the Twist cis-regulatory element, which includes Dorsal binding sites and Snail binding sites, was used to drive reporter expression (Jiang et al., 1991). Our result confirms that Dorsal binding sites alone are not sufficient to generate the broad lateral expression. When Zelda sites and T motif are used to direct a reporter, early expression is ubiquitous (data not shown), and expression is essentially ubiquitous at nuclear cycle 14, with slight repression in ventral regions of the embryo and some obvious anterior-posterior modulation (Fig. 4B). Although we do not know what factor binds T motif, the protein Zelda is ubiquitously expressed (Liang et al., 2008), suggesting that the expression we see is largely due to Zelda activation. Some of the predicted Snail sites, overlap with the predicted binding domain for Zelda; this likely accounts for the subtle ventral repression observed.
We multimerized the conserved sequence block containing Dorsal, Zelda, and T motif sites as well as one Snail site (delineated by gray box in Fig. 2D) to generate a synthetic cis-regulatory construct which was used to drive reporter expression. This synthetic reporter drives early ubiquitous expression (data not shown), which at nuclear cycle 14 refines to a broad lateral stripe of expression (Fig. 4C). Since mutagenizing the T motif did not appear to affect the width of the broad lateral stripe (see Fig. 3H), we tested whether Zelda and Dorsal could function without the T motif. When these two sites, Zelda and Dorsal, are multimerized, they also direct early ubiquitous reporter expression, and furthermore a broad lateral stripe is generated at nuclear cycle 14 (Fig. 4D).
If Zelda functions as an early temporal activator, this activating role might be replaceable by other activators. To test this idea, we designed other synthetic regulatory elements to direct expression in a broad lateral domain. We used the binding sites from a segment of the brinker (brk) regulatory element (Markstein et al., 2004) which contained Dorsal and Snail sites (Fig. 5A). Interestingly, the expression of this synthetic reporter encompasses a broad lateral domain, where as the entire brk cis-regulatory sequence only generated expression in a ventral-lateral domain. We identified that a site for a ubiquitous maternal activator, D-STAT (Yan et al., 1996), was introduced in the process of generating the synthetic element. Therefore, we hypothesized that this STAT site, in combination with Dorsal and Snail sites, may be responsible for directing broad lateral reporter expression. To test this hypothesis directly, STAT sites were used in place of Zelda sites in a similar synthetic background (as Fig. 4D); expression was found to be broad, but occasionally exhibits anterior-posterior modulation (Fig. 5B and data not shown). This is not surprising considering the suggestion that STAT activity is modulated along the anterior-posterior axis by phoshorylation (Shi et al., 2008). This result suggests that a more general mechanism for creating expanded expression domains that are Dorsal-dependent may rely on interactions between Dorsal and other coactivators. For instance, multiple ubiquitous or broadly expressed activators may be competent to interact with Dorsal in order to support expression within the broad lateral domain in question here (see Discussion).
Fig. 5. Identification of a novel regulatory element that functions in the neurogenic ectoderm.
(A) Dorsal sites, Snail sites and a STAT site drive expression in a broad lateral stripe. A ~20 bp element derived from the cis-regulatory module controlling expression of the brinker (brk) gene was multimerized; one STAT site was introduced in the course of cloning. (B) The conserved Dorsal site (grey box Fig. 2D) was used in a synthetic with a Snail site and two STAT sites. Expression is broad, but occasionally stripy.
In situ hybridization patterns using riboprobes specific for the genes Neu3 (C) and LacZ (D) are depicted.
(C) Neu3 is expressed in a broad lateral domain at cellularization.
(D) Expression of LacZ supported by the identified Neu3 cis-regulatory element is depicted. A schematic of the binding sites found in the putative cis-regulatory element is shown at the bottom (E). Scale bars represent 100bp.
Also of note is the fact that we observed that all of the synthetic cis-regulatory elements we generated have expanded expression domains at the anterior and posterior poles. Such expression is not seen when the sog cis-regulatory module drives reporter expression, nor when sog mRNA expression is observed (Fig. 4, compare with Fig. 2B and 3). This result supports the idea that other transcription factor(s), functioning along the anterior-posterior axis, work to refine sog expression. In this particular case, the factor may function downstream of the terminal signaling cascade.
Flexibility of regulatory inputs provides insight for finding additional neurogenic ectoderm-specific regulatory elements
Previous attempts to identify cis-regulatory elements that function in the neurogenic ectoderm focused on the identification of clusters of high-affinity Dorsal binding sites; the hypothesis was that multiple high-affinity Dorsal binding sites could support expression even where levels of nuclear Dorsal are quite low as is the case in dorsal regions of the neurogenic ectoderm (Markstein et al., 2002; Stathopoulos et al., 2002). This approach was successful in finding both sog and ths cis-regulatory elements. Yet the cis-regulatory elements that drive expression of other genes expressed in a broad lateral expression domain (Neu3: Fig. 5, SoxN: Supplemental Figure 4, and pyramus:Stathopoulos et al., 2002) could not be found in this manner. We hypothesized that one reason that the respective cis-regulatory elements have remained elusive is that multiple mechanisms may exist to support activation within a broad lateral domain of the early embryo.
In vivo binding data for Dorsal, Twist, and Snail transcription factors has facilitated the prediction of hundreds of cis-regulatory regions based on genome-wide occupancy of these factors (Zeitlinger et al., 2007; A. Ozdemir and A. Stathopoulos, unpublished observation). Clusters of high-affinity Dorsal binding sites were not identified within the genomic sequences defined by the ChIP-chip analyses near any of the genes in question. However, we scanned the DNA sequences which were bound by the transcription factors in the ChIP studies and found several candidate regions that contained Dorsal as well as Zelda binding sites in proximity to the genes SoxNeuro (SoxN), pyramus, and Neu3. Although we tested four putative cis-regulatory elements, we validated only one regulatory region.
A ~2kB fragment of genomic sequence from within an intron of the Neu3 gene supports expression of a reporter gene in a domain similar to that of Neu3 mRNA expression (Fig. 5D, compare with 5C). This regulatory region contains two weak Dorsal binding sites, a STAT site, three Zelda sites, as well as several bHLH binding sites (Fig. 5E). A comparison of putative homologous regulatory regions revealed little conservation of sequence (see Supplemental Fig. 5 for sequences and alignment). Dorsal sites are present in many of the homologous sequences from other Drosophilids, though the relative positions of these sites have changed, and their PWM scores were poor. One Zelda site appears to be conserved, both in sequence and position, but the distance between this site and the nearest Dorsal site is 198 bp, which is further than the distance in the replacement experiment (Fig. 3K, L) suggestion that they are not able to function to generate broad lateral expression in this regulatory element. The STAT sites are also present in some of the other Drosophilid species, but their location is variable. This analysis demonstrates that multiple, high-affinity Dorsal binding sites are not required to support expression in a broad lateral domain within the early embryo, but instead provides further evidence suggesting that additional activators are functioning to drive expression in this domain.
DISCUSSION
Even limited sequence conservation within cis-regulatory modules can provide insights into the underlying regulatory logic
Through a comparative analysis of orthologous sog cis-regulatory modules from twelve Drosophilid species, we identify core regulatory elements conserved in these sequences. Considerable binding site turnover has occurred during the approximately 40 million years of evolution, yet some sequences are conserved (see Fig. 2). This observation supported the hypotheses we investigated in this work which are, 1) that conserved sequences are functionally required and, 2) that variable architectures might generate the same or similar patterns of expression. Surprisingly, despite the opportunity for binding site turnover during the course of evolution, the sog regulatory regions from D. virilis can still be interpreted faithfully when used to drive reporter expression in D. melanogaster. We conclude from these experiments, despite flexibility in the cis-regulatory element structure, regulatory logic has been conserved during evolution of the cis-regulatory module sequences to support sog expression.
Though this comparative analysis identified limited sequence homology, we allowed what sequence conservation was present to guide our efforts to examine the core regulatory elements required for patterning the neurogenic ectoderm. Using site-directed mutagenesis to eliminate sites within the sog cis-regulatory sequence, we obtain results which suggest that Dorsal functions together with the ubiquitous activator Zelda to control sog expression within the neurogenic ectoderm (Fig. 3). Furthermore, we constructed synthetic cis-regulatory elements, consisting of Dorsal and Zelda or Dorsal and D-STAT sites, which are both able to support expression in the broad lateral domain of Drosophila early embryo (Fig. 4 and Fig. 5). From these results we conclude that broad lateral expression is achieved by a combination of Dorsal sites and sites for the ubiquitous activator Zelda, which suggests that a more general mechanism to create broad expression may involve interactions between Dorsal and other broadly expressed transcription factors.
Dorsal functions with distinct transcriptional activators to support expression along the dorsal-ventral axis
Our mutagenesis and mutant analysis results demonstrate that Dorsal and Zelda support expression of sog along the dorsal-ventral axis (Figure 3 and data not shown). In the absence of Dorsal protein, expression of sog is gone; however when Dorsal binding sites were mutagenized, weak ventral-lateral reporter expression remains that could be due to unknown Dorsal binding sites that were not detected by our PWM searches or due to input from another transcription factor (Fig. 3D). In the absence of Zelda binding sites or in Zelda mutants, expression is slightly broader than when Dorsal sites are eliminated (Fig. 3F, compare with 3D; and Liang et al., 2008). This residual expression could be due to Dorsal and/or other transcription factor (e.g. bHLH) functioning to direct expression, in a Zelda-independent manner, within the ventral-neurogenic ectoderm; however, our data suggests that Twist is not likely involved, as the domain of sog expression along the dorsal-ventral axis is not severely affected in twist mutants (data not shown).
Previous genetic studies have demonstrated that Dorsal is required for specification of the presumptive neurogenic ectoderm, but binding sites for Dorsal alone are not sufficient to generate expression within the broad lateral domain of embryos. Dorsal has been shown to function synergistically with Twist to pattern the presumptive mesoderm and ventral neurogenic ectoderm (Jiang and Levine, 1993). Here, we present evidence that Dorsal and Zelda function synergistically to regulate expression that is able to encompass the entire presumptive neurogenic ectoderm domain. Some method of cooperativity likely exists between Dorsal and Zelda, at the level of DNA binding or downstream, and is responsible for extending the expression domain into dorsal-lateral regions of the embryos, where the levels of nuclear Dorsal are low.
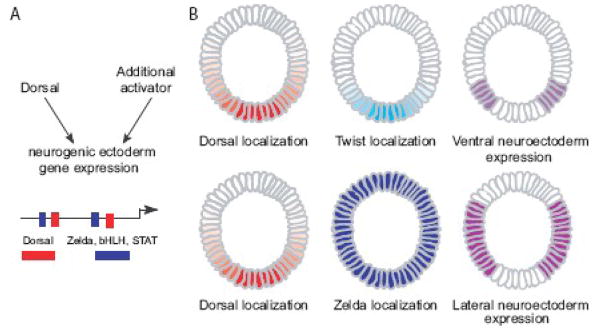
We propose that Dorsal functions as a spatial regulator in the neurogenic ectoderm and that additional transcription factors like Zelda, act as co-activators to regulate the precise onset of expression (see Fig. 6A). Furthermore, we suggest that multiple ubiquitous or broadly expressed activators may function with Dorsal to support expression in a broad lateral domain (e.g. Zelda, STAT, and bHLH transcription factors such as Daughterless (Da), see Fig. 6A). We have demonstrated that STAT binding sites can also function together with Dorsal to drive expression in a broad lateral domain. Further support for this idea includes the observation that sog as well as ths exhibit broad expression early (see Fig. 2A and Supplemental Fig. 4A). Sites for Zelda are also present in the ths cis-regulatory module, and these sites likely direct the almost-ubiquitous early expression of ths observed. Interaction of Dorsal with distinct co-activators may not only regulate the spatial domain of expression supported, but also the temporal output. Zelda along with Dorsal or a Dorsal target initiates the earliest zygotic expression detected; perhaps interactions between Dorsal and other activators facilitate expression within a broad lateral domain (or other defined pattern) at later time-points. We assert that gene expression is achieved at the intersection of the Dorsal nuclear gradient and the additional activator which could either be ubiquitous in the case of Zelda or localized in the case of Twist (Fig. 6B)
Fig. 6. Model of transcription factor participation in patterning the presumptive neurogenic ectoderm.
(A) We propose that Dorsal and Zelda both activate expression in the presumptive neurogenic ectoderm. Zelda functions to initiate ubiquitous expression while Dorsal functions primarily as a regulator of spatial expression. Neither alone is sufficient to support expression of genes like sog or ths during cellularization. We suggest that other ubiquitous or broadly expressed activators activators may function with Dorsal in a general manner to regulate expression within different domains of the presumptive neurogenic ectoderm of Drosophila early embryo. (B) Schematic of Dorsal and Twist functioning together to generate expression in the ventral neurogenic ectoderm. We propose that Dorsal and Zelda function in an analogous manner to generate broad lateral expression.
Flexibility in organization and composition of binding sites can complicate identification of co-expressed genes
Even equipped with this new knowledge, other cis-regulatory modules that support co-expression of genes SoxN, pyramus and Neu3 have proven difficult to identify. To date, SoxN and pyramus regulatory elements remain unidentified. Flexible regulatory structures could account for some of the obscurity that has been encountered in the identification of cis-regulatory modules that support expression of genes within Drosophila early embryos. Flexibility in binding site composition, orientation and number of sites has also been demonstrated in the regulation of co-expressed genes in Ciona by extensive co-expression analyses (Brown et al., 2007). Possibly the observed flux in binding site composition and arrangement provides a mechanism that facilitates the introduction of mutations, which may be selected when a fitness advantage is provided to the developing embryo.
Recently, a second regulatory element for sog located upstream of the gene was identified which also drives expression in a broad lateral stripe in the presumptive neurogenic ectoderm of cellularized embryos (Hong et al., 2008; A. Ozdemir and A. Stathopoulos, unpub. obs). This novel regulatory element as well as the known regulatory element, the intronic enhancer examined in this study, probably function together to control the full expression pattern of sog in the developing embryo. While both cis-regulatory sequences contain Dorsal and Zelda binding sites, the novel enhancer contains many more bHLH sites (L. Liberman, unpub. obs.), which is in stark contrast to the the intronic sog regulatory element, which contains only one bHLH site and exhibits very little change of expression in twist mutant embryos. This new regulatory element presents further evidence that there exist multiple solutions for the developmental problem of producing spatially and temporally regulated expression. Future experiments will address whether these early embryonic enhancers controlling the expression of the sog gene within similar domains use the same mechanism (i.e. Dorsal + Zelda cooperativity) to support expression in a broad lateral stripe or whether different mechanisms are used.
Conclusion and implications for vertebrate biology
Evolutionary comparisons of sequences from diverged species can be very useful for the dissection of underlying cis-regulatory logic, as we have shown here; yet the important variable is that the proper comparisons of sequences must be made (i.e. species of appropriate evolutionary distance) and this is not always easy to define. In vertebrate systems, analyses of cis-regulatory modules usually focus on modules identified by methods that select for high degrees of conservation, which inherently have a low amount of flexibility. Recently, the identification of ultra-conserved regions, defined as greater than 200 base-pairs of conservation within non-coding DNA sequence, was used as a criterion to identify cis-regulatory modules in the mouse (e.g. Visel et al., 2008). Arguments have been made that deciphering the underlying regulatory logic from evolutionary comparisons of sequences, when conservation is too high, is hard to interpret. However, we contend that the relevant comparisons are context-dependent. In our analysis of the sog and Neu3 cis-regulatory modules, we found only limited sequence conservation was identified in comparisons of homologous sequences isolated from D. melanogaster and other Drosophilids. In the case of the sog early embryonic regulatory element, we analyzed in this study, 71 (of 395) base-pairs of non-contiguous sequence exhibits conservation. The degree of conservation that was retained however was useful for dissecting the underlying regulatory logic.
Identifying regulatory regions with flexible structure is more challenging than scanning for a stringent set of binding sites, but it may also reveal alternative mechanisms for specification that were not previously considered. Our prediction is that studies that dissect the flexibility of cis-regulatory modules may one day provide insights to facilitate dissection of vertebrate regulatory elements in general, including ones that exhibit flexibility of sequence. It seems plausible that stringently conserved regulatory elements control gene expression of certain classes of genes, like those required for certain essential processes. Flexible regulatory architectures may provide a mechanism for generating variability throughout evolution. Ultimately it will prove useful to make evolutionary comparisons with both highly conserved sites and flexible architectures to determine how each contributes to establishment or maintenance of gene regulation.
Supplementary Material
Acknowledgments
We are grateful to Christine Rushlow for sharing unpublished information regarding Zelda, to Kate Senger and Mike Levine for sharing SELEX data, and to Eric Davidson for helpful discussions. We thank Brant Peterson and Dmitri Papatsenko for help with the evolutionary analysis of binding sites and the alignment presentation. Desirea Mecenas provided excellent technical assistance and helpful insights regarding ubiquitous activators. We are grateful to Titus Brown for his support generating position weight matrices and for introducing us to the program Cartwheel. We thank Greg Reeves, who was particularly helpful with the mutant analysis, and Sarah Payne for providing comments on the manuscript. We also thank Junhyong Kim (University of Pennsylvania) for advice regarding the in situ hybridization conditions in other Drosophilid species. This work was supported by grants to A.S. from the NIH (R01 GM077668) and the Searle Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Development. 1995;9(21):2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific {varphi}C31 integrases. Proceedings of the National Academy of Sciences. 2007;104:3312. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–60. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Brown CT, Xie Y, Davidson EH, Cameron RA. Paircomp, FamilyRelationsII and Cartwheel: tools for interspecific sequence comparison. BMC Bioinformatics. 2005;6:70. doi: 10.1186/1471-2105-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Winslow GM, Twombly VJ, Scott MP. Genes that control dorsoventral polarity affect gene expression along the anteroposterior axis of the Drosophila embryo. Development. 1987;99:327–32. doi: 10.1242/dev.99.3.327. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–18. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic regulatory systems: development and evolution. Academic Press; San Diego, Calif: 2001. [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA. A Gene-Centered C. elegans Protein-DNA Interaction Network. Cell. 2006;125:1193. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Francois V, Solloway M, O’Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 1994;8:2602–16. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G, Levine M. Evolution of the dorsal-ventral patterning network in the mosquito, Anopheles gambiae. Development. 2007;134:2415–24. doi: 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhathakurta D, Schriefer LA, Hresko MC, Waterston RH, Stormo GD. Identifying muscle regulatory elements and genes in the nematode Caenorhabditis elegans. Pac Symp Biocomput. 2002:425–36. doi: 10.1142/9789812799623_0040. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS. Shadow Enhancers as a Source of Evolutionary Novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes and Development. 1991;5:1881. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- Keranen SV, Fowlkes CC, Luengo Hendriks CL, Sudar D, Knowles DW, Malik J, Biggin MD. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: dynamics. Genome Biol. 2006;7:R124. doi: 10.1186/gb-2006-7-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Ip YT, Levine M, Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991;254:118–22. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- Lee YM, Park T, et al. Twist-mediated Activation of the NK-4 Homeobox Gene in the Visceral Mesoderm of Drosophila Requires Two Distinct Clusters of E-box Regulatory Elements. J Biol Chem. 1997;272(28):17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- Levis R, Hazelrigg T, Rubin GM. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–61. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–3. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Levine M. Decoding cis-regulatory DNAs in the Drosophila genome. Curr Opin Genet Dev. 2002;12:601–6. doi: 10.1016/s0959-437x(02)00345-3. [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proceedings of the National Academy of Sciences. 2002;99:763. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–94. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Mauhin V, Lutz Y, Dennefeld C, Alberga A. Definition of the DNA-binding site repertoire for the Drosophila transcription factor SNAIL. Nucleic Acids Res. 1993;21:3951–7. doi: 10.1093/nar/21.17.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–99. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–33. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–35. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Small S. Developmental mechanisms and cis-regulatory codes. Current Opinion in Genetics & Development. 2006;16:165. doi: 10.1016/j.gde.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine M. Computational identification of regulatory DNAs underlying animal development. Nat Meth. 2005a;2:529. doi: 10.1038/nmeth0705-529. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine M. Gene Regulatory Networks Special Feature: Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proceedings of the National Academy of Sciences. 2005b;102:4966. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Levine M. A rationale for the enhanceosome and other evolutionarily constrained enhancers. Curr Biol. 2007;17:R955–7. doi: 10.1016/j.cub.2007.09.035. [DOI] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell. 2004;7:229–40. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–99. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Rusch J, Levine M. Regulation of a dpp target gene in the Drosophila embryo. Development (Cambridge) 1997;124:303–311. doi: 10.1242/dev.124.2.303. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10:489–96. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341:331–5. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Whole-genome expression profiles identify gene batteries in Drosophila. Dev Cell. 2002;3:464–5. doi: 10.1016/s1534-5807(02)00300-3. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Whole-genome analysis of Drosophila gastrulation. Current Opinion in Genetics & Development. 2004;14:477. doi: 10.1016/j.gde.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic Regulatory Networks and Animal Development. Developmental Cell. 2005;9:449. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell. 2006;17:2356–65. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–60. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–7. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–30. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–90. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D. Computational Models for Neurogenic Gene Expression in the Drosophila Embryo. Current Biology. 2006;16:1358. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.