Abstract
The molecular mechanisms that cause organismal aging are a topic of intense scrutiny and debate. Dietary restriction extends the life span of many organisms, including yeast, and efforts are underway to understand the biochemical and genetic pathways that regulate this life span extension in model organisms. Here we describe the mechanism by which dietary restriction extends yeast chronological life span, defined as the length of time stationary yeast cells remain viable in a quiescent state. We find that aging under standard culture conditions is the result of a cell-extrinsic component that is linked to the pH of the culture medium. We identify acetic acid as a cell-extrinsic mediator of cell death during chronological aging, and demonstrate that dietary restriction, growth in a non-fermentable carbon source, or transferring cells to water increases chronological life span by reducing or eliminating extracellular acetic acid. Other life span extending environmental and genetic interventions, such as growth in high osmolarity media, deletion of SCH9 or RAS2, increase cellular resistance to acetic acid. We conclude that acetic acid induced mortality is the primary mechanism of chronological aging in yeast under standard conditions.
Keywords: aging, dietary restriction, longevity, chronological life span, acetic acid, programmed cell death
Introduction
All living systems undergo physiologic decline with age, and aging in humans is a non-modifiable risk factor for the development of many diseases. Despite decades of study and many recent advances, there has yet to emerge a consensus regarding the primary molecular cause(s) of aging. Free radical-induced oxidative damage, telomere erosion, secretion of factors by senescent cells, depletion of stem cells, mitochondrial dysfunction, DNA mutation, genomic instability, epigenetic changes and proteotoxicity have all been proposed as causal factors in aging.1-7 None of these has yet been demonstrated as a primary cause of aging in people.
The best characterized molecular mechanism of aging in any organism is the accumulation of extrachromosomal rDNA circles (ERCs) in yeast mother cells, which is one contributing cause of replicative aging in Saccharomyces cerevisiae.8 ERCs are formed by homologous recombination between adjacent rDNA repeats, and mutations that reduce rDNA recombination, such as deletion of FOB1 or overexpression of SIR2, can increase replicative life span.9,10 ERCs do not appear to cause aging in non-dividing yeast cells (referred to as chronological aging),11 and there is no evidence that aging in multicellular eukaryotes is influenced by ERCs. Thus, ERCs are likely to be a private mechanism of replicative aging in budding yeast and related organisms.
Dietary restriction (DR), defined by a reduction in nutrient availability without malnutrition, has been demonstrated to increase life span in evolutionarily divergent organisms, including yeast, nematodes, fruit flies and rodents.12,13 The mechanism(s) by which DR promotes longevity remain unknown, although recent studies have suggested that the nutrient-responsive target of rapamycin (TOR) kinase is likely to play an important role in mediating the physiological response to DR in yeast, worms and flies.14-23 Interestingly, the replicative life span of yeast cells with reduced ERC levels is dramatically extended by DR (reduced glucose in the medium) or by decreased TOR signaling,17,21 indicating that a non-ERC component of yeast replicative aging may be more relevant to aging in multicellular eukaryotes.20
Although yeast replicative aging has been more widely studied,24 substantial advances have been made in characterizing the genetic and environmental factors that modulate yeast chronological longevity.25-27 Chronological life span (CLS) is generally determined by growing cells to stationary phase in synthetic complete (SC) medium, maintaining them in a quiescent state, and monitoring viability over time.25 Viability, in this case, is defined as the ability to re-enter the cell cycle upon return to nutrient-replete conditions. Like replicative life span, CLS can be increased by DR, which is accomplished by reducing the glucose concentration from 2% to 0.5% or lower in the initial culture medium, or by transferring yeast cells to water after they have exhausted available nutrients.28-30
In addition to DR, several genetic factors involved in nutrient and stress response have been shown to modulate CLS. A screen of transposon-mutagenized yeast uncovered the nutrient-responsive Sch9 kinase and adenylate cyclase (Cyr1) as negative regulators of CLS.31 Deletion of RAS2, which acts upstream of Cyr1 also increases CLS, likely by downregulating the activity of the cAMP-dependent protein kinase (PKA).32 CLS extension from mutation of Cyr1, Sch9 or Ras2, as well as DR, involves multiple stress-responsive transcription factors and is thought to be mediated by increased expression of antioxidant enzymes and decreased oxidative stress.23,33,34 This hypothesis is also supported by prior reports that mitochondrial superoxide dismutase (Sod2) is required for normal CLS,35,36 required for full life span extension from mutation of Cyr1, Sch9 or Ras2,37,38 and sufficient to increase CLS when overexpressed simultaneously with Sod1.37
In addition to Sch9 and PKA, TOR signaling has emerged as a key factor determining CLS.39 TOR and Sch9, which is a functional ortholog of ribosomal S6 kinase,40 act cooperatively with PKA to regulate multiple downstream processes, including autophagy, protein synthesis, mitochondrial function and stress resistance in response to nutrient availability and other environmental signals.41 Thus, it is reasonable to speculate that reduced TOR signaling is likely to mediate CLS by a mechanism similar to Sch9 and PKA. Recently, Bonawitz et al.14 reported that reduced TOR signaling promotes chronological longevity by enhancing respiration and mitochondrial gene expression. Under conditions of high glucose availability, yeast produce ATP primarily via fermentation to ethanol; however, when glucose availability is low, yeast switch to a respiratory form of metabolism. Thus, a metabolic shift from fermentation to respiration would be expected under conditions of DR as well. Consistent with this, Piper and colleagues42 previously showed that pre-adaptation to respiratory growth can also promote longer CLS. This mechanism of CLS extension, at least in the case of reduced TOR signaling, is at least partially independent of Sod2.14
As part of an ongoing effort to comprehensively characterize the environmental and genetic determinants of CLS, we have examined the molecular basis for CLS extension in response to DR by glucose depletion. Our group and others have previously reported that, similar to the case for replicative life span,43-46 DR increases CLS by a mechanism that is independent of Sir2 and its homologs.29,30,47 In addition to reducing glucose availability, CLS can also be extended by growing cells in a non-fermentable carbon source or by increasing the osmolarity of the medium with either sorbitol or sodium chloride.29,30 Here we propose a unifying molecular mechanism of yeast chronological aging that accounts for these observations and also explains many of the previously reported genetic and environmental methods for increasing CLS.
Results
Medium composition determines chronological life span
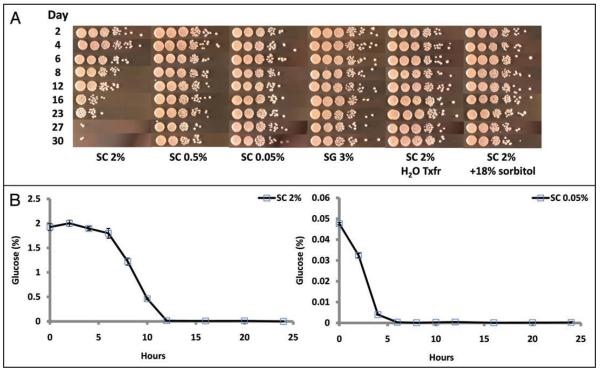
Several environmental conditions have been described which extend yeast CLS, relative to the standard culture conditions of growth in SC 2% glucose. These conditions include reducing the starting glucose concentration four- to forty-fold (SC 0.5 and 0.05% glucose, respectively), growth in a non-fermentable carbon source such as 3% glycerol (SG 3%), transferring stationary phase yeast seeded in SC 2% glucose to water at day 2 of culture, or growth in a high-osmolarity medium such as SC 2% supplemented with 18% sorbitol, a non-metabolizable monosaccharide (Fig. 1A).
Figure 1.
Growth medium conditions that extend chronological life span. (A) Relative to growth in SC 2%, reducing the glucose present in the medium, use of glycerol as the carbon source, transfer to water, or addition of sorbitol increases cell survival. On the indicated days, the aging culture was serially diluted ten-fold in rich YPD medium, and 5 ul of each dilution was spotted onto YPD agar plates and incubated for 48 hours at 30°C. A single representative biological experiment for each condition is depicted; triplicate biological replicates were performed. SC 2%, SC 0.5% and SC 0.05% = synthetic complete medium at the indicated glucose concentration; SG 3% = synthetic complete 3% glycerol; SC 2% H2O Txfr = SC 2% cells washed and transferred to distilled water at day 2; SC 2% + 18% sorbitol = SC 2% supplemented with 18% sorbitol. (B) Glucose is depleted in logarithmically growing cultures. The diploid strain BY4743 was grown overnight in YPD, and back-diluted 1:100 (starting density = ∼1.4 × 106 cells/ml) in SC 2% or SC 0.05% medium. Error bars are standard deviation of biological duplicates.
Metabolic pre-adaptation to respiration, accomplished by growth in glycerol or overexpression of the Hap4 transcription factor, has been proposed as one model to account for increased CLS.42 This model is consistent with CLS extension by DR, as cells grown in either SC 0.5%48 or SC 0.05% glucose show increased transcriptional expression of several genes known to be upregulated during the metabolic shift from fermentation to respiratory metabolism (Suppl. Fig. S1).49 It is not obvious, however, whether this model can explain CLS extension from other environmental interventions such as transfer to water at day 2 or in response to high osmolarity, and the mechanism by which pre-adaptation to respiratory growth might increase CLS has yet to be described.
Given that CLS can be modulated by changing the glucose concentration of the medium, we wished to determine the kinetics of glucose utilization in the context of the CLS experiment. Glucose consumption was measured under our standard aging conditions for control (SC 2%) and DR (SC 0.05%) cultures. In control cultures, glucose was rapidly depleted and was undetectable after 12 hours of growth (Fig. 1B). As expected, glucose depletion occurred more rapidly in DR cultures and was undetectable after 6 hours of growth. The observation that yeast can survive in glucose depleted media under both control and DR conditions for several days suggests that very early events during the exponential growth phase or the diauxic shift in low glucose medium can have long-lasting effects on the rate at which the population loses viability.
One parameter that is directly influenced by initial glucose concentration is the maximal cell density of the aging culture. Maximum viable cell density reaches 2.71 × 108 cells/ml and 4.86 × 107 cells/ml in SC 2% and SC 0.05% medium, respectively (Table 1). However, lower maximal cell density alone is not a predictor of extended CLS, as cells grown in SG 3% or SC 2% + 18% sorbitol reach a 48 hour cell density similar to the SC 2% cultures, but live substantially longer (Table 1, Fig. 1A).
Table 1.
48 hour cell density of chronologically aging yeast under varying environmental conditions
| BY4743 | Density (cells/mL) |
| SC 2% | 2.71 × 108 (±3.0%) |
| SC 0.5% | 1.59 × 108 (±0.3%) |
| SC 0.05% | 4.86 × 107 (±2.4%) |
| SG 3% | 1.59 × 108 (±3.4%) |
| SC 2% + 18% Sorb | 2.23 × 108 (±5.0%) |
| SC 2% pH 6.0 | 3.37 × 108 (±1.7%) |
| DBY746 | Density (cells/mL) |
| WT (SC 2%) | 1.95 × 108 (±1.7%) |
| sch9 (SC 2%) | 2.00 × 108 (±2.1%) |
Wild type BY4743, DBY746 or sch9Δ cells were grown for 48 hours in the indicated media, and the cell density was calculated from the A600 measurement, based on a conversion factor of 2 × 107 cells/OD unit. Parentheses denote standard deviation of three biological replicates.
A cell extrinsic factor determines the rate of chronological aging
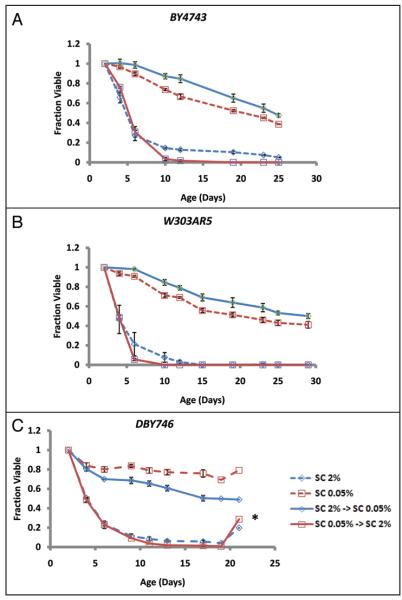
The fact that initial glucose concentration of the medium had a large effect on CLS even though glucose was undetectable in both control and DR cultures after 48 hours suggested one of two possibilities: either initial glucose abundance defines a long-lasting metabolic state that determines the subsequent rate of chronological aging (cell-intrinsic) or initial glucose abundance influences the external environment in a manner that determines the subsequent rate of chronological aging (cell-extrinsic). To differentiate between these two possibilities, cells were cultured in control (SC 2%) or DR (SC 0.05%) conditions for 48 hours, pelleted by centrifugation, and cell-free supernatants were collected. Control and DR cells were then resuspended in cell-free supernatant from a 48 hour SC 0.05% culture or SC 2% culture, respectively. Strikingly, resuspension of control cells in supernatant from the DR culture was sufficient to increase CLS to the same extent as cells maintained in SC 0.05% for the entire experiment (Fig. 2A). Likewise, resuspension of DR cells in supernatant from control cells completely suppressed the life span extension from DR, and phenocopied the life span of control cells.
Figure 2.
Chronological aging is caused by a cell-extrinsic factor. (A) BY4743, (B) W303AR5 or (C) DBY476 were grown in SC 2% or SC 0.05% medium for 2 days then resuspended in supernatant from isogenic cells grown in either in SC 2% or SC 0.05% medium for 2 days. In each case, cells grown for 2 days in SC 2% then transferred to cell free pre-conditioned medium from 2 day old SC 0.05% yeast (SC 2% → SC 0.05%) lived significantly longer than cells maintained in SC 2% medium (SC 2%). Cells grown for 2 days in SC 0.05% then transferred to cell free pre-conditioned medium from 2 day old SC 2% yeast (SC 0.05% → SC 2%) lived significantly shorter than cells maintained in SC 0.05% medium. The asterisk in (C) indicates culture re-growth, a phenomenon known as gasping that is observed in chronologically aging cultures when a small fraction of the population re-enters the cell cycle. Error bars are standard deviation of three biological replicates.
Since different yeast strains have been shown to have very different aging properties, at least with respect to replicative life span (reviewed in refs. 50–52), we wished to confirm that a similar cell-extrinsic mechanism of CLS extension by DR occurs in other yeast strains. As was the case for BY4743, cell-free supernatant from a 48 hour SC 0.05% culture was sufficient to increase the CLS of 48 hour control cells in both W303AR5 and DBY746 cells (Fig. 2B and C). Likewise, cell-free supernatant from 48 hour SC 2% cells prevented life span extension in 48 hour DR cells in both strain backgrounds. BY4743, W303AR5 and DBY746 are commonly used for both replicative and chronological aging studies in yeast and represent a diversity of genetic backgrounds. Thus, we conclude that CLS extension from DR is due to altered abundance of a cell-extrinsic aging factor and that this mechanism of chronological aging is likely to be general among different strain backgrounds. Further, since glucose is already depleted within 12 hours in both control and DR cultures (Fig. 1B), the aging factor cannot be glucose, although accumulation of the aging factor is influenced by the initial glucose concentration of the aging culture.
Acidification of the medium promotes chronological aging
Under standard glucose culture conditions yeast cells accumulate fermentation products, such as ethanol, from sugars in aerobic culture.53 This accompanies an increase in medium acidity, presumably through secretion of organic acids during fermentative metabolism. We therefore asked whether the cell extrinsic chronological aging factor might be related to differential acidification of the media in control versus DR cultures. The initial pH of the culture medium was unaffected by the glucose concentration; however, significant acidification of the medium was observed for BY4743 cells grown in SC 2% after 24 hours, reaching a final pH of 2.82 after 96 hours of outgrowth (Table 2, Suppl. Fig. S2). Unlike SC 2%, outgrowth in SC 0.5% had only a slight effect on medium pH (pH 3.67) (Table 2). Surprisingly, SC 0.05% cultures became alkaline over the first 48 hours and achieved a final pH of 5.80. Growth in 3% glycerol (SG 3%) led to an attenuation of acidification comparable to 0.5% glucose cultures. The reduced acidification of the culture environment in response to DR was not specific to BY4743 cells; a similar effect was observed in isogenic MATα haploid cells (BY4742), as well as in W303AR5 and DBY746 cells (Table 2, Suppl. Fig. S2). Thus, acidification of the culture medium is a general response that (1) correlates with starting glucose concentration, (2) is attenuated by DR and (3) correlates inversely with CLS.
Table 2.
pH of aging cultures is influenced by media composition
| SC 2% | SC 0.5% | SC 0.05% | SG 3% | SC 2% H2O Txfr | SC 2% + 18% Sorb | |
| Initial | 3.97 | 3.97 | 3.96 | 3.93 | N/a | 3.89 |
| BY4743 | 2.82 (±0.07) | 3.67 (±0.06) | 5.80 (±0.19) | 3.74 (±0.09) | 5.87 (±0.12) | 2.67 (±0.08) |
| BY4742 | 2.87 (±0.04) | 4.21 (±0.04) | 5.96 (±0.01) | |||
| W303AR5 | 2.54 (±0.07) | 3.67 (±0.03) | 6.01 (±0.02) | |||
| DBY746 | 2.90 (±0.09) | 3.91 (±0.03) | 4.75 (±0.39) | |||
| DBY746 sch9Δ | 3.31 (±0.09) |
Wild type cells were inoculated into the indicated medium and pH was measured after 96 hours of aging. Data is presented as mean pH of three biological replicates with standard deviation in parentheses.
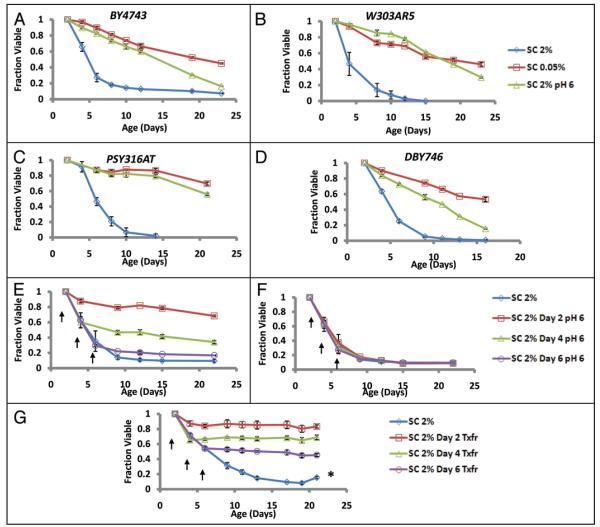
The differences observed in media pH in control versus DR cultures indicates that, although glucose is rapidly depleted from the medium in both cases, DR yeast cells are aging in a substantially different environment than control cells. This observation combined with the cell extrinsic nature of the chronological aging factor led us to consider the possibility that CLS extension observed from DR could be due to the less acidic environment experienced by these cells. To test this possibility, we asked whether the addition of a pH buffer to the culture medium could influence the CLS of yeast cells. In four different strain backgrounds, cells inoculated into SC 2% supplemented with either of two different pH 6.0 buffers (a citrate phosphate buffer, or a low salt MES buffer, see Materials and Methods), showed significant increases in CLS, relative to cells aged in unbuffered SC 2% (Figs. 3A—D; S3A and B). Cell survival in buffered SC 2% was comparable to that of unbuffered SC 0.05% medium. Unlike the case for cells grown in SC 0.05% medium, however, cells grown in buffered SC 2% media reached a similar maximal density to those in unbuffered standard medium (2.71 × 108 vs. 3.37 × 108 cells/ml, Table 2). These observations are consistent with a prior report that CLS can be increased by addition of base (NaOH) to the culture during aging.32
Figure 3.
Buffering the aging culture at pH 6 extends CLS. Yeast strains (A) BY4743, (B) W303AR5, (C) PSY316AT or (D) DBY746 were inoculated into SC 2%, SC 0.05%, or SC 2% supplemented with a citrate phosphate buffer at pH 6.0 (SC 2% pH 6, see Materials and Methods). Buffering the SC 2% medium increased CLS to an extent similar to DR. Error bars indicate the standard deviation of three biological replicates. (E) Addition of a pH 6.0 citrate phosphate buffer in SC at day 2, day 4, or day 6 of chronological age is sufficient to increase CLS, relative to (F) cells treated with an equal volume of unbuffered basic medium. (G) Transfer of cells from the aging culture to water at day 2, day 4 or day 6 also significantly increases CLS. The asterisk in (G) indicates gasping (see Fig. Legend 2). Arrows indicate age at treatments. Error bars indicate the standard deviation of three biological replicates.
Since the pH of SC 2% cultures becomes acidified within 24 hours (Fig. S2), one possibility is that exposure to acidic conditions early in life leads to a long-term increase in mortality. Alternatively, it may be the case that mortality is high only while cells are exposed to low pH. The latter hypothesis is attractive, as this could explain the observation that transferring cells to water after 48 hours increases CLS (Fig. 1). To further examine this question, we asked whether raising the pH of the culture would be sufficient to increase CLS after 2, 4 or 6, days of age in SC 2% medium. In each case, subsequent addition of a concentrated pH 6 citrate phosphate buffer led to an apparently immediate increase in survival (Fig. 3E). The slight volume change due to addition of buffering components cannot account for this effect, as addition of a volume matched control of unbuffered basic medium fails to increase CLS (Fig. 3F). Likewise, transfer of cells grown in SC 2% medium to water at day 2, 4 or 6 had a similar protective effect (Fig. 3G). Thus, we conclude that removal of the cells from a low pH environment rapidly decreases mortality and improves survival of chronologically aging yeast cells.
Dietary restriction decreases extracellular production of organic acids
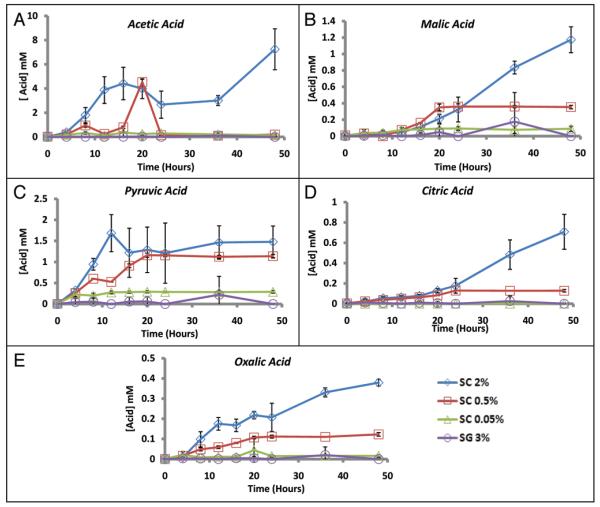
Since organic acids are known to accumulate in yeast cultures, we hypothesized that the relatively higher pH of DR cultures might be due to altered secretion of acidic metabolic by-products. Using high performance liquid chromatography (HPLC), we were able to identify and quantify the accumulation of several organic acids, including acetic, malic, citric, pyruvic, and oxalic acids in cultures grown in the presence of 2% glucose (Figs. 4; S4A and B). Consistent with our hypothesis, each of these acidic metabolites accumulated to a lesser extent in SC 0.5% and SC 0.05% cultures, relative to SC 2% cultures (Fig. 4). Similar to the organic acid profile for SC 0.05%, acid metabolites did not accumulate in cultures whose sole carbon source was 3% glycerol (Figs. 4 and S4C). The accumulation of acetic acid differed from the other organic acids in that a decrease in acetate abundance was observed following the initial increase. This is likely due to the re-utilization of acetate as a carbon source when glucose is depleted,54 and was observed in all four biological replicates with HPLC and supported using an enzymatic acetic acid assay detection kit (data not shown). Malic acid is excreted into the environment, but not re-absorbed as a citric acid cycle intermediate, presumably because intracellular malate is replenished by anapleurotic reactions.55 Regardless of these differential patterns of acid accumulation, in each case DR led to a decreased abundance of the acid product in the culture environment.
Figure 4.
Quantification of organic acid production during chronological aging. Organic acids were quantified by HPLC at the indicated age-points for yeast cells grown in SC 2%, SC 0.5%, SC 0.05% or SG 3%. Quantification was performed for (A) acetic, (B) malic, (C) pyruvic, (D) citric and (E) oxalic acids. The two peaks observed in the acetic acid profile were reproducible across multiple biological replicates and independent experiments and verified using an enzymatic acetic acid assay (Boehringer Mannheim, see Materials and Methods, and data not shown). Error bars indicate standard deviation of biological replicates (n = 4 for SC 2C%, SC 0.05% and SG 3%; n = 2 for SC 0.5%).
Acetic acid is toxic to yeast and sufficient for chronological aging
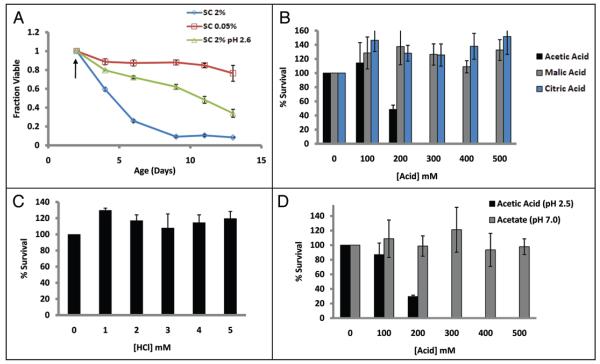
We considered two possibilities for the acidic cell-extrinsic aging factor: (1) low pH is sufficient to induce chronological aging or (2) one (or more) specific component(s) of the aged culture medium induces cellular senescence. As an initial test to determine if an acidic extracellular environment alone was sufficient to cause cell death, BY4743 cells were aged in SC 2%, SC 0.05%, or pre-grown for 48 hours in SC 0.05% and buffered to pH 2.6 using a concentrated citrate phosphate buffer. Buffering the media to pH 2.6 had only a modest inhibitory effect on DR, demonstrating that an acidic environment alone is not sufficient to suppress the CLS extension associated with DR (Fig. 5A).
Figure 5.
Acetic acid is specifically toxic to yeast cells. (A) BY4743 cells were aged in SC 2%, SC 0.05%, or pre-grown for 48 hours in SC 0.05% then buffered to pH 2.6 with the addition of a concentrated buffer (see Materials and Methods). Arrow indicates age at buffer addition. Error bars indicate the standard deviation of three biological replicates. (B) Acetic acid significantly reduced survival of 4 day old yeast cells cultured in SC 2%, but neither malic nor citric acid had a similar effect, even though the pH was similar in each case (Table S1). (C) Addition of hydrochloric acid at a concentration sufficient to decrease pH to a level comparable to 500 mM acetic acid did not reduce survival of 4 day old yeast cells. (D) Only acetic acid (pH 2.5) and not the conjugate base (pH 6.0) caused cell death in BY4742 cells. Error bars indicate standard deviation of three technical replicates.
We next examined the relative resistance of yeast cells to general acid stress (hydrochloric acid) or to acid stress in the presence of different organic acids identified in the aged culture medium. Cells were challenged with varying concentrations of hydrochloric, acetic, malic and citric acids for 200 minutes, and a dilution was plated onto rich agar medium. Surprisingly, only treatment with acetic acid showed a deleterious effect on cell survival, while the other acids tested did not result in substantial mortality at any concentration tested, even though the pH of the solutions became acidic in each case (Fig. 5B and C; Suppl. Table 1). Treatment with acetate (pH 7.0) had no effect on cell viability, indicating that only acetic acid, and not the conjugate base, is toxic (Fig. 5D). These data suggest that neither acetate nor acidic environment alone is sufficient to rapidly kill yeast cells, but the combination of both is toxic. Further, this toxicity is apparently specific to acetic acid, as other organic acids do not have a similar effect.
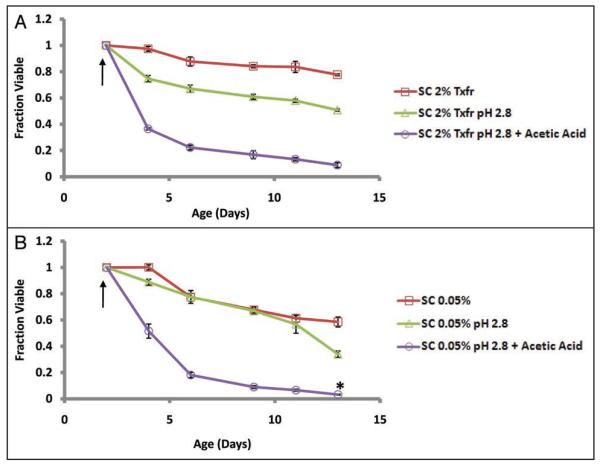
In order to determine the relevance of acetic acid toxicity in chronological aging, we next asked whether the addition of acetic acid at physiologically relevant levels is sufficient to prevent CLS extension by transfer to water at day 2. Cells from day 2 SC 2% cultures were transferred to low pH (2.8) water, and acetic acid was maintained at 10 mM for the next 36 hours to replicate the extracellular levels of this organic acid in chronologically aging cells in SC 2% (see Materials and Methods). Relative to cells maintained in water alone, cells maintained in water supplemented with 10 mM acetic acid had a significantly reduced CLS, similar to that of cells aged in SC 2% (Fig. 6A). Addition of acetic acid was also sufficient to prevent life span extension from DR (Fig. 6B). Together, these data demonstrate that (1) acetic acid in the environment of aging cells is dramatically reduced by DR or transfer to water, and (2) restoration of acetic acid to the level observed in cells aged under standard conditions is sufficient to fully suppress the CLS extension associated with these interventions.
Figure 6.
Acetic acid is sufficient to cause chronological aging. (A) 2-day old BY4743 cultures were transferred to water adjusted to pH 2.8 with HCl and maintained at a concentration of 10 mM acetic acid. Acetic acid concentration was monitored every 1–2 hrs for the first 36 hours and additional acetic acid was provided as needed to maintain a concentration of 10 mM (see Suppl. Fig. S8A). The total molar amount of acetic acid added was 108.6 mM. (B) 2-day old BY4743 cultures grown in SC 0.05% were first lowered to pH 2.8 with HCl and then supplemented with 10 mM acetic acid. Acid was monitored similarly as (A) and added as needed over the first 36 hours (see Suppl. Fig. S8B). Mortality curve for SC 0.05% pH 2.8 + acetic acid was normalized for culture growth observed during the course of adding the acid (see Suppl. Fig. S8C) by dividing the viability by the ratio of culture ODs (pH 2.8 + acetic acid/pH 2.8 alone). Error bars indicate the standard deviation of 3 biological replicates for untreated and pH 2.8 cultures, and 6 biological replicates of pH 2.8 cultures maintained at 10 mM acetic acid.
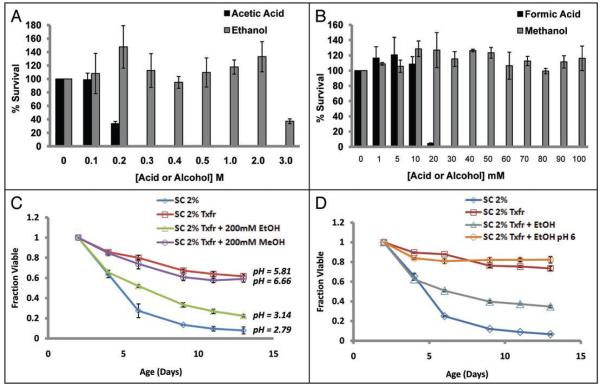
It has been previously reported that ethanol produced during fermentative growth is a cause of chronological aging in yeast.47 Ethanol produced during fermentation can be utilized as a carbon source for respiratory metabolism in the post-diauxic phase, and we observe this phenomenon in different strain backgrounds in our standard aging cultures (Suppl. Fig. S5A and B). Additionally, the sequence of ethanol metabolism requires the oxidation of ethanol to acetaldehyde and the subsequent conversion to acetate in an ATP-consuming reaction. To determine if ethanol could also be an extracellular chronological aging factor, stationary phase yeast were transiently treated (200 minutes) with either ethanol or acetic acid and viability was determined. Ethanol only affected survival at the highest concentration tested (3 M), while acetic acid had exhibited toxicity at concentrations 10-fold lower than that of ethanol (Fig. 7A). Unlike ethanol, S. cerevisiae are unable to ferment methanol to formic acid; however, a similar relative toxicity was observed in cells transiently treated with either methanol or formic acid. Methanol was not toxic at any concentration tested, while formic acid displayed even greater toxicity than acetate, likely due to its smaller size and greater ability to enter the cell (Fig. 7B). These data support the hypothesis that acetic acid, not ethanol, is the toxic product produced during chronological aging.
Figure 7.
Metabolism of ethanol to acetic acid. (A and B) Stationary phase Saccharomyces are more resistant to the ethanol and methanol than to the corresponding metabolic acid by-products, acetic acid and formic acid acidic. (C) 2-day old cultures transferred to water containing 200 mM ethanol have a shorter CLS than cultures transferred to methanol-containing water, and a concomitant acidification of the culture medium only occurs in cells receiving ethanol. (D) The short CLS of ethanol treated cultures can be rescued by buffering the culture medium to pH 6. Error bars indicate the standard deviation of three biological replicates.
Although transient treatment with ethanol or methanol showed no toxicity to yeast cells at concentrations where acetic and formic acids showed complete toxicity, we wished to directly test whether conversion of ethanol to acetic acid is sufficient to cause chronological aging. The CLS of SC 2% cells transferred to water containing 200 mM of either ethanol or methanol was determined. We reasoned that cells maintained in the presence of ethanol would convert the ethanol to acetic acid and show death kinetics similar to normally aging cells, while cells maintained in the presence of methanol would survive longer, since they would be unable to convert the methanol to formic acid. Consistent with this prediction, acidification and loss of cell viability similar to normal chronological aging was only observed in ethanol treated cells (Fig. 7C). Furthermore, the reduced viability of ethanol treated yeast could be rescued by buffering the ethanol medium to pH 6 (Fig. 7D), thus alleviating the acid stress. Conversion of ethanol to acetic acid was verified both by the change in pH and by quantification of ethanol and acetic acid in the culture supernatant 24 hours after addition of ethanol (Suppl. Fig. S5C and D). Together, these results strongly suggest that acetic acid causes chronological aging in yeast, and ethanol indirectly contributes to chronological aging by being metabolized to acetic acid.
Resistance to acetic acid induced aging is associated with long life span
Based on our model that acetic acid is the primary cause of chronological aging in yeast, we reasoned that interventions that substantially increase CLS could act by either (1) reducing the amount of acetic acid present in the environment (via higher medium pH or reduced secretion of acetic acid/acetate) or (2) increasing the intrinsic resistance of the cells to acetic acid. DR, growth in a non-fermentable carbon source such as glycerol, or transfer to water can all be explained by reduced acetic acid in the environment (Fig. 4). We therefore set out to determine the mechanism of CLS extension associated with three additional longevity-enhancing interventions: growth in high osmolarity medium, deletion of SCH9, and deletion of RAS2.
Cells grown in SC 2% supplemented with 18% sorbitol or 300 mM NaCl are long-lived relative to control cells and have a CLS comparable to cells grown in DR medium (Fig. 1A).29,30 Unlike the case with DR, medium acidification occurs to a similar extent in control and high osmolarity medium after 96 hours of culture growth (Table 2 and Suppl. Fig. S2). Therefore, reduced accumulation of acidic metabolites is unlikely to explain CLS extension by high osmolarity.
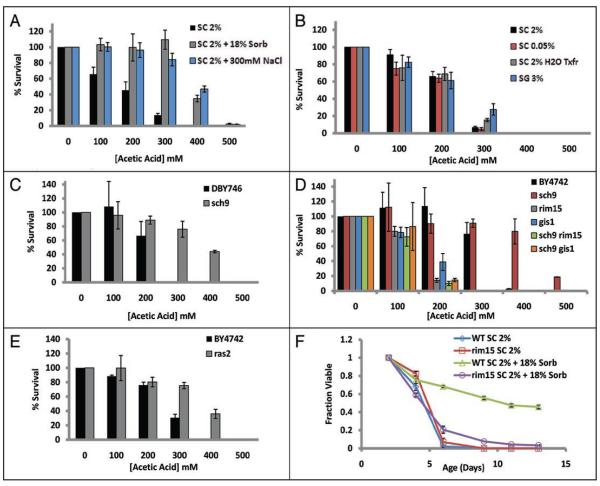
We next tested whether BY4743 cells grown in 18% sorbitol or 300 mM NaCl are resistant to acetic acid-induced death. Indeed, relative to cells cultured in SC 2% medium, cells cultured in high osmolarity medium show a significantly enhanced survival in the presence of acetic acid (Fig. 8A). As expected, cells grown in DR medium, glycerol medium, or transferred to water after 48 hours do not show resistance to acetic acid (Fig. 8B). Thus, high osmolarity increases CLS by a mechanism distinct from DR, specifically by increasing resistance to acetic acid.
Figure 8.
High osmolarity, deletion of SCH9, or deletion of RAS2 increase CLS by enhancing resistance to acetic acid. (A) Four day old BY4743 cells grown in SC 2% supplemented with 18% sorbitol are significantly more resistant to acetic acid, compared to control cells. (B) Growth in reduced glucose medium, growth in 3% glycerol, or transfer to water does not significantly increase resistance to acetate. Deletion of SCH9 increases resistance to acetic acid in (C) DBY746 and (D) BY4742 cells. Acetic acid resistance associated with deletion of SCH9 requires both Rim15 and Gis1. (E) BY4742 ras2Δ cells are resistant to acetic acid, relative to parental wild type cells. (F) CLS life span extension by growth in high osmolarity is dependent upon the protein kinase RIM15. Error bars indicate the standard deviation of three technical replicates.
Deletion of SCH9 is the best-characterized genetic model of increased CLS. Sch9 is known to regulate the response to glucose56-58 and acts in the same or overlapping genetic pathway(s) as TOR, PKA and DR to increase yeast replicative life span.17,21 Based on this, we reasoned that chronologically aging sch9Δ cells might phenocopy the reduced production of acetate associated with DR, and that this contributes to the long CLS of these cells. Somewhat surprisingly, however, the pH of aging DBY746 sch9Δ cultures drops from 3.97 to 3.31 after 96 hours (Table 2). This acidification occurs despite the fact that sch9Δ cells are slow growing (Suppl. Fig. S7). The slight reduction in acidification that occurs in the sch9Δ strain relative to the parental background suggests that a partial effect on life span extension may be explained by an attenuated secretion of fermentation products in the medium, and is consistent with reports that SCH9 has a role in repressing respiratory metabolism in the presence of glucose.59
In addition to extended CLS, deletion of SCH9 is associated with enhanced resistance to a variety of stresses, including oxidative and thermal stress.31 We therefore tested whether sch9Δ cells are also resistant to acetic acid stress. Similar to cells treated with 18% sorbitol or 300 mM NaCl, DBY746 sch9Δ cells show a significant increase in survival after treatment with acetic acid (Fig. 8C). The life span extension observed by deletion of SCH9 has been demonstrated by epistasis experiments to be dependent on the glucose-repressible protein kinase RIM15,31 and partially dependent on the transcription factor GIS1.23 Consistent with these longevity data, RIM15 and GIS1 were required for enhanced acetic acid resistance in BY4742 sch9Δ cells (Fig. 8D). Based on these observations, we conclude that RIM15 and GIS1-dependent resistance to acetic acid induced cell death is likely the primary mechanism by which deletion of SCH9 increases CLS.
Deletion of the gene RAS2 has been shown to increase CLS, and RAS2 acts upstream of PKA to repress stress resistance.23,37 Similar to sch9Δ cells, ras2Δ cells showed an increased survival following exposure to acetic acid (Fig. 8E). These data suggest that deletion of SCH9 and RAS2 both extend CLS in defined medium by increasing cellular resistance to acetic acid produced during fermentation.
Since acetic acid resistance by SCH9 deletion was shown to be dependent upon RIM15 and GIS1, we reasoned that these genes might also be involved in acid resistance associated with high osmolarity. Consistent with this idea, RIM15 is required for the corresponding increase in CLS observed in 18% sorbitol medium (Fig. 8F). Thus, we conclude that CLS extension from growth in high osmolarity medium, deletion of SCH9, or deletion of RAS2 can be explained, at least in part, by enhanced resistance to acetic acid induced death.
Discussion
Yeast has proven to be a useful model organism for studying the genetic modulation of longevity.26,27 A molecular mechanism of yeast replicative aging was described several years ago,8 although other mechanisms exist and have yet to be characterized. Here we describe a molecular mechanism of chronological aging caused by extracellular acetic acid, which accumulates in the culture medium as a by-product of fermentative metabolism. Although several other organic acids also accumulate in the culture medium during chronological aging, only acetic acid is sufficient to cause chronological aging. We conclude that a variety of interventions known to increase CLS, including DR, transfer to water, growth in glycerol, high osmolarity, deletion of SCH9, and deletion of RAS2, promote longevity by either reducing acetic acid accumulation or increasing resistance to acetic acid-induced death (Fig. 9). These findings strongly support the hypothesis that acetic acid toxicity is the predominant cause of chronological aging under the standard conditions used in a majority of prior studies.
Figure 9.
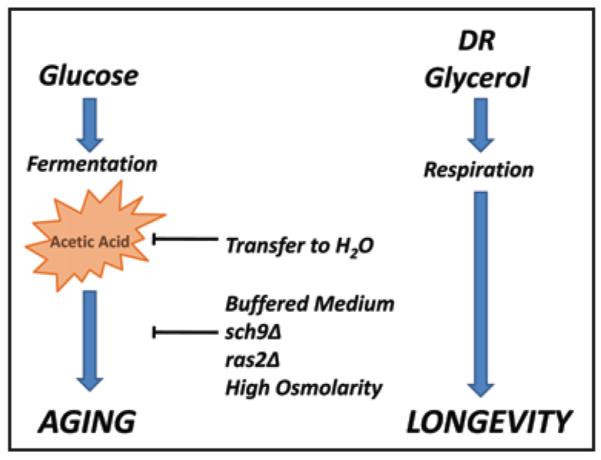
Model for acetic acid as a cause of chronological aging in yeast. Chronological life span can be increased by either (1) increasing cellular resistance to acetic acid produced as a by-product of fermentative metabolism or (2) by reducing the amount of acetic acid produced via a shift toward respiratory metabolism. This model explains many known modifiers of CLS.
Decreased acetic acid production results from a metabolic shift toward respiration
The observation that DR, or replacing the glucose carbon source with glycerol, leads to a reduced accumulation of acetic acid during chronological aging can be explained by a metabolic shift from fermentation to respiration. Acetic acid is produced as a by-product of fermentative metabolism in yeast, due to conversion of acetaldehyde to acetate.53 A similar metabolic shift toward respiration has been reported for cells with reduced TOR signaling14 or cells overexpressing the HAP4 transcription factor,48 and both of these interventions results in increased CLS.19,42 Thus, it seems likely that CLS extension from these interventions, like DR, occurs through decreased production of acetic acid in the aging culture (Fig. 9). Our results are also consistent with prior work showing that ethanol production correlates inversely with CLS,47 since metabolic utilization of ethanol results in production of acetic acid. In addition, they are consistent with a recent report that overexpression of the alcohol dehydrogenase Adh1p increases CLS,60 as this would drive the biochemical pathway towards ethanol production and away from acetic acid metabolism. Furthermore, our data show that deletion of ADH2, the alcohol dehydrogenase which primarily converts ethanol to acetaldehyde, also increases CLS (Suppl. Fig. S7A and B), supporting the assertion that genetic interventions which drive metabolism away from acetic acid production increase CLS.
It is interesting to note that, like DR, overexpression of HAP4 also increases replicative life span.48 It was initially proposed that this increase in replicative life span was due to activation of Sir2 in response to elevated respiration; however, more recent data has called this model into question.44 Given that high osmolarity61 and deletion of SCH962 also increase replicative life span, the possibility that acetic acid induced aging might contribute to replicative aging warrants further examination. However, a key difference between the two yeast aging assays is that yeast cells aging chronologically in liquid culture do so in spent media at relatively high density whereas replicatively aging yeast live in relative isolation on solid media.63 Thus, it seems unlikely that extracellular accumulation of acetic acid occurs to any great extent during replicative aging.
Acetic acid induced aging and programmed cell death
The mechanism(s) by which acetic acid kills yeast cells is an area of active study. It is generally agreed that the acetate anion is not readily taken up from the environment by yeast cells, but that protonated acetic acid can cross the plasma membrane resulting in acidification of the intracellular space.64,65 This explains our observation that buffering the pH of the aging culture is sufficient to increase CLS, even after 2, 4 or 6 days, and also explains why exposure to acetic acid is not toxic to yeast cells when the growth medium is buffered to pH 6.0. While we have shown that acetic acid is sufficient to cause chronological aging, we note that production of other organic acids during chronological aging is likely to enhance acetic acid toxicity by further lowering the pH. Thus, while acetic acid is a proximal cause of chronological aging, malic, citric, pyruvic and other organic acids are likely to play a secondary contributing role.
At concentrations appreciably higher than those observed in chronologically aging cultures, acetic acid can initiate a cell death pathway that has been likened to programmed cell death (PCD).66 PCD in response to acetic acid molecularly and morphologically resembles apoptosis, insofar as it requires cytochrome c release and depolarization of the mitochondrial membrane potential.67 Interestingly, aging yeast cells show markers consistent with apoptotic cell death and deletion of the yeast metacaspase gene YCA1 modestly increases CLS.68,69 It is therefore reasonable to speculate that acetic acid induced chronological aging induces PCD. Whether PCD occurs in all chronologically senescent cells or in only a fraction of cells remains unknown. It has also been speculated that PCD is associated with adaptive outgrowth in a minority of the cell population, which may suggest that alternative pathways of acetic acid induced cell death are at play in a majority of the senescent cells.32,70,71
A predominant hypothesis in the field has been that chronological aging is caused by oxidative damage resulting from reactive oxygen species. This hypothesis is supported by experimental evidence. For example, some chronologically long-lived cell types, such as sch9Δ, cyr1-1, and superoxide dismutase overexpressing cells, are also resistant to oxidative stress.31,37 Likewise, markers associated with oxidative stress have been observed in chronologically aging cells.68 Our data are not inconsistent with these observations, but suggest an alternative hypothesis: namely that oxidative stress is a secondary effect of acetic acid induced cell death, perhaps occurring as part of the PCD process, rather than a cause of chronologic aging. It is also possible that oxidative damage accumulates as part of the normal chronological aging process, and perhaps even contributes to acetic acid sensitivity in aging cells. Regardless, our data clearly demonstrate that the primary cause of chronological aging under standard conditions is extracellular and can be directly attributed to acetic acid.
Relevance of yeast chronological aging to aging in other organisms
Given the nature of our findings, it will be important for future studies to carefully explore the relevance of the yeast chronological aging paradigm as a model for dietary restriction specifically, and aging generally, in multicellular eukaryotes. How acetic acid induced cell death could contribute to aging of higher organisms is not immediately apparent. Nonetheless, it may be the case that underlying similarities exist. For example, accumulating evidence suggests that age-associated changes in the extracellular milieu occur as a consequence of cell senescence and can contribute to cellular dysfunction and disease in people.72 While the precise molecular mechanisms accounting for these changes may not be shared between yeast and humans, the cellular responses could be quite similar.
In this study, we have explored only the standard method for measuring CLS that has been used in a majority of prior studies. Other methods for chronologically aging yeast cells have been described or can be envisioned, including aging cells in rich media such as YPD (2% yeast extract, 1% peptone, 2% glucose), aging cells in buffered media, or transferring aged cells to water and maintaining them at high temperature. Whether one or more of these alternative methods will ultimately prove to be a better model of aging in multicellular eukaryotes than the standard conditions remains to be determined.
In support of the idea that yeast CLS shares features with aging in other organisms, we note again that several genetic and environmental factors that modulate CLS also modulate aging in other systems. DR, Sch9 (S6 kinase), and TOR all play a similar role in modulating longevity in replicatively aging yeast cells, worms and flies.73 Decreased PKA activity also increases yeast replicative life span74 and has been recently associated with increased life span in mice.75 It may be that these similarities can be accounted for by a general increase in stress resistance. Alternatively, there may be underlying metabolic or molecular changes that modulate longevity in evolutionarily divergent organisms through conserved pathways. Until the detailed mechanisms by which these factors increase life span in different organisms is known, it will be important to continue to characterize the molecular causes of aging in diverse systems.
Materials and Methods
Strains and growth conditions
Saccharomyces cerevisiae diploid strain BY4743 (MATa/α his3Δ1 leu2Δ0 ura3Δ0), and the haploid strains BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), W303AR5 (MATa ura3-1 leu2-3, 112 trp1-1 his3-11, 15 can1-100 rad5-535::RAD5 rDN1::ADE2), PSY316AT (MATα ura3-52 leu2-3, 112 his3-Δ300 ade2-101 lys2-801) and DBY746 (MATα leu2-3, 112 his3Δ1 trp1-289 ura3-52 GAL+) were used throughout this study. DBY746 and the isogenic sch9Δ mutant were a kind gift from V. Longo and P. Fabrizio. Chronological aging assays were performed using outgrowth data from a Bioscreen C MBR machine as previously described.29 All aging cultures were initiated by seeding a single colony on YPD agar into 5 mL of rich medium (YPD) overnight. A 1:100 dilution of this culture was then made into synthetic complete (SC) medium, containing glucose or glycerol at the indicated concentrations. Basic medium is 1.7 g/L Yeast Nitrogen Base (-AA/-AS) (BD Difco™) and 5 g/L (NH4)2SO4. Components of the SC medium used in this study have been described elsewhere in careful detail.29 All strain auxotrophies were compensated for with four-fold excesses of amino acids. Cultures were grown and aged in a roller drum enclosed in a water-jacketed incubator at 30°C.
For incubations in water, cultures were centrifuged at 3,000 × g for five minutes in a refrigerated Sorvall HS-4 swinging bucket rotor. The spent medium was discarded, and the cell pellets were washed twice with water before resuspension in a volume of water equal to initial culture volume. For experiments using conditioned medium, yeast was inoculated in either SC 2% or SC 0.05% media and the cells were pelleted at 3,000 × g for five minutes. The conditioned medium was harvested and filtered through 0.22 μm filters. The cells were washed twice in water before resuspension in switched medium.
For growth in buffered medium, either a citrate phosphate buffer (64.2 mM Na2HPO4 and 17.9 mM citric acid, pH 6.0), or a low salt 0.1 M MES buffer adjusted to pH 6.0 was added to the medium prior to inoculation. In cases where buffer was added at later age points, a 10X solution of the citrate phosphate buffer in basic medium was added at the appropriate volume. For the pH 2.6 citrate phosphate buffer, a 10X concentrated buffer was added to bring the final concentration of the aging medium to 10.8 mM Na2HPO4 and 44.6 mM citric acid.
Glucose, ethanol and acetic acid enzymatic detection assays
Glucose/Glucose Oxidase Assay Kit was purchased from Invitrogen Molecular Probes (cat# A22189). At designated time points, an aliquot was removed from the aging culture, the cells were pelleted at 3,000 × g for 1 min, and the supernatant was filtered through a ≤0.45 μm filter and stored at -80°C until analysis. The supernatant was diluted 1:1,500–1:2,000 for 2% glucose, and 1:30 for 0.05% glucose, and assayed twice per biological replicate (n = 2) on a Molecular Devices UVMax kinetic microplate reader at 560 nm absorbance.
Enzymatic detection of acetic acid was performed using Boehringer Mannheim r-biopharm Acetic Acid Kit (cat # 10 148 261 035), following the manufacturer’s instructions, diluting samples 1:2 prior to assaying concentration. Ethanol detection was performed using BioVision, Inc., (cat # K620-100) following the manufacturer’s instructions, diluting samples 1:50 (for SC 0.05%) or 1:250 (for SC 2% or addition of 200 mM EtOH) prior to assaying concentrations. All samples were filtered sterilized through ≤0.45 μm filter and stored at -80°C until analysis.
Analysis of gene expression by real time PCR
Cell lysates were made from logarithmically growing BY4743 cells in SC 2% and SC 0.05% (OD = 0.4–0.5) as previously described.76 RNA was purified from 5 A260 units of the cell lysate using Qiagen RNeasy Mini Kit (cat # 74104) following the manufacturer’s instructions. Reverse transcription was performed using Invitrogen SuperScript III reverse transcriptase (cat # 18080-044) using oligo d-T primer. Quantitative PCR was performed using an iCycler (Bio-Rad, Hercules, CA) and the iQ SYBR-green Supermix (cat # 170-8882) detection reagent. All starting quantities were normalized to the yeast gene PRP8, and the fold change is expressed relative to cells grown in SC 2%. RNA was purified from two biological samples each from SC 2% and SC 0.05% cultures, and all samples were analyzed in triplicate. Primer sequences for QPCR expression analysis are as follows: CYT1 5′-TCA TGC ATC CAT TAG AAG AGG TTA C-3′ and 5′-TCA TCA GGT TCG TCA TCG TAT TC; ALD2 5′-ACG ATG AAG ATG TTA CCG TTC C-3′ and 5′-TGT GAA CTG CTT TTG TTT GAA GAT A-3′; CIT1 5′-GTG GTA ACG TTT CTG CCC ATA C-3′ and 5′-AAC CAG CGG CCA AAG ATA AG-3′; FBP1 5′-AAT GGT AGC CGC TTG CTA TG-3′ and 5′-TCG CCC AAG TTT GTG TCT AAG-3′; ACS1 5′-TGG CCA AGG CTA TTC CAT TAC-3′ and 5′-CTT GCG AAC GCC CAT AGA G-3′; HAP4 5′-CGC ATC ACC ATG ACG AGT TAG-3′ and 5′-GTA CCG GCA CTA CCG TCA ATA C-3′; PRP8 5′-GCG GCC TTT ATT TAT GGT ATG TC-3′ and 5′-CTC CGA TGT CAG GGA TGT TG-3′.
High performance liquid chromatography and organic acid analysis
Cells were grown according to our CLS aging protocols, and at the indicated time points, an aliquot of 500 ul was removed from the culture tube. Cells were pelleted at 3,000 × g for 1 min, and the supernatant was removed and filtered through a 0.22 μm syringe filter. The samples were frozen at -80°C until analysis.
An Aminex HPX-87H column (BioRad cat # 125-0140) was used with a Varian Pro Star UV detector and pump. Organic acids were detected at abs 210 nm, with 4 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min at room temperature. Organic Acid Analysis Standard (BioRad cat# 125-0586) at varying concentrations was used for the purposes of assigning elution times and generating a standard curve of organic acid concentration. 50 μl of undiluted supernatant was injected once per biological replicate. Varian Star Chromatography Workstation was used for identifying elution times of the various acids and performing integrations, with all peaks and integrations verified by the user. Acetic acid concentrations were verified using an enzymatic acetic assay (Boehringer Mannheim, cat # 10 148 261 035), following the manufacturer’s instructions.
Organic acid treatments
Cultures were grown in the indicated medium to 96 hour stationary phase. Concentrated organic acids were added at either a 1:5 or 1:10 ratio of acid/culture volume to 500 μL. 0 mM treatments were carried out by adding a volume matched control of water to the culture. Cells were incubated for 200 mins at 30°C with regular mixing. After incubation, cultures were diluted 10,000 fold in liquid YPD and plated for colony forming units (CFUs). The plates were incubated at 30°C for 3 days and CFUs were counted. Percent CFUs were normalized to the 0 mM treatment. pH was measured after the 200 minute incubation with litmus paper to determine the acidity of each treatment.
Experiments to determine whether addition of physiologic levels of acetic acid (10 mM) at low pH is sufficient to accelerate chronologic aging in DR cultures were performed in the following manner. In either low glucose (SC 0.05%) or cells transferred to water, the culture pH was adjusted to pH 2.8 with HCl at 48 hours for both the control and acetic acid add-back cultures, and then 5 M acetic acid was added to bring the culture to a final concentration of 10 mM. The culture was monitored at one or two-hour intervals for 36 hours using the Boehringer Mannheim r-biopharm Acetic Acid Kit (see above) and the appropriate volume of 5 M acetic acid was added to maintain the concentration at 10 mM (see Suppl. Fig. S8 for measurements and additions). In SC 0.05%, but not in cultures transferred to water, addition of acetic acid initially increased cell density (Suppl. Fig. 8C). In cases where cell density increased as a result of treatment, the life span curve was normalized to the fold increase.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. C. Harwood, J. McKinlay and J. Hurley for technical assistance and expertise with HPLC experiments, Drs. D. Morris, D. Brackett and V. MacKay for assistance with quantitative PCR, A. Huh and D. Chandler-Brown for technical assistance, and Dr. V. Longo for helpful discussion. C.R.B. was supported in part by a grant to the University of Washington from the Howard Hughes Medical Institute through the Med into Grad Initiative, and by National Institutes of Health Training Grant P30 AG013280. This work was supported by a Pilot Project grant to M.K. from the University of Washington Nathan Shock Center for Excellence in the Basic Biology of Aging, NIH Grants R01AG025549 to B.K.K. and 1R21AG031965-01A1 to M.K., and an award to B.K.K. and M.K. from the Ellison Medical Foundation. M.K. is an Ellison Medical Foundation New Scholar in Aging.
Abbreviations
- DR
dietary restriction
- ERC
extrachromosomal rDNA circle
- CLS
chronological life span
- RLS
replicative life span
- SC
synthetic complete medium
- SG
synthetic complete glycerol medium
- HPLC
high performance liquid chromatography
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/BurtnerCC8-8-Sup.pdf
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–82. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 3.Campisi J. Senescent cells, tumor suppression and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Dolle ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002;12:1732–8. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. doi: 10.1111/j.1474-9726.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- 6.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 7.Kaeberlein M, Kennedy BK. Protein translation, 2007. Aging Cell. 2007;6:731–4. doi: 10.1111/j.1474-9726.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 8.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–42. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 9.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–55. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 10.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:9100–5. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–22. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Weindruch RH, Walford RL. The Retardation of Aging and Disease by Dietary Restriction. Thomas; Springfield IL.: 1988. [Google Scholar]
- 14.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–77. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 17.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 18.Kapahi P, Zid B. TOR pathway: linking nutrient sensing to life span. Sci Aging Knowledge Environ. 2004;2004:34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–84. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–70. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 23.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, et al. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor and Sch9. PLoS Genet. 2008;4:13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- 26.Kaeberlein M. Longevity and aging in the budding yeast. In: Conn PM, editor. Handbook of models for human aging. Elsevier Press; Boston: 2006. pp. 109–20. [Google Scholar]
- 27.Piper PW. Long-lived yeast as a model for ageing research. Yeast. 2006;23:215–26. doi: 10.1002/yea.1354. [DOI] [PubMed] [Google Scholar]
- 28.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 29.Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. J Gerontol A Biol Sci Med Sci. 2008;63:113–21. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- 30.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–62. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 31.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–90. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 32.Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–67. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng C, Fabrizio P, Ge H, Longo VD, Li LM. Inference of transcription modification in long-live yeast strains from their expression profiles. BMC Genomics. 2007;8:219. doi: 10.1186/1471-2164-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng C, Fabrizio P, Ge H, Wei M, Longo VD, Li LM. Significant and systematic expression differentiation in long-lived yeast strains. PLoS ONE. 2007;2:1095. doi: 10.1371/journal.pone.0001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longo VD, Ellerby LM, Bredesen DE, Valentine JS, Gralla EB. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J Cell Biol. 1997;137:1581–8. doi: 10.1083/jcb.137.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–80. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 37.Fabrizio P, Liou LL, Moy VN, Diaspro A, Valentine J Selverstone, Gralla EB, et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longo VD. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp Gerontol. 2003;38:807–11. doi: 10.1016/s0531-5565(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 39.Chen JC, Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr Genet. 2006;49:281–93. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- 40.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–74. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev. 2006;127:733–40. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. Faseb J. 2000;14:2135–7. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 44.Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, et al. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–14. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 47.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–67. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 48.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez P, Culotta VC, et al. Calorie restriction extends Saccharomyces cerevisiae life span by increasing respiration. Nature. 2002;418:344–8. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 49.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 50.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, et al. Substrate-specific Activation of Sirtuins by Resveratrol. J Biol Chem. 2005;280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 51.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 52.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 53.Fraenkel DG. Carbohydrate metabolism. Cold Spring Harbor Press; Cold Spring Harbor: 1982. [Google Scholar]
- 54.Wang CH, Labbe RF, Christensen BE, Cheldelin VH. Utilization of C14 labeled pyruvate and acetate by yeast. J Biol Chem. 1952;197:645–53. [PubMed] [Google Scholar]
- 55.Schwartz H, Radler F. Formation of L(-)malate by Saccharomyces cerevisiae during fermentation. Appl Microbiol Biotechnol. 1988;27:553–60. [Google Scholar]
- 56.Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, et al. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–13. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 57.Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, et al. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol. 2005;55:862–80. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 58.Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–18. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 59.Lavoie H, Whiteway M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell. 2008;7:1127–35. doi: 10.1128/EC.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reverter-Branchat G, Cabiscol E, Tamarit J, Sorolla MA, de la Torre M Angeles, Ros J. Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1. Microbiology. 2007;153:3667–76. doi: 10.1099/mic.0.2007/009340-0. [DOI] [PubMed] [Google Scholar]
- 61.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22:8056–66. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–42. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- 63.Steinkraus KA, Kaeberlein M, Kennedy B. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology. 2001;147:2635–42. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- 65.Mollapour M, Piper PW. Hog1p mitogen-activated protein kinase determines acetic acid resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:1274–80. doi: 10.1111/j.1567-1364.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 66.Ludovico P, Sousa MJ, Silva MT, Leao C, Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–15. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- 67.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2598–606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, et al. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–7. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, et al. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39:1166–73. [PubMed] [Google Scholar]
- 70.Longo VD, Mitteldorf J, Skulachev VP. Programmed and altruistic ageing. Nat Rev Genet. 2005;6:866–72. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- 71.Skulachev VP. Programmed death in yeast as adaptation? FEBS Lett. 2002;528:23–6. doi: 10.1016/s0014-5793(02)03319-7. [DOI] [PubMed] [Google Scholar]
- 72.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 73.Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–8. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 75.Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–58. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 76.MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3:478–89. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.