Summary
Among dominant neurodegenerative disorders, Huntington’s disease (HD) is perhaps the best candidate for treatment with small interfering RNAs (siRNAs) [1–9]. Invariably fatal, HD is caused by expansion of a CAG repeat in the Huntingtin gene, creating an extended polyglutamine tract that makes the Huntingtin protein toxic [10]. Silencing mutant Huntingtin mRNA should provide therapeutic benefit, but no siRNA strategy can yet distinguish among the normal and disease Huntingtin alleles and other mRNAs containing CAG repeats [11]. siRNAs targeting the disease isoform of a heterozygous single-nucleotide polymorphism (SNP) in Huntingtin provide an alternative [12–16], because such siRNAs should preserve expression of normal Huntingtin, which likely contributes to neuronal function [17–19]. We sequenced 22 predicted SNP sites in 225 human samples corresponding to HD and control subjects. We find that 48% of our patient population is heterozygous at a single SNP site; one isoform of this SNP is associated with HD. Several other SNP sites are frequently heterozygous. Consequently, five allele-specific siRNAs, corresponding to just three SNP sites, could be used to treat three-quarters of the United States and European HD patient populations. We have designed and validated selective siRNAs for the three SNP sites, laying the foundation for allele-specific RNAi therapy for HD.
Results
Current strategies for designing single nucleotide-selective siRNAs rely on SNPs that produce a purine:purine mismatch between the siRNA guide strand and the counter-selected mRNA target [12, 20]. Only 4 of the 12 nucleotide mismatches satisfy this criterion. Even when purine:purine mismatches are available, single-mismatch siRNAs vary in their selectivity, ranging in one study, for example, from 4.3- to 133-fold discrimination between the fully complementary targeted RNA and the mismatched, counter-selected RNA [12].
Sequencing and analysis of Huntingtin SNP sites in HD and control patients
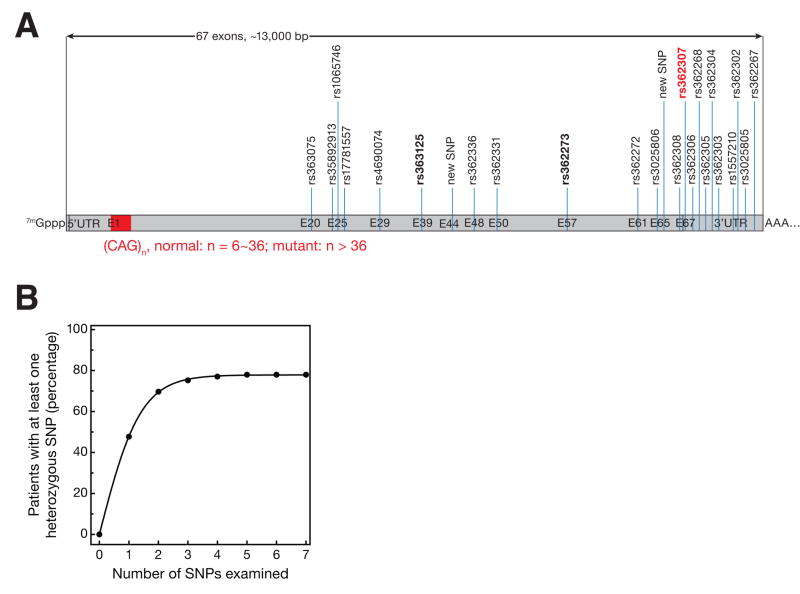
We sequenced twelve PCR amplicons spanning 22 known SNP sites in Huntingtin using genomic DNA from 109 Huntington’s disease patients and 116 non-HD controls (Figure 1A). The sequenced DNA encompassed six complete coding exons and the portion of exon 67 that contains the stop codon and part of the 3′ untranslated region (UTR). Twenty-two of the SNP sites were reported in the SNPper database [21, 22]. Four of these reported SNP sites were present only as a single isoform in our population. We identified an additional two sites by resequencing exons 2–67 in the Huntingtin locus from six HD patient samples. Table 1 reports the frequency of heterozygosity for each SNP site for patient and control DNA.
Figure 1.
Analysis of SNPs in the human Huntingtin mRNA. (A) We sequenced PCR amplicons from genomic DNA from 109 HD patients and 116 controls spanning 22 SNP sites within the Huntingtin mRNA. The SNP at nucleotide 9,633 (rs362307, shown in red) is associated with HD and sites for which we have designed siRNAs are in bold. (B) The maximum percentage of patients to have at least one heterozygous SNP using any combination of 1 to 7 SNPs was calculated using the experimentally determined frequency of heterozygosity for the SNP sites in our study. Three SNPs cover ~75% of the patient population analyzed here.
Table 1.
Frequency of Heterozygosity for 24 SNP sites in the Huntingtin mRNA. The SNP sites for which we tested siRNAs are in bold; the SNP associated with HD is shaded.
| Location in mRNA (position, nt) | Reference Number | Percent heterozygosity | |
|---|---|---|---|
| Controls | HD patients | ||
| ORF, exon 20 (2822) | rs363075 | G/A, 10.3% (G/G, 89.7%) | G/A, 12.8% (G/G, 86.2%; A/A 0.9%) |
| ORF, exon 25 (3335) | rs35892913 | G/A, 10.3% (G/G, 89.7%) | G/A, 13.0% (G/G, 86.1%; A/A, 0.9%) |
| ORF, exon 25 (3389) | rs1065746 | G/C, 0% (G/G 100%) | G/C, 0.9% (G/G 99.1%) |
| ORF, exon 25 (3418) | rs17781557 | T/G, 12.9% (T/T, 87.1%) | T/G, 1.9% (T/T, 98.1%) |
| ORF, exon 29 (3946) | rs4690074 | C/T, 37.9% (C/C, 50.9%; T/T, 11.2) | C/T, 35.8% (C/C, 59.6%; T/T, 4.6%) |
| ORF, exon 39 (5304) | rs363125 | C/A, 17.5% (C/C, 79.0%; A/A, 3.5%) | C/A, 11.0% (C/C, 87.2%; A/A, 1.8%) |
| ORF, exon 44 (6150) | exon 44 (new) | G/A, 0% (G/G, 100%) | G/A, 2.8% (G/G, 97.2%) |
| ORF, exon 48 (6736) | rs362336 | G/A, 38.7% (G/G, 49.6%; A/A, 11.7%) | G/A, 37.4% (G/G, 57.9%; A/A, 4.7%) |
| ORF, exon 50 (7070) | rs362331 | T/C, 45.7% (T/T, 31.0%; C/C, 23.3%) | T/C, 39.4% (T/T, 49.5%; C/C, 11.0%) |
| ORF, exon 57 (7942) | rs362273 | A/G, 40.3% (A/A, 48.2%; G/G, 11.4%) | A/G, 35.2% (A/A, 60.2%; G/G, 4.6%) |
| ORF, exon 61 (8501) | rs362272 | G/A, 37.1% (G/G, 51.7%; A/A, 11.2%) | G/A, 36.1% (G/G, 59.3%; A/A, 4.6%) |
| ORF, exon 65 (9053) | rs3025806 | A/T 0% (C/C, 100%) | A/T 0% (C/C, 100%) |
| ORF, exon 65 (9175) | exon 65 (new) | G/A, 2.3% (G/G, 97.7%) | G/A, 0% (G/G, 100%) |
| ORF, exon 67 (9523) | rs362308 | T/C, 0% (T/T, 100%) | T/C, 0% (T/T, 100%) |
| 3′ UTR, exon 67 (9633) | rs362307 | C/T, 13.0% (C/C, 87.0%) | C/T, 48.6% (C/C, 49.5%; T/T 1.9%) |
| 3′ UTR, exon 67 (9888) | rs362306 | G/A, 36.0% (G/G, 52.6%; A/A, 11.4%) | G/A, 35.8% (G/G, 59.6%; A/A, 4.6%) |
| 3′ UTR, exon 67 (9936) | rs362268 | C/G, 36.8% (C/C, 50.0%; G/G 13.2%) | C/G, 35.8% (C/C, 59.6%; G/G, 4.6%) |
| 3′ UTR, exon 67 (9948) | rs362305 | C/G, 20.2% (C/C, 78.1%; G/G 1.8%) | C/G, 11.9% (C/C, 85.3%; G/G, 2.8%) |
| 3′ UTR, exon 67 (10060) | rs362304 | C/A, 22.8% (C/C, 73.7%; A/A, 3.5%) | C/A, 11.9% (C/C, 85.3%; AA, 2.8%) |
| 3′ UTR, exon 67 (10095) | rs362303 | C/T, 18.4% (C/C, 79.8%; T/T, 1.8%) | C/A, 11.9% (C/C, 85.3%; T/T, 2.8%) |
| 3′-UTR, exon 67 (10704) | rs1557210 | C/T, 0% (C/C 100%) | C/T, 0% (C/C 100%) |
| 3′ UTR, exon 67 (10708) | rs362302 | C/T, 4.3% (C/C, 95.7%) | C/T, 0% (C/C, 100%) |
| 3′-UTR, exon 67 (10796) | rs3025805 | G/T, 0% (G/G 100%) | G/T, 0% (G/G 100%) |
| 3′ UTR, exon 67 (11006) | rs362267 | C/T, 36.2% (C/C, 52.6%; T/T,11.2%) | C/T, 35.5% (C/C, 59.8%; T/T, 4.7%) |
Of the 24 SNPs, rs362307 at nt 9,633 (exon 67) of the mRNA was significantly associated with HD (p = 0.0000523). After Bonferroni correction for multiple testing, the association remained significant (p = 0.000890). More than 48% of the HD patients we examined—which are believed to be representative of the US and European patient pool—were heterozygous at this site (Table 1). The U isoform of the rs362307 SNP comprised 26% of Huntingtin alleles among the patients we tested, but in only 6% of alleles among the controls. This finding suggests that a single, allele-specific siRNA selectively targeting the U mRNA isoform of this SNP could be used to treat nearly half of this patient population. To confirm our statistical analysis, we used a previously reported method to determine the rs362307 SNP isoform linked to the CAG repeat expansion allele [23] for 16 patient blood samples. Eight out of the 16 patients were heterozygous at this site; of the eight, the U isoform was linked to the expanded CAG repeat for seven patients (Table S1). We conclude that the U isoform of this SNP is associated with the disease allele of Huntingtin mRNA.
An additional two SNPs achieve patient coverage > 75%
Eight other SNP sites were each heterozygous in > 33% of our patient population, but did not show a statistically significant association with HD. Because no particular isoform of these SNPs is associated with HD in our patient population, each SNP site requires two distinct, isoform-selective siRNAs. We calculated the maximum coverage (i.e., the number of patients with at least one heterozygous SNP site) for all possible combinations of 1 to 7 SNPs. Adding two additional SNP sites covered ~75% of our patient population. Using four or more SNP sites as potential targets for siRNA therapy is not predicted to provide much additional benefit: using even seven SNP sites achieves < 80% coverage, but would require 13 isoform-selective siRNAs (Figure 1B).
Development of allele-specific siRNAs
The HD-associated SNP site at position 9,633 of the Huntingtin mRNA does not fall into the category of SNPs predicted to be readily amenable to selective targeting, because it does not create a purine:purine mismatch between siRNA and mRNA [12, 20]. However, our analysis of Huntingtin SNPs in HD patients and controls (Figure 1B and Table 1) suggests that a practicable RNA silencing therapy for HD requires an siRNA that targets the disease isoform at this site, but spares the normal Huntingtin mRNA. To this end, we designed siRNAs targeting the U isoform of the position 9,633 SNP. We tested both the efficacy and selectivity of the siRNAs in cultured human HeLa cells co-transfected with the siRNA and luciferase reporters containing in their 3′ UTRs either the U or C isoform of the sequence containing the SNP. In our hands, such luciferase reporter assays are good predictors of the efficacy and selectivity of siRNAs for endogenous mRNA targets. Figures S1 and S2 present an example using a pair of siRNAs—one fully matched and one bearing a position 10 (P10) mismatch—that targets a SNP site (rs363125) in endogenous Huntingtin mRNA in HeLa cells. Previous work has shown that such SNP selective siRNAs can reduce mutant Huntingtin levels while leaving normal Huntingtin intact [24].
siRNAs whose guide strand was fully matched to the U isoform, which is associated with HD, but mismatched at position 10 or position 16 to the C isoform, were functional, but failed to discriminate between U and C reporter mRNAs (Figure S3A). (siRNAs that bear purine:pyrimidine mismatches to their counter-selected targets generally show poor discrimination [12].) We also tested single mismatches at positions 2 through 9 (Table S2), but found that all of these were less specific than the most selective position 10 + seed mismatch. Double-mismatch strategies based on a position 16 mismatch with the counter-selected isoform had very low activity (Table S2).
Previous work has shown that adding a second mismatch can improve the ability of siRNA to discriminate between alleles [25]. We reasoned that adding a mismatch in the seed sequence of the siRNA might sufficiently destabilize our siRNA so that the doubly mismatched siRNA would lose its ability to silence the wild-type Huntingtin mRNA, while pairing at the SNP site would allow the singly mismatched siRNA to retain silencing activity for the disease allele. Therefore, we tested doubly mismatched siRNAs combining a seed mismatch with a position 10 mismatch. We prepared siRNAs predicted to mismatch at position 10 with the normal Huntingtin mRNA and also bearing an additional mismatch to both normal and disease alleles at one of the six seed positions (2–7). Mismatches at positions 5 or 6, combined with a position 10 mismatch with the counter-selected isoform, resulted in a reduction or loss of silencing of the SNP-mismatched target, while retaining good activity against the SNP-matched target (Figure S3B).
Table 2 reports “discrimination ratios”—the ratio of the IC50 of the siRNA for the counter-selected target to the IC50 of the targeted mRNA. The P10 (SNP) + P5 siRNA (IC50P10 mismatch >20; IC50P10 match = 0.62 ± 0.43 nM) had a discrimination ratio > 32 and at 20 nM—the highest concentration tested—reduced expression of the counter-selected reporter by only 33%. The P10 + P6 siRNA achieved no appreciable reduction in expression of the mismatched reporter, even at 20 nM (IC50P10 mismatch > 20 nM), but was less effective against the matched reporter (IC50P10 match = 1.5 ± 0.31 nM), yielding a lower discrimination ratio. We often observed such a trade-off between the efficacy and the selectivity of SNP-specific siRNAs. We also designed and tested an siRNA targeting the C isoform; while it was less active than the siRNA targeting the U isoform, it selectively targeted the P10-matched allele (IC50P10 mismatch > 20; IC50P10 match = 3.2 ± 2.2 nM; Figure S3C and Table 2).
Table 2.
Validation of siRNAs designed to distinguish between matched and mismatched SNP isoforms. IC50 values are given as the average ± standard deviation for at least three independent experiments. The IC50 is reported as > 20 nM for siRNAs that failed to achieve half-maximal inhibition at the highest concentration tested.
| Reference Number |
siRNA guide strand | SNP position |
primary mismatch |
secondary mismatch position |
secondary mismatch |
IC50 (nM) |
Discrimination ratio |
|
|---|---|---|---|---|---|---|---|---|
| Match | Mismatch | |||||||
| rs363125 | 5′-agcguugaaguacugucccca-3′ | 10 | G:A | none | none | 0.17 ± 0.11 | 0.27 ± 0.25 | 1.6 |
| rs363125 | 5′-agcguugaauuacugucccca-3′ | 10 | U:C | none | none | 0.18 ± 0.09 | 0.22 ± 0.07 | 1.2 |
| rs363125 | 5′-ucuucuagcguugaaguacug-3′ | 16 | G:A | none | none | 0.36 ± 0.24 | >20 | >55 |
| rs363125 | 5′-ucuucuagcguugaauuacug-3′ | 16 | U:C | none | none | 0.74 ± 0.40 | >20 | >27 |
| rs362307 | 5′-cacaagggcgcagacuuccaa-3′ | 10 | G:U | none | none | 0.36 ± 0.04 | 0.77 ± 0.16 | 2.1 |
| rs362307 | 5′-uacaagggcacagacuuccaa-3′ | 10 | A:C | none | none | 0.16 ± 0.09 | 0.14 ± 0.10 | 0.87 |
| rs362307 | 5′-gcagggcacaagggcgcagac-3′ | 16 | G:U | none | none | 0.73 ± 0.12 | 0.72 ± 0.12 | 0.99 |
| rs362307 | 5′-ucagggcacaagggcacagac-3′ | 16 | A:C | none | none | 0.19 ± 0.02 | 0.20 ± 0.06 | 1.1 |
| rs362307 | 5′-cgcaagggcacagacuuccaa-3′ | 10 | A:C | 2 | G:U | 1.0 ± 0.35 | 1.9 ± 0.27 | 1.9 |
| rs362307 | 5′-cauaagggcacagacuuccaa-3′ | 10 | A:C | 3 | U:G | 3.0 ± 1.8 | 3.5 ± 1.6 | 1.2 |
| rs362307 | 5′-caccagggcacagacuuccaa-3′ | 10 | A:C | 4 | C:U | 1.0 ± 0.22 | 1.6 ± 1.2 | 1.6 |
| rs362307 | 5′-cacacgggcacagacuuccaa-3′ | 10 | A:C | 5 | C:U | 0.62 ± 0.43 | >20 | >32 |
| rs362307 | 5′-cacaauggcacagacuuccaa-3′ | 10 | A:C | 6 | U:C | 1.5 ± 0.31 | >20 | >13 |
| rs362307 | 5′-cacaagugcacagacuuccaa-3′ | 10 | A:C | 7 | U:C | 1.3 ± 0.51 | 5.9 ± 1.9 | 4.5 |
| rs362307 | 5′-cacaauggcgcagacuuccaa-3′ | 10 | G:U | 6 | U:C | 3.2 ± 2.2 | >20 | >6 |
| rs362273 | 5′-guugaucuguagcagcagcuu-3′ | 10 | U:G | none | none | 0.09 ± 0.14 | 0.01 ± 0.006 | 0.11 |
| rs362273 | 5′-guugaucugcagcagcagcuu-3′ | 10 | C:A | none | none | 0.12 ± 0.06 | 0.44 ± 0.11 | 3.7 |
| rs362273 | 5′-cucgggguugaucuguagcag-3′ | 16 | U:G | none | none | 0.01 ± 0.002 | 0.007 ± 0.002 | 0.70 |
| rs362273 | 5′-cucgggguugaucugcagcag-3′ | 16 | C:A | none | none | 0.01 ± 0.003 | 0.004 ± 0.001 | 0.41 |
| rs362273 | 5′-ucugaucuguagcagcagcuu-3′ | 10 | U:G | 2 | C:A | 0.01 ± 0.002 | 0.06 ± 0.008 | 5.9 |
| rs362273 | 5′-uucgaucuguagcagcagcuu-3′ | 10 | U:G | 3 | C:A | 0.02 ± 0.003 | 0.29 ± 0.04 | 15 |
| rs362273 | 5′-uuuuaucuguagcagcagcuu-3′ | 10 | U:G | 4 | U:C | 0.03 ± 0.006 | 0.37 ± 0.11 | 11 |
| rs362273 | 5′-uuugcucuguagcagcagcuu-3′ | 10 | U:G | 5 | C:U | 0.02 ± 0.003 | 0.59 ± 0.08 | 31 |
| rs362273 | 5′-uuugaccuguagcagcagcuu-3′ | 10 | U:G | 6 | C:A | 0.02 ± 0.002 | 0.06 ± 0.015 | 2.7 |
| rs362273 | 5′-uuugauuuguagcagcagcuu-3′ | 10 | U:G | 7 | U:G | 0.006 ± 0.001 | 0.10 ± 0.02 | 17 |
| rs362273 | 5′-uuugcucugcagcagcagcuu-3′ | 10 | C:A | 5 | C:U | 0.15 ± 0.04 | 0.74 ± 0.11 | 4.9 |
To cover 75% of HD patients requires siRNAs targeting additional SNPs. Because no specific nucleotide isoform of these SNP sites is associated with HD, selective siRNAs are needed for both isoforms. Our long-term strategy would be to screen patients to determine the SNP isoform associated with the expanded CAG repeat Huntingtin allele [23] and select the corresponding for therapy. As a first step toward this goal, we tested its ability to target one isoform of the SNP while minimizing silencing of the other isoform. For the SNP site rs363125, which lies at nt 5,304 (exon 39) and occurs as either an A or a C, a single mismatch was sufficient to provide a high degree of selectivity for the fully matched target for both the A (>27-fold discrimination; IC50mismatch > 20 nM; IC50match = 0.74 ± 0.40 nM) and C (IC50mismatch > 20nM; >55-fold discrimination; IC50match = 0.36 ± 0.24 nM) isoforms (Figure 2A, Figure S4, and Table 2). For a second SNP, rs362273, which lies at nt 7,942 (exon 57) in the Huntingtin mRNA and occurs as either an A or a G, the P10 (SNP) + P5 siRNA design targeting the A isoform of the SNP provided ~30-fold selectivity (IC50P10 mismatch = 0.59 ± 0.08 nM; IC50P10 match = 0.02 ± 0.003 nM), whereas the siRNA targeting the G isoform (IC50P10 mismatch = 0.74 ± 0.11 nM; IC50P10 match = 0.15 ± 0.04 nM) gave ~4.9-fold selectivity (Figure 2B, Figure S5 and Table 2).
Figure 2.
Representative data for the development of isoform-specific siRNAs targeting two additional SNP sites. (A) siRNAs mismatched at position 16 discriminated between luciferase reporter mRNAs bearing either the C or the A isoform of the rs363125 SNP site. (B) siRNAs bearing a mismatch to the SNP site at position 10 and an additional position 5 mismatch discriminated between the G and A isoforms of the rs362273 SNP site.
Discussion
Targeted reduction of mutant Huntingtin mRNA is considered an ideal strategy for treating HD. The primary obstacles to the development of such a therapy have been concerns about the number of siRNAs that would require testing in clinical trials. It is not clear if drug regulatory agencies will permit patient-specific siRNAs to be used in humans without large-scale clinical trials. Such trials are, of course, not possible if only small numbers of patients share a common SNP isoform. Our results suggest that there is sufficient heterozygosity at a small number of SNP sites among American and European HD patients to support SNP-specific siRNA therapy. Targeting just three SNPs with five siRNAs should cover the majority of HD patients in the population studied here. This is possible because of the presence of several highly heterozygous SNPs and because a single SNP isoform for SNP rs362307 is associated with HD. One siRNA targeting this HD-associated isoform should target the mutant Huntingtin allele in nearly 50% of our patient population. We have developed an siRNA that selectively targets the disease-associated isoform of this SNP in cultured human cells. In the near future, pre-clinical testing of this siRNA for efficacy, selectivity, and safety is clearly of the highest importance.
What of the ~25% of patients predicted to be beyond the reach of the five siRNAs developed here? Unfortunately, our analysis predicts that a very large number of siRNAs will be required to provide siRNA therapy for this sub-population. Adding an additional four siRNAs (for a total of nine siRNAs corresponding to 5 SNP sites) only increases the treatable patient population by 3%. A further increase in the number of siRNAs provides no real additional benefit.
Finally, we find that by using potentiating mismatches in the seed sequence, isoform-selective siRNAs can be designed for SNP sites predicted to be poor candidates for the development of allele-selective siRNAs. Our data suggest that a single siRNA directed against a SNP isoform associated with HD could be used to treat nearly half the US and European HD population. Clearly, an siRNA directed against this SNP isoform, such as the siRNA presented here, merits thorough pre-clinical validation to test its promise as a candidate therapy for HD.
Supplementary Material
Acknowledgments
We thank members of the Zamore and Aronin laboratories for encouragement, helpful discussions and comments on the manuscript. This work was supported, in part, by grants from the NIH to NA, MD, and PDZ (NS38194), to the Diabetes and Endocrinology Research Center (P30DK032520-25), and from the CHDI (High Q Foundation) to NA and PDZ,.
Footnotes
Author contributions
ELP, PDZ, and NA conceived the experiments. ELP, JS, SW, PDZ, and NA designed the experiments. ELP, LK, SW, and WL performed the experiments. ELP, LK, JS, PDZ, and NA analyzed the results. MD, BL, and J-PV contributed reagents. ELP and PDZ prepared the figures, and ELP, PDZ and NA wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 2.Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 3.Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington’s disease by siRNA. Neurosci Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Xia X, Zhou H, Huang Y, Xu Z. Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutic benefit in vivo. Neurobiol Dis. 2006;23:578–586. doi: 10.1016/j.nbd.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 6.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 9.Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 10.Group THDCR. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell . 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 11.Caplen NJ, Taylor JP, Statham VS, Tanaka F, Fire A, Morgan RA. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum Mol Genet. 2002;11:175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA That Distinguish between Genes That Differ by a Single Nucleotide. PLoS Genet. 2006;2:1307–1318. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding H, Schwarz DS, Keene A, Affar el B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 14.Dahlgren C, Zhang HY, Du Q, Grahn M, Norstedt G, Wahlestedt C, Liang Z. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller VM, Gouvion CM, Davidson BL, Paulson HL. Targeting Alzheimer’s disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auerbach W, Hurlbert MS, Hilditch-Maguire P, Wadghiri YZ, Wheeler VC, Cohen SI, Joyner AL, MacDonald ME, Turnbull DH. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 18.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 19.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 20.Dykxhoorn DM, Schlehuber LD, London IM, Lieberman J. Determinants of specific RNA interference-mediated silencing of human beta-globin alleles differing by a single nucleotide polymorphism. Proc Natl Acad Sci U S A. 2006;103:5953–5958. doi: 10.1073/pnas.0601309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riva A, Kohane IS. A SNP-centric database for the investigation of the human genome. BMC Bioinformatics. 2004;5:33. doi: 10.1186/1471-2105-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riva A, Kohane IS. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Kennington LA, Rosas HD, Hersch S, Cha JH, Zamore PD, Aronin N. Linking SNPs to CAG repeat length in Huntington’s disease patients. Nat Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Hum Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.