Abstract
Regions of heterochromatin are often found at the periphery of the mammalian nucleus, juxtaposed to the nuclear lamina. Genes in these regions are likely maintained in a transcriptionally silent state, although other locations at the nuclear periphery associated with nuclear pores are sites of active transcription. As primary components of the nuclear lamina, A- and B-type nuclear lamins are intermediate filament proteins that interact with DNA, histones and known transcriptional repressors, leading to speculation that they may promote establishment of repressive domains. However, no direct evidence of a role for nuclear lamins in transcriptional repression has been reported. Here we find that human lamin A, when expressed in yeast and cultured human cells as a fusion protein to the Gal4 DNA-binding domain (DBD), can mediate robust transcriptional repression of promoters with Gal4 binding sites. Full repression by lamin A requires both the coiled-coil rod domain and the C-terminal tail domain. In human cells, other intermediate filament proteins such as lamin B and vimentin are unable to confer robust repression as Gal4-DBD fusions, indicating that this property is specific to A-type nuclear lamins. These findings indicate that A-type lamins can promote transcriptional repression when in proximity of a promoter.
Keywords: Lamin A/C, Laminopathies, Transcription repression
Introduction
The eukaryotic nucleus is comprised of many different sub-organelle structures, including the nucleolus, the nuclear lamina, and multiple protein ‘bodies’. The genome housed within the nucleus is also organized into subdomains, such as heterochromatin and chromosome territories [1]. While it is generally accepted that the nuclear lamina functions to maintain the shape of the nucleus, it has also been proposed to play a role in the regulation of gene expression. The major components of the nuclear lamina are A- and B-type lamins [2]. The nuclear lamins are intimately associated with other nuclear lamina-associated proteins forming a scaffold-like protein network juxtaposed to the inner nuclear membrane. Nuclear lamins have been implicated in the establishment of heterochromatic regions at the nuclear periphery, although the mechanism(s) by which this occurs remains largely unknown [3].
A-type and B-type nuclear lamins are intermediate filament (IF) proteins unique to metazoans [4,5]. Like all other IF-family proteins, the nuclear lamin protein consists of three distinct domains: a central alpha-helical “rod” domain flanked by a globular amino-terminal “head” and carboxy-terminal “tail” domain. The IF monomers dimerize by the winding of the rod domains into a coiled-coil [5]. Two dimers associate laterally to form a tetramer which comprises the basic subunit for the assembly of IF polymers. Subsequent filament assembly varies between IF types with nuclear lamins preferentially forming polar head-to-tail associations in vitro. In vivo, nuclear lamin assembly is still poorly understood.
The A-type lamins are derived from the alternative splicing of a single gene, LMNA. The two major sub-types, lamin A and lamin C, are identical except at their C-termini. Unlike B-type lamins which are universally expressed, A-type lamin expression is generally restricted to differentiating or differentiated cells [6], suggesting a role for these proteins in maintaining the integrity of differentiated cell types. Mutations in the LMNA gene have been linked to several rare but debilitating developmental diseases: skeletal and cardiac myopathies, partial lipodystrophies, peripheral neuropathy, and progeroid-like syndromes, collectively termed laminopathies [7-9]. Most cases of laminopathies are associated with a single point mutation in one allele of LMNA and are autosomal dominant. The mechanisms by which mutations in LMNA result in apparently unrelated diseases affecting highly specialized tissues are still not clear.
There is increasing evidence that A-type lamins are involved in various nuclear processes through their interaction with specific nuclear factors [10-12]. One model to explain disease progression proposes that mutations in LMNA affect nuclear structure and function which consequently lead to aberrant gene expression in differentiating and/or differentiated tissues. A-type lamins may therefore coordinate the activity of differentiation- and/or tissue-specific transcription factors [13-16]. Both A- and B-type lamins are thought to have a role in establishing regions of silent chromatin at the nuclear periphery [3]. A-type lamins have been reported to interact with a number of transcriptional regulatory proteins [17-21], including the retinoblastoma protein [19,20], a known repressor that colocalizes with A-type lamins in internal nuclear foci [22,23]. A-type lamins also interact with histones and with DNA in a sequence non-specific manner [24,25].
Interactions with specific transcription factors and other lamin A-associated proteins would serve to bring lamins in close proximity to promoters. However, a direct role for A-type lamins in the regulation of gene expression has not been demonstrated. Here, we show that lamin A can function as a transcriptional repressor when tethered to a promoter using Gal4 fusion proteins in both yeast and human cells. Maximal repression requires both the rod and C-terminal globular domain of lamin A. We propose a model in which lamin A establishes regions of heterochromatin in the nucleus through the formation of higher order structures via the rod domain and interaction with transcriptional co-repressors through the C-terminal globular domain.
Materials and methods
Plasmid construction
The yeast Gal4-DNA binding domain (Gal4DBD) (1-147 aa) fusion constructs were made by recombination cloning to the pGBT-CYH vector (CEN TRP1 CYH2 ADHp-GAL4DBD). The coding sequences of human lamins A, C and B1 and human vimentin were PCR amplified using primers that added tails with 40mer homology to the pGBT-CYH vector. The domains of lamin A and vimentin were interchanged to create intermediate filament chimeras by using internal primers to the respective cDNA with 40mer tails homologous to the cDNA of other protein. Primer sequences are available on request. The linearized vector was co-transformed with the PCR products for recombination cloning in yeast.
The mammalian Gal4DBD vector was made by the insertion of Gal4DBD cDNA sequence from pGBT-CYH into the HindIII/BamHI restriction sites of pcDNA3 (Invitrogen). Primers that added 5′ BamHI and 3′ EcoRI restriction sites were used to PCR amplify the coding sequences of human lamins A, C and B1 and human vimentin. The cDNA inserts were cloned into BamHI/EcoRI restriction sites of pcDNA3.GAL4DBD vector. Lamin A/vimentin intermediate filament chimeras were made by PCR subcloning the yeast constructs.
Yeast strains and beta-galactosidase repression assay
The yeast two-hybrid bait strain PJ69-4alpha contains three GAL4-responsive reporters (relevant genotype: MATα LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) [26]. The strain MaV103 (relevant genotype: MATa GAL1::lacZ@URA3 GAL1::HIS3@LYS2) was used for quantitative lacZ reporter assays [27]. A GAL4-responsive lacZ reporter was integrated at the URA3 locus [28]. The plasmids JK1621 (4XLexop-UAS-lacZ reporter) and pLG312ΔS (UAS-lacZ reporter) were integrated to MaV103 for the LexA based repression assays [29,30].
The expression of beta-galactosidase from the lacZ reporter was assayed by hydrolysis of O-nitrophenyl-β-d-galactopyranoside (ONPG) as described previously [31]. Yeast strains were transformed with Gal4DBD- or LexA-fusion constructs and grown in selective media to log-phase. Cells were harvested by centrifugation and permeabilized with chloroform. Each assay performed in triplicate from at least three independent transformants. The results are expressed as fold repression over the empty DBD vector. Protein extracts for western blot analysis were made from the harvested cells by glass bead disruption in SDS-PAGE sample buffer (100 mM Tris, pH 6.8, 2% SDS, 2% β-mercaptoethanol, 10% glycerol). Expression of the Gal4DBD fusion constructs was detected by a mouse monoclonal antibody against Gal4DBD (Santa Cruz Biotechnology Inc., RK5C1) at a dilution of 1:5000. Rabbit polyclonal antibody against lamin A/C (Cell Signaling, 2032) was used to detect the lamin A protein at a dilution of 1:1000.
Mammalian cell culture and luciferase repression assay
The luciferase reporter G5GC6-Luc was used in the mammalian repression assays [32]. 293 T cells were transiently transfected with 1 μg of Gal4DBD fusion plasmid and 1 μg of luciferase reporter plasmid. 0.5 μg of β-galactosidase expression plasmid was cotransfected as a reference plasmid to normalize for transfection efficiency. The 293 T cells were transfected by calcium phosphate precipitation and the cells were at approximately 30% confluency in monolayer cultures in 6 well plates (BD Falcon 353046). Cells were harvested 36-48 h post transfection with luciferase/lacZ lysis buffer (25 mM Tris, pH7.8, 1 mM EDTA, 10% glycerol, 1% Triton-X and protease inhibitors). Luciferase activity in the cell lysates was assayed using britelite luminescence assay reagent (PerkinElmer Biosciences) and measured using a Victor3 Multi-label plate reader (PerkinElmer Biosciences). β-galactosidase activity was assayed by hydrolysis of ONPG and A420 measured. The relative luciferase units were calculated by normalizing the luminometer counts per second to the A420. The results are presented as fold repression over the empty DBD vector. Expression of the Gal4-DBD fusion constructs was detected by the mouse anti-Gal4DBD at a dilution of 1:5000. Lamin A was detected by rabbit anti-pan lamin A/C at a dilution of 1:1000. Nuclear and cytoplasmic fractions were prepared using ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas Inc.) according to the manufacturer’s protocols.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using a modification of conditions previously described [33]. 293 T cells were transfected as above on 10 cm tissue culture plates. 36 h after transfection, the media was aspirated and attached cells fixed with 5 ml of 1% formaldehyde (in serum-free media) for 10 min at room temperature. The reaction was quenched with 340 μl 2 M glycine. Cells were washed twice with cold phosphate-buffered saline (PBS) and harvested with 1 ml PBS into a clean microfuge tube. The fixed cells were centrifuged and resuspended in 1 ml cold ChIP buffer (0.1% deoxycholate, 1 mM EDTA, 50 mM HEPES pH 7.5, 140 mM NaCl, 1% Triton-X, 1%NP-40, 10% glycerol and protease inhibitors). The cell suspension was sonicated briefly on ice and centrifuged at full speed for 1 min. The lysate was pre-cleared with 20 μl bed volume of protein-G agarose beads (Roche) for 1 h at 4 °C. Pre-cleared chromatin was incubated with 1 μg Gal4DBD antibody for 4 h at 4 °C. Protein-G agarose beads were then added (20 μl bed volume) and incubated at 4 °C for 1 h. The immunoprecipitated complexes were collected by centrifugation and washed three times with ChIP buffer. The pellets were resuspended in 100 μl Tris-EDTA with 10 μg of proteinase K and incubated at 65 °C overnight. The immunoprecipitated DNA was phenol:chloroform extracted and ethanol precipitated. The DNA pellets were resuspended in 50 μl of water and PCR was performed using 5 μl of immunoprecipitated DNA for 22 cycles at 94 °C for 20 s, 50 °C for 20 s, and 72 °C for 20 s using the primer set previously described [33]. Three independent experiments were performed and the representative data are shown.
Indirect immunofluorescence
Immunofluorescence was performed on formaldehyde fixed cells as previously described [34]. Gal4DBD-fusion proteins were detected using mouse monoclonal antibody against Gal4DBD (Santa Cruz Biotechnology Inc., RK5C1). All images were acquired using Zeiss Axiovert 200 with ApoTome (Carl Zeiss MicroImaging, Inc.). Images have equal exposure times and are processed similarly. Representative cells were chosen for figures.
Results
Lamin A represses transcription in yeast
In an effort to identify novel lamin A-interacting proteins, we initiated a yeast two-hybrid screen using lamin A as bait. Although lamins are not present in yeast, the single-celled eukaryote has been used commonly to examine the function of mammalian transcription factors including those with no yeast counterparts [35-39]. With this goal in mind, the lamin A bait protein was constructed by fusing human lamin A to the Gal4 DNA-binding domain (Gal4DBD) and under the control of the constitutive ADH1 promoter. The yeast two-hybrid strain PJ69-4α contains three Gal4 responsive reporters [26], one of which, the GAL1-HIS3 promoter, has leaky expression. This expression is permissive for growth on media lacking histidine. For two hybrid analysis, the His3 inhibitor 3-aminotriazole (3-AT) was used at varying levels to modulate the levels of HIS3 expression necessary for yeast growth.
Surprisingly, we discovered that yeast strains expressing Gal4DBD-lamin A were unable to grow in the absence of histidine, indicative of this fusion protein having the capacity to act as a transcriptional repressor (Fig. 1A). A strain expressing the Gal4DBD alone did not confer a growth defect on -His relative to strains with no Gal4 derivative (not shown). We examined this in a second yeast two-hybrid strain, MaV103, again finding that growth was inhibited, this time in the presence of 5 mM 3-AT [40]. Further, expression of Gal4DBD-lamin A did not confer histidine auxotrophy in strains with HIS3 expression mediated by its normal endogenous promoter (not shown). These findings indicate that human lamin A may function as a transcriptional repressor in yeast.
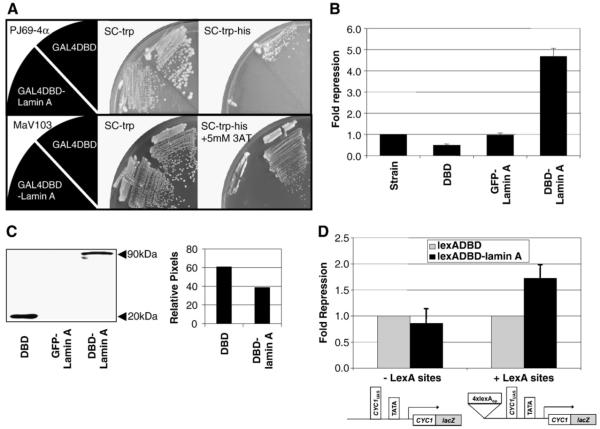
Fig. 1.
Lamin A represses transcription in yeast. (A) Plasmids expressing Gal4DBD or Gal4DBD-lamin A was transformed into the yeast 2-hybrid strains PJ69-4α (top panel) or MaV103 (lower panel). Colonies with the plasmids were selected and streaked onto synthetic drop-out plates and incubated for 3 days at 30 °C. SC-trp, synthetic drop-out plate lacking tryptophan. SC-trp-his, synthetic drop-out plate lacking tryptophan and histidine. SC-trp-his+5 mM 3AT, synthetic drop-out plate lacking tryptophan and histidine supplemented with 5 mM 3-aminotriazole. (B) Beta-galactosidase activity from an integrated lacZ reporter was quantitatively measured in yeast strains expressing Gal4DBD, GFP-lamin A or Gal4DBD-lamin A. Fusion proteins were expressed from ARS/CEN plasmids under ADH1 promoter. Results are shown as fold repression over reporter strain (n=3, mean±SD). (C) Western blot analysis was performed to verify the expression of the Gal4DBD-fusion proteins. Protein samples were separated by using SDS-4-20% PAGE and detected by immunoblotting with mouse anti-Gal4DBD. The relative protein levels were determined by counting pixel levels using NIH ImageJ and represented as a percentage of total pixels counted. (D) Repression of integrated lexA responsive lacZ reporters. JK1621 is identical to LG312ΔS except for the presence of 4xlexA operators 5′ to the UAS. LexADBD-fusion proteins were expressed from ARS/CEN plasmids under ADH1 promoter. Beta-galactosidase activity was quantitatively measured and results are shown as fold repression over LexADBD alone (n>3, mean±SD).
To quantitate levels of repression, we examined the effects of targeting Gal4DBD-lamin A to a highly expressing, integrated promoter driving expression of a lacZ reporter. β-galactosidase activity was measured and normalized to cell densities for strains expressing no Gal4 protein, Gal4DBD, Gal4DBD-lamin A or GFP-lamin A (Fig. 1B). Gal4DB and Gal4DB-lamin A were expressed at near equivalent levels (Fig. 1C). In this assay, we find that Gal4DBD-lamin A confers approximately five-fold repression relative to the strain with no Gal4 derivative and ten-fold relative to Gal4DBD. Expression of GFP-lamin A did not result in repression even though this construct was expressed at high levels (data not shown), indicating that the protein must be recruited to the promoter in order to establish repression. The level of repression by lamin A in yeast was comparable to that of other known mammalian transcription factors, such as pRB [37].
We verified that lamin A could repress transcription using a third, LexA-responsive reporter system (Fig. 1D). Two strains were used for this experiment: JK1621, which has 4 lexA operator sites and LG312ΔS which does not. β-galactosidase activity was compared in each strain expressing either LexADBD-lamin A or LexADBD. In this system, we found a modest (1.5-2 fold), reproducible reduction in lacZ expression levels in JK1621 expressing LexADBD-lamin A relative to LexADBD alone. Repression by lamin A was dependent on the presence of the lexA operator sites, since no difference in expression of lacZ was observed between LexADBD-lamin A and LexADBD in LG312ΔS. We do not know why lamin A-dependent repression was less robust in the lexA system but one possibility may be that Gal-dependent promoters in yeast have easier access to the nucleus and/or nuclear periphery (see Discussion).
All domains of lamin A are important for repression in yeast
The lamin A protein, like all IF proteins, is characterized by 3 distinct domains: a central alpha-helical “rod” domain flanked by globular N- and C-terminals. To map the repression domain(s) of lamin A, the rod domain and the C-terminal domain of lamin A respectively, were fused to Gal4DBD and tested in repression assays (Fig. 2A). The rod- and C-terminal domain fusions were able to confer repression, although at reduced levels relative to full length lamin A despite comparable expression levels (not shown). This finding suggests that these lamin A domains contribute independently to the transcriptional regulatory activity of lamin A. We did not examine the N-terminal domain alone for repression activity since it is quite small (only 33 amino acids), but we did find that the N-terminus of lamin A modestly increases repression mediated by the rod domain. This may be because the N-terminus of lamin A helps to stabilize higher order lamin structures (see Discussion).
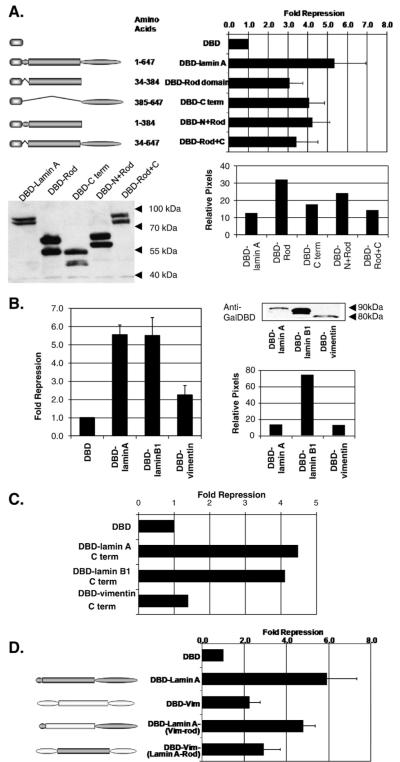
Fig. 2.
All domains of lamin A are required for full repression activity in yeast. (A) Repression of lacZ reporter in yeast by Gal4DBD-fusion proteins of lamin A domains. The graph shows the fold repression of the fusion proteins over GAL4DBD empty vector alone. The data shown is representative of at least three independent transformants. (B) Gal4DBD-intermediate filament fusion proteins were expressed from ARS/CEN plasmids under ADH1 promoter. Results shown are fold repression over GAL4DBD empty vector alone (n=3, mean±SD). Western blot analysis was performed to verify the expression of fusion proteins. Protein samples were separated by using SDS-4-20% PAGE and detected by immunoblotting with mouse anti-Gal4DBD. The relative protein levels were determined by counting pixel levels using NIH ImageJ and represented as a percentage of total pixels counted. (C) Repression of lacZ reporter in yeast by Gal4DBD-intermediate filament protein C-terminus domains. The graph presents the fold repression of the fusion proteins over GAL4DBD empty vector alone. The data shown are representative of at least three independent transformants. (D) Repression of lacZ reporter in yeast by Gal4DBD-fusion proteins of intermediate filament chimeras. Chimeras were made by interchanging the domains between lamin A and vimentin. Results shown are fold repression over GAL4DBD empty vector alone (n=3, mean±SD).
To ascertain if the repression activity is unique to A-type lamins or if any intermediate filament protein tethered to a promoter can repress, lamin B1 (a B-type nuclear lamin) and vimentin (a cytoplasmic IF protein) were fused with Gal4DBD and tested in repression assays (Fig. 2B). Both lamin B1 and vimentin are of similar length and organization to lamin A and share sequence homology particularly in the rod domain [41]. We find that Gal4DBD-lamin B1 repressed transcription to a level comparable to lamin A; however, its expression was approximately 10-fold higher than that of lamin A making direct comparison of repression levels difficult. Vimentin exhibited minimal repression activity, despite being targeted to the nucleus through the NH2-terminus of the Gal4DBD [42]. Similarly, we tested the repression ability of the C-terminal domains of lamin B1 and vimentin. Consistent with their respective full length proteins, the C-terminus of lamin B1 possessed repression activity albeit reduced, whereas the C-terminus of vimentin did not (Fig. 2C). These results suggest that full length nuclear lamin proteins have the ability to repress transcription in yeast and this repression may be a specific feature of nuclear intermediate filaments.
Since vimentin-dependent repression was similar to that conferred by the lamin A rod domain alone, we considered the possibility that the vimentin rod domain was comparable to that of lamin A but that it lacked repression-specific functions of the C-terminal globular domain of lamin A. We tested this hypothesis by creating chimeras between lamin A and vimentin and examining their transcription repression activities (Fig. 2D). Consistent with our prediction, a Gal4DBD-vimentin fusion in which its rod domain was replaced with that of lamin A repressed to a similar extent to Gal4DBD-vimentin and Gal4DBD-lamin A rod. Similarly, a Gal4DBD-lamin A fusion in which its rod domain was replaced with that of vimentin conferred stronger repression although perhaps marginally reduced relative to full length laminA. Together, these findings suggest that lamin A has two repression functions in yeast: one dependent on the rod domain and shared by rod domains of other intermediate filament proteins and one specific to the C-terminal globular domain of nuclear lamins.
A-type lamins repress transcription in mammalian cells
The transcriptional machinery is highly conserved between yeast and mammalian systems. To test whether repression by heterologous expression of human lamin A in yeast is relevant to its role in mammalian cells, we performed similar experiments in human cell culture to determine whether lamin A could repress transcription when targeted to a promoter. Gal4DBD-fusion proteins were co-transfected in 293 T cells with a luciferase reporter plasmid containing Gal4 binding sites [32] and a lacZ vector (without Gal4 binding sites) to normalize for transfection efficiency. This reporter has six copies of a GC box, which serves as a binding site for the Sp1 transcriptional activator. Cells were harvested 36-48 h after transfection and both luciferase and β-galactosidase activities were measured in the cell lysates (Fig. 3A). Western blot analysis was performed to verify the expression of lamin A fusion proteins (Fig. 3B). After normalization to transfection efficiency, luciferase activity in extracts from Gal4DBD fusion proteins was normalized to Gal4DBD alone to determine repression specific to each intermediate filament protein. Similar to studies in yeast, we find that lamin A can confer approximately 4-5 fold repression when targeted to a promoter driving luciferase expression. GFP-lamin A did not affect luciferase expression, indicating as in yeast, that repression is a direct consequence of targeting lamin A to the promoter.
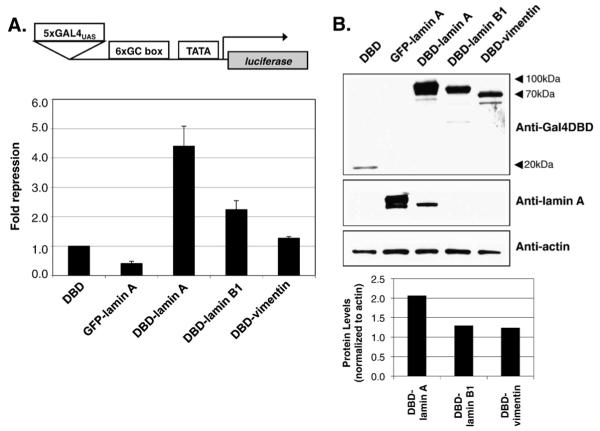
Fig. 3.
Lamin A represses transcription of reporter gene in mammalian cells. (A) Repression of a luciferase reporter in 293 T cells. cDNA encoding Gal4DBD alone, GFP-LMNA or Gal4DBD-IF fusion proteins were transiently cotransfected with a luciferase reporter. Luciferase activity in the cell lysates were measured and normalized to β-galactosidase activity for transfection efficiencies. Results are expressed as fold repression over Gal4DBD alone (n>5, mean±SD). (B) Western blot analysis of the transiently transfected fusion proteins. Protein samples were separated by using SDS-8% PAGE and detected by immunoblotting with mouse anti-Gal4DBD or rabbit anti-pan lamin A. The relative protein levels were determined by counting pixel levels using NIH ImageJ and normalized to beta-actin loading control.
We observed modest repression activity with other intermediate filaments, lamin B1 and vimentin in mammalian cells, consistent with our findings in yeast. We also compared repression by lamin C to that of lamin A and found that lamin C possessed repression activity comparable with that of lamin A (see Fig. 5). These two alternative splice products of the LMNA gene share in common the N-terminal 566 amino acids, only diverging in their C- terminus where lamin A has 98 unique amino acids and lamin C has six. This finding suggests that the C terminal 98 amino acids of lamin A are not required for full repression.
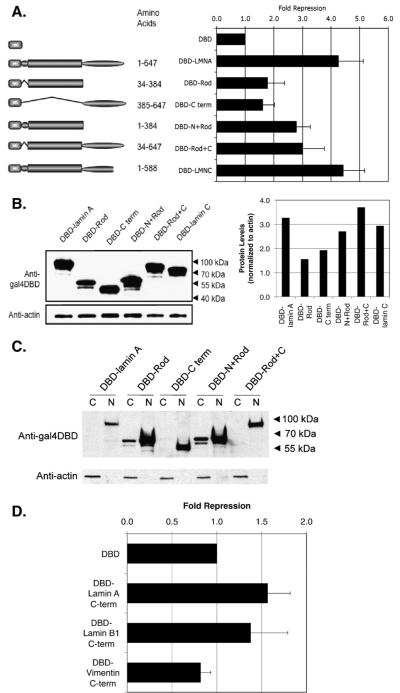
Fig. 5.
The presence of the N- or C-terminus is required for enhanced repression activity in mammalian cells. (A) Gal4DBD fusions of lamin A domains were cotransfected with a luciferase reporter and the luciferase activity in the cell lysates were measured. The graph presents the fold repression of the fusion proteins over GAL4DBD empty vector alone. The data shown are representative of at least three independent experiments. (B) Western blot analysis of the transiently transfected fusion proteins. Protein samples were separated by using SDS-4-20% PAGE and detected by immunoblotting with mouse anti-Gal4DBD. The relative protein levels were determined by counting pixel levels using NIH ImageJ and normalizing to beta-actin loading control. (C) Cytoplasmic (C) and nuclear (N) fractions were prepared and samples were separated by SDS-4-20% PAGE and detected by immunoblotting with mouse anti-Gal4DBD. (D) Repression of luciferase reporter by Gal4DBD-intermediate filament protein C-terminal domains. The graph presents the fold repression of the fusion proteins over GAL4DBD empty vector alone. The data shown are representative of at least three independent experiments.
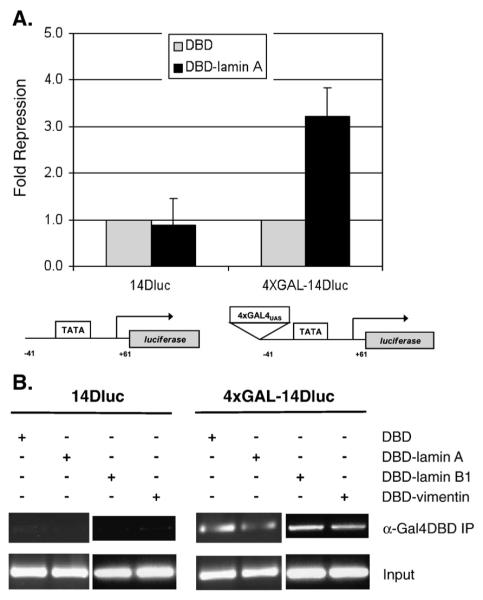
To verify that lamin A-mediated repression is dependent on the occupancy of the promoters, we used a different set of two luciferase reporters: “14D-Luciferase” a constitutive luciferase reporter and “4xGAL-14D-Luciferase” which is identical to “14D-Luciferase” but has 4xGAL4 binding sites 5′ of the reporter [33]. The Gal4 sites are integrated into the SV40 promoter, which confers strong transcription through recruitment of an array of transcription factors. We compared the effect of expressing Gal4DBD-lamin A versus Gal4DBD on the luciferase activity of the two reporters (Fig. 4A). Repression by lamin A was dependent on the presence of the Gal4 binding sites, since no difference in luciferase activity was observed between Gal4DBD-lamin A and Gal4DBD in 14D-Luciferase. We observed a lamin A-dependent repression in 4xGAL-14D-Luciferase. A chromatin immunoprecipitation (ChIP) was performed to confirm that the Gal4DBD proteins occupied the Gal4 binding sites (Fig. 4B). Notably, chromatin immunoprecipitation indicates that Gal4DBD-vimentin can interact with the Gal4 binding sites in this reporter, demonstrating that this cytoplasmic intermediate filament protein has access to the nucleus when fused to Gal4DBD.
Fig. 4.
Lamin A-mediated repression is dependent on the tethering of lamin A to the promoter. (A) Repression of luciferase reporters 14DLuc and 4xGAL-14DLuc. The reporters are identical to except for the presence of 4xGAL4 binding sites 5′ to the TATA box in 4xGAL-14DLuc. cDNA encoding Gal4DBD alone or Gal4DBD-lamin A was transiently cotransfected with the respective luciferase reporters. Luciferase activity in the cell lysates were measured and normalized to β-galactosidase activity for transfection efficiencies. Results are expressed as fold repression over Gal4DBD alone (n=3, mean±SD). (B) Chromatin immunoprecipitation of luciferase reporters 14DLuc and 4xGAL-14DLuc. Chromatin was immunoprecipitated from the cell lysates with antibodies to Gal4DBD. Binding of Gal4DBD-fusion proteins to the promoter region were detected by PCR using primers that amplified the 5′ end of the luciferase gene. Input lanes represent PCR from 5% of the pre-immunoprecipitated DNA.
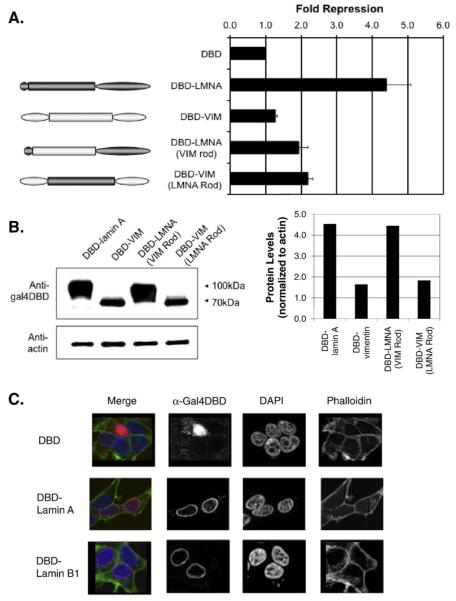
Full length of lamin A/C is required for robust transcriptional repression
Using a similar strategy to that employed in yeast, we created fusions between Gal4DBD and lamin A domains. Consistent with our yeast studies, maximal repression was observed with only the full length intact lamin A protein (Fig. 5A). Results with individual domains or combinations thereof were similar with one notable exception. While full-length lamin A conferred 4-5 fold repression, the rod domain and C-terminus alone showed only weak repression activity. The Gal4DBD-lamin A rod fusion had weak repressive activity unless fused to the N-terminus or to the C-terminus. Relative expression levels of transfected constructs were determined by western blot analysis (Fig. 5B). We routinely observe in human cells that transient transfection of lamin A constructs containing the C-terminal domain result in higher expression than those without. Whether this reflects an ability of the C-terminal domain to stabilize the protein is unknown. However, it raises the possibility that reduced repression by the rod domain relative to the rod+C-terminal domain may be a consequence of reduced expression levels. As in yeast, the N-terminus+rod fusion represses transcription better than the rod domain alone, although this could be through enhanced stability as well. The small N-terminus is known to assist in lamin A association into higher order structures, potentially explaining its importance (see Discussion). We also considered the possibility that some lamin A mutants may not have access to the nucleus, even though they are expressed as a fusion protein to Gal4DBD. To test this, we fractionated cells into nuclear and cytoplasmic fractions and determined the relative expression level of lamin A mutants (Fig. 5C). Although we detect a minority of Gal4DBD-rod and Gal4DBD-N+rod in the cytoplasm, all fusion proteins are enriched in the nuclear fraction.
For comparison, we measured the repression activities of the C-terminal domains of lamin B1 and vimentin (Fig. 5D). We found that like the C-terminus of lamin A, the C-terminal domains of lamin B1 and vimentin did not repress appreciably.
Finally, we determined whether the vimentin rod domain can substitute for the lamin A rod domain for repression in human cells, as it does in yeast. Our findings indicate that both the lamin A-vimentin rod chimera and the vimentin-lamin A rod chimera confer only modest transcriptional repression (2 fold) relative to full length lamin A(4-5folds) (Fig. 6A). Western blot analyses of relative expression are provided for comparison (Fig. 6B). Taken together, these data are consistent with our previous observations that both the rod domain and C-terminal domain of lamin A have repression activity.
Fig. 6.
The vimentin rod domain cannot substitute for the lamin A rod for repression in mammalian cells. (A) Repression of luciferase reporter by Gal4DBD- fusion proteins of intermediate filament chimeras. Chimeras were made by interchanging the domains between lamin A and vimentin. The graph presents the fold repression of the fusion proteins over GAL4DBD empty vector alone. The data shown are representative of at least three independent experiments. (B) Western blot analysis was performed to verify the expression of fusion proteins. Protein samples were separated by using SDS-4-20% PAGE and detected by immunoblotting with mouse anti-Gal4DBD. The relative protein levels were determined by counting pixel levels using NIH ImageJ and normalizing to beta-actin loading control. (C) Indirect immunofluorescence of 293 T cells expressing Gal4DBD-fusion proteins. Gal4DBD-fusion proteins (red) were detected with antibodies against the Gal4-DNA binding domain. Phalloidin (green) labels actin filaments indicating the cytosol. DAPI (blue) labels DNA indicating the nucleus. Images were acquired using equal exposure times and image processing.
We considered the possibility that lamin A-dependent transcriptional repression was a consequence of localization of reporters to the periphery of the nucleus. In mammalian cells, both lamin A and lamin B are known to be enriched at the nuclear periphery. However, the possibility existed that fusion of either protein to Gal4DBD altered its subnuclear distribution. Indirect immunofluorescence with antibodies to Gal4DBD was performed to test this possibility (Fig. 6C). Upon transient transfection, we find as expected that the Gal4DBD-lamin A and lamin B proteins were mostly detected in the nucleus and that both are enriched at the nuclear periphery. Since repression by lamin A is more robust than that conferred by lamin B, we conclude that peripheral localization is not sufficient for lamin A-dependent repression, instead relying on functional properties unique to lamin A. Gal4DBD-vimentin was found to be localized in a more distributed fashion in the nucleus and also in the cytoplasm (not shown). In the discussion, we consider possible mechanisms of lamin A-dependent repression and their potential significance to the functions of lamin A in maintaining healthy differentiated tissues.
Discussion
In summary, we have examined the effects of targeted recruitment of lamin A, lamin A domains and other intermediate filament proteins to promoters in yeast and mammalian cells, finding similar although non-identical results in both systems. Taken in sum, we conclude that lamin A has two repression domains, one in the rod and the other in the C-terminus, which can confer transcriptional repression. However, the full length lamin A protein is required for maximal repression in both systems.
The rod domain of lamin A is required for dimerization of the coiled-coil and for formation of higher order intermediate filament polymers. One mechanism by which transcriptional repression may occur is through generation of filament-like structures at promoters that serve as an impediment to other transcription factors. Both the N- and C-terminus have been shown to be important for homotypic associations leading to higher order structures, likely providing one reason why the N- and/or C-terminus, when fused to the rod domain, increases the basal repression activity of the rod domain in human cells. As our understanding of the mechanisms by which lamin A forms filaments in vivo improve, it may become possible to assess more specifically the role of lamin A filament formation in transcriptional repression, and it should be noted that given our current understanding, a more specific activity of the lamin A rod domain in transcriptional repression cannot be excluded.
Another non-exclusive mechanism by which A-type lamins might repress transcription is through re-localization of promoters to the nuclear periphery into heterochromatic regions not permissive to transcription. The nuclear periphery is associated with transcriptional silencing in both yeast and mammalian cells [43-45]. While there are no yeast orthologs of nuclear lamins, we find that GFP-lamin A fusion proteins localize specifically to the nuclear periphery in yeast cells in what appears to be a higher order structure (data not shown). Our localization data regarding GFP-lamin B1 are also consistent with prior reports showing localization of chicken lamin B correctly targeted to the inner nuclear membrane but also regions of the cytoplasm when expressed in yeast [46]. Therefore, lamin A targeting to promoters may re-direct them to the nuclear periphery in yeast as well as in mammalian cells. Presumably, this would require lamin A assembly into intermediate filaments structures and therefore this mechanism could underlie repression linked to the rod domain. In mammalian cells, relocalization of promoters to the periphery is likely insufficient to mediate repression, since both the Gal4DBD-lamin A and Gal4DBD-lamin B fusions are enriched at the periphery and only Gal4DBD-lamin A confers robust repression. Therefore, although peripheral localization may be one component, functions unique to lamin A must also exist.
The C-terminal domain of lamin A represses transcription both in yeast and human cells in the absence of the rest of the protein. This region of lamin A is not capable of forming higher order structures, thus we speculate that repression by this domain may occur through a different mechanism, perhaps direct interaction with a transcriptional co-repressor conserved between yeast and humans. In addition to lamins, yeast lacks many lamin-associated proteins [10-12], including LEM domain proteins and BAF. Therefore, while these proteins are likely to contribute to the establishment of heterochromatin in mammalian cells and may also contribute to A-type lamin regulation at specific promoters, they are probably not required for the transcriptional repression we observed (at least in yeast). Under endogenous settings, lamin A is likely to be recruited to promoters through interaction with DNA binding transcription factors, for instance E2F-retinoblastoma protein (pRB) complexes [16,23]. Therefore the repression activities of lamin A may synergize with those of repressors such as pRB to strongly inhibit transcription.
A recent study has uncovered potential roles for lamin A at specific reporters. Reddy et al. devised an approach to inducibly tether genes at the inner nuclear membrane (46). When genes were relocalized to this site, repression was observed although it is unknown whether lamin A is required for this activity. Interestingly, they found that in fibroblasts inactive immunoglobulin loci localize to the peripheral lamina, making them candidates for targets of lamin A-mediated repression.
During cellular differentiation, many gene expression changes occur such as the activation of lineage-specific genes and the repression of progenitor-specific genes. Coincidentally, the induction of lamin A/C expression during development correlates with differentiation [47]. Our data suggests that A-type lamins may play a part in regulating nuclear factors responsible for cell-type differentiation, through the negative regulation of transcription. Missense mutations throughout LMNA are linked to a myriad of developmental diseases, collectively termed laminopathies [7-9]. In most cases, the alterations in lamin A/C function linked to the particular mutation are not known, and it is speculated that different diseases arise through different defects in lamin A/C function. Therefore, it is possible that a subset of LMNA-disease mutations impairs lamin A/C-mediated transcriptional repression, and it will be important to test this possibility in future studies.
In summary, our findings suggest a novel activity for lamin A in the regulation of gene expression and shed light upon potential mechanisms by which A-type lamins may inhibit transcription. By identifying the promoters associated with human lamin A/C in vivo, it will become possible to test the importance of these repressive activities in endogenous settings.
Acknowledgments
We would like to thank Ryan A. Lemke and Trang M. Pham for technical assistance. We thank the members of the Kennedy lab for helpful discussions concerning studies presented in the manuscript. Research on A-type lamin function is funded by the National Institutes of Health grant R01AG024287 granted to B.K.K.
REFERENCES
- [1].Kosak ST, Groudine M. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- [2].Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J. Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shaklai S, Amariglio N, Rechavi G, Simon AJ. Gene silencing at the nuclear periphery. FEBS J. 2007;274:1383–1392. doi: 10.1111/j.1742-4658.2007.05697.x. [DOI] [PubMed] [Google Scholar]
- [4].Goldman RD, Goldman AE, Shumaker DK. Nuclear lamins: building blocks of nuclear structure and function. Novartis Found. Symp. 2005;264:3–16. discussion 16-21, 227-230. [PubMed] [Google Scholar]
- [5].Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. FEBS J. 2007;274:1354–1361. doi: 10.1111/j.1742-4658.2007.05694.x. [DOI] [PubMed] [Google Scholar]
- [6].Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- [7].Mattout A, Dechat T, Adam SA, Goldman RD, Gruenbaum Y. Nuclear lamins, diseases and aging. Curr. Opin. Cell Biol. 2006;18:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- [8].Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat. Rev., Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- [9].Vlcek S, Foisner R. Lamins and lamin-associated proteins in aging and disease. Curr. Opin. Cell Biol. 2007;19:298–304. doi: 10.1016/j.ceb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- [10].Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- [11].Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- [12].Dorner D, Gotzmann J, Foisner R. Nucleoplasmic lamins and their interaction partners, LAP2alpha, Rb, and BAF, in transcriptional regulation. FEBS J. 2007;274:1362–1373. doi: 10.1111/j.1742-4658.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- [13].Nitta RT, Jameson SA, Kudlow BA, Conlan LA, Kennedy BK. Stabilization of the retinoblastoma protein by A-type nuclear lamins is required for INK4A-mediated cell cycle arrest. Mol. Cell. Biol. 2006;26:5360–5372. doi: 10.1128/MCB.02464-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006;20:486–500. doi: 10.1101/gad.1364906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Melcon G, Kozlov S, Cutler DA, Sullivan T, Hernandez L, Zhao P, Mitchell S, Nader G, Bakay M, Rottman JN, Hoffman EP, Stewart CL. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- [16].Dorner D, Vlcek S, Foeger N, Gajewski A, Makolm C, Gotzmann J, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha regulates cell cycle progression and differentiation via the retinoblastoma-E2F pathway. J. Cell Biol. 2006;173:83–93. doi: 10.1083/jcb.200511149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- [18].Dreuillet C, Tillit J, Kress M, Ernoult-Lange M. In vivo and in vitro interactions between human transcription factor MOK2 and nuclear lamin A/C. Nucleic Acids Res. 2002;30:4634–4642. doi: 10.1093/nar/gkf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ozaki T, Saijo M, Murakami K, Enomoto H, Taya Y, Sakiyama S. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- [20].Shan B, Zhu X, Chen P-L, Durfee T, Yang Y, Sharp D, Lee W-H. Molecular cloning of cellular genes encoding retinoblastoma-associated proteins: identification of a gene with properties of the transcription factor E2F. Mol. Cell Biol. 1992;12:5620–5631. doi: 10.1128/mcb.12.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ivorra C, Kubicek M, Gonzalez JM, Sanz-Gonzalez SM, Alvarez-Barrientos A, O’Connor JE, Burke B, Andres V. A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev. 2006;20:307–320. doi: 10.1101/gad.349506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johnson BR, Nitta RT, Frock RL, Mounkes L, Barbie DA, Stewart CL, Harlow E, Kennedy BK. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9677–9682. doi: 10.1073/pnas.0403250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stierle V, Couprie J, Ostlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–4828. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- [26].James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vidal M, Braun P, Chen E, Boeke JD, Harlow E. Genetic characterization of a mammalian protein-protein interaction using a reverse two-hybrid system. Proc. Natl. Acad. Sci. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- [29].Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- [30].Guarente L, Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box”. Proc. Natl. Acad. Sci. U. S. A. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- [32].Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- [33].Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu Y, Leung CG, Lee DC, Kennedy BK, Crispino JD. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia. 2006;20:1261–1269. doi: 10.1038/sj.leu.2404252. [DOI] [PubMed] [Google Scholar]
- [36].Lee MS, D’Amour KA, Papkoff J. A yeast model system for functional analysis of beta-catenin signaling. J. Cell Biol. 2002;158:1067–1078. doi: 10.1083/jcb.200204063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kennedy BK, Liu OW, Dick FA, Dyson N, Harlow E, Vidal M. Histone deacetylase-dependent transcriptional repression by pRB occurs independently of interaction through the LXCXE binding cleft. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8720–8725. doi: 10.1073/pnas.151240898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kennedy BK. Mammalian transcription factors in yeast: strangers in a familiar land. Nat. Rev., Mol. Cell Biol. 2002;3:41–49. doi: 10.1038/nrm704. [DOI] [PubMed] [Google Scholar]
- [39].Malcov M, Cesarkas K, Stelzer G, Shalom S, Dicken Y, Naor Y, Goldstein RS, Sagee S, Kassir Y, Don J. Aym1, a mouse meiotic gene identified by virtue of its ability to activate early meiotic genes in the yeast Saccharomyces cerevisiae. Dev. Biol. 2004;276:111–123. doi: 10.1016/j.ydbio.2004.08.026. [DOI] [PubMed] [Google Scholar]
- [40].Vidal M, Brachmann R, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissocation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev., Mol. Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- [42].Silver PA, Keegan LP, Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc. Natl. Acad. Sci. U. S. A. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinucleolar localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- [44].Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- [45].Heilbronn LK, de Jonge L, Frisard MI, DeLany JP, Larson-Meyer DE, Rood J, Nguyen T, Martin CK, Volaufova J, Most MM, Greenway FL, Smith SR, Deutsch WA, Williamson DA, Ravussin E. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295:1539–1548. doi: 10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Smith S, Blobel G. Colocalization of vertebrate lamin B receptor (LBR) in nuclear envelopes and in LBR-induced membrane stacks of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10124–10128. doi: 10.1073/pnas.91.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]