Abstract
Piwi family proteins are essential for germline development and bind piwi-interacting RNAs (piRNAs 1 2 3). The grandchildless gene aub of Drosophila melanogaster encodes the piRNA-binding protein Aub that is essential for formation of primordial germ cells (PGCs) 4. Here we report that mouse, Xenopus laevis and Drosophila melanogaster Piwi family proteins contain symmetrical dimethylarginines (sDMAs). We find that Piwi proteins are expressed in X. laevis oocytes and we identify numerous X. laevis piRNAs. We report that the Drosophila homolog of protein methyltransferase 5 (dPRMT5, csul/dart5), which is also the product of a grandchildless gene 5, 6, is required for arginine methylation of Drosophila Piwi, Ago3 and Aub proteins, in vivo. Loss of dPRMT5 activity leads to reduction of piRNAs and in particular of Ago3 and Aub protein levels and accumulation of retrotransposons in the Drosophila ovary. Our studies explain the relationship between aub and dPRMT5 (csul/dart5) genes by demonstrating that dPRMT5 is the enzyme that methylates Aub. Our findings underscore the significance of sDMA modification of Piwi proteins in the germline and suggest an interacting pathway of genes that are required for piRNA function and PGC specification.
Piwi family proteins are expressed in the germline and bind ∼26 to ∼30 nucleotide (nt) piRNAs (reviewed in1 2 3). Drosophila express three Piwi proteins: Aub 4, Piwi 7 and Ago3 8 9. Mice also express three Piwi proteins termed Miwi, Mili/PiwiL2 and Miwi2/PiwiL4 10. Tens of thousands of distinct piRNAs have been described and most of them are species-specific 1 2 3. In Drosophila, Piwi proteins and piRNAs (also known as rasiRNAs –repeat associated small interfering RNAs11) silence transposons in the germline 12 2 10 13. A similar function has been found for a subset of mouse 14 and zebrafish piRNAs 15. An amplification loop of piRNAs has been described 8 9 but how primary piRNAs are generated is unknown 3. Arginine methylation is an important post-translational modification that is mediated by two types of protein methyltransferases (PRMTs): type I enzymes catalyze asymmetric dimethylation of arginines (aDMA) and type II enzymes catalyze symmetric arginine dimethylation (sDMA) (Figure 1d) 16. sDMA modifications occur in sequence motifs composed of arginines flanked by glycines (GRG) or alanines (GRA or ARG) that are often found as repeats 17 16. PRMT5 and its cofactors MEP50/WD45 and pICln form the methylosome that methylates Sm proteins 18, 19.
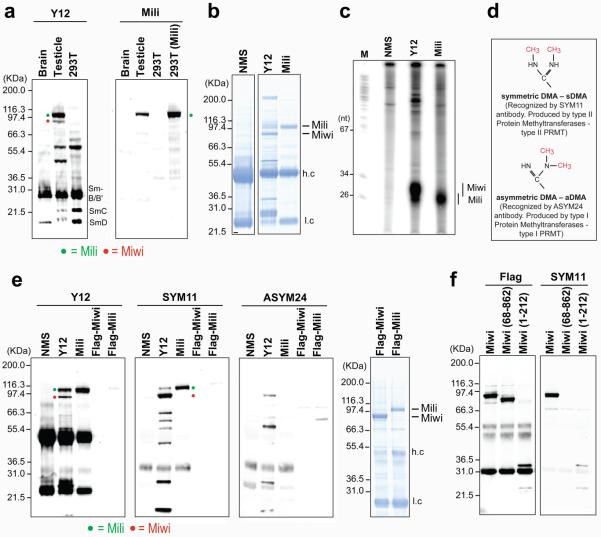
Figure 1. Miwi and Mili proteins contain symmetrical dimethylarginines (sDMAs).
(a) Western blots; 293T(Mili) refers to 293T cells with forced expression of Flag-Mili.
(b) Protein immunoprecipitates from mouse testis (NMS: non-immune mouse serum). Mass spectrometry identified Miwi and Mili proteins in the Y12 and Mili protein in the anti-Mili immunoprecipitates (Supplementary Table 1). (h.c) heavy, (l.c) light antibody chain.
(c) RNA-immunoprecipitation from mouse testis.
(d)Arrangement of methyl groups in dimethylarginines.
(e) Immunoprecipitates from mouse testis or purified recombinant proteins (Flag-Miwi, Flag-Mili) were probed on Western bots with indicated antibodies. Right panel: coomasie-stained gel of purified recombinant proteins.
(f) Full-length or truncated Flag-Miwi (68-862 or 1-212) expressed in 293T cells were immunopurified with anti-Flag and probed with anti-Flag or SYM11 antibodies.
We produced a highly specific monoclonal antibody (17.8) that recognizes Mili by Western blot (Figure 1a, right panel), immunoprecipitation (Figure 1b, c and Supplementary Table 1) and immunofluorescence microscopy (Supplementary Figure 1). By serendipity we discovered that the widely used Y12 monoclonal antibody 20 recognizes mouse Mili and Miwi proteins and their bound piRNAs (Figure 1a, b, c). The Sm proteins of spliceosomal small nuclear ribonucleoproteins (snRNPs) constitute the main antigen for Y12 20 21. We did not identify piRNAs in immunoprecipitates of snRNPs, heterogeneous ribonucleoproteins (hnRNPs) or of the Survival of Motor Neurons (SMN) complex using various antibodies (Supplementary Figure 2). By Northern blot analysis we found that piR-1, but not miR-16, an abundant miRNA, was found in Y12 immunoprecipitates (Supplementary Figure 3), suggesting that Y12 recognizes Piwi but not Ago proteins.
The epitope that Y12 recognizes on Sm proteins consists of symmetrically dimethylated arginines, in the glycine-arginine rich regions of the proteins 21. We reasoned that Y12 likely reacted with sDMA-containing epitopes in Mili and Miwi and we searched for arginine residues that could be symmetrically methylated. Intriguingly, we found that most animal Piwi proteins contain sDMA motifs that are typically clustered close to the amino terminus (Supplementary Table 2), while no animal Ago proteins contained such motifs (data not shown). However, we found that four of ten Arabidopsis Ago proteins contained sDMA motifs (Supplementary Table 2).
To test whether Miwi and Mili contain sDMAs, we used SYM11 and ASYM24 antibodies, which specifically recognize proteins containing sDMA-glycine or aDMA-glycine repeats, respectively (Figure 1d) 22. As shown in Figure 1e, SYM11, as well as Y12, reacted strongly with endogenous Miwi and Mili, while ASYM24 showed only faint reactivity towards endogenous Miwi. In contrast, recombinant Flag-Mili or Flag-Miwi purified from baculovirus-infected Sf9 cells, were not recognized by Y12 or SYM11 (or ASYM24) (Figure 1e) despite loading of large amounts of recombinant proteins on the gels (Figure 1e, right panel). This is entirely consistent with the finding that recombinant human Sm proteins expressed in Sf9 cells also do not contain sDMAs because Sf9 cells do not express type II PRMTs and thus cannot produce sDMA modifications 21. These findings indicate that Mili and Miwi proteins contain sDMAs. The putative sDMA motifs of Miwi are concentrated very close to the amino terminus with the exception of one GRG triplet (Supplementary Table 2). We transfected Flag-tagged full-length Miwi and two truncated forms of Miwi (aa 68-862 or 1-212) in 293T cells, purified the proteins by Flag immunoprecipitation and subjected them to western blot with SYM11 antibody. As shown in Figure 1f, SYM11 antibody recognizes the amino - terminus of Miwi protein.
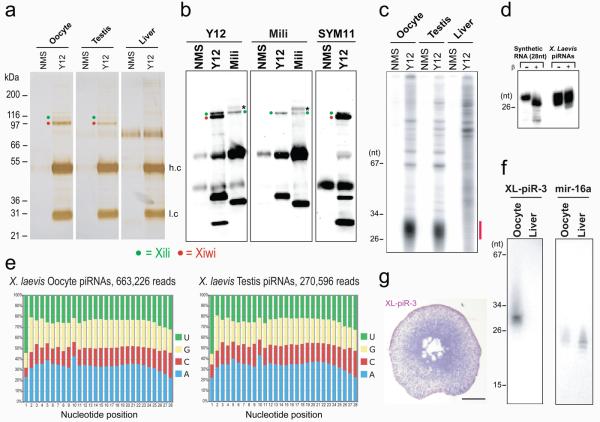
Next we asked whether the sDMA modification was conserved in Piwi family proteins from other species. A stumbling block in studying the molecular functions of Piwi proteins and piRNAs has been the lack of suitable cell culture systems. We reasoned that Xenopus laevis oocytes might express Piwi proteins and piRNAs and thus prove very useful not only to confirm that sDMAs of Piwi proteins are conserved but also as a model to study the function of Piwi proteins and piRNAs. By searching the Gurdon EST database at Xenbase 23 we identified three Xenopus Piwi proteins which we named Xili, Xiwi and Xiwi2 (Supplementary Figure 4). All three Xenopus Piwi proteins contain putative sDMA motifs (Supplementary Table 2). Immunoprecipitations with Y12 from X. laevis oocytes (defolliculated, mixed Dumont stages I-VI), testis and liver revealed the presence of two proteins at ∼95 kDa and ∼110 kDa specifically in the Y12 immunoprecipitates from oocytes and testis (Figure 2a) that we identified by mass spectrometry as Xiwi and Xili respectively (Supplementary Table 3). As shown in the western blots in Figure 2b, Y12 recognized both Xiwi and Xili, while anti-Mili (17.8) reacted only with Xili. In addition, both Xiwi and Xili were recognized by SYM11, indicating that Xiwi and Xili contain sDMAs.
Figure 2. Xenopus laevis Piwi proteins with bound piRNAs are immunoprecipitated by Y12 and contain sDMAs.
(a) Protein immunoprecipitates from indicated X. laevis tissues; Xili and Xiwi were identified by mass spectrometry (Supplementary Table 3).
(b) Immunoprecipitates from X. laevis oocytes were probed on Western blots with indicated antibodies. Band with asterisk is bovine IgG from tissue culture supernatant of anti-Mili hybridoma.
(c) RNA-immunoprecipitations from X. laevis.
(d) Periodate oxidation and β-elimination of X. laevis piRNAs isolated from Y12 immunoprecipitates.
(e) Nucleotide composition of X. laevis piRNAs.
(f) Northern blot for XL-piR-3
(g) In situ hybridization for XL-piR-3 in X. laevis oocyte; bar = 100μm
We isolated and analyzed X. laevis piRNAs from Y12 immunoprecipitates. As shown in Figure 2c, ∼26-29 nt piRNAs are present in the Y12 immunoprecipitates and their 3′-termini are not eliminated by periodate oxidation (Figure 2d) and are thus likely 2′-O-methylated, as seen in piRNAs from other species 24 25 26 27 15.
We next performed deep sequencing of X. laevis piRNAs from Y12 immunoprecipitates of oocytes and testis. The sequences and analysis are presented in the Supplement. The nucleotide composition of X. laevis piRNAs is shown in Figure 2e and shows enrichment of Uridine in the first nucleotide position and of Adenine in the tenth nucleotide position. There is also enrichment for piRNAs whose first 10 nucleotides are complementary to the first 10 nucleotide of other piRNAs (Supplement). These features indicate that a fraction of X. laevis piRNAs target transposon transcripts and that they also participate in a piRNA amplification loop, as has been described for Drosophila and zebrafish piRNAs and prepachytene mouse piRNAs 8 9 15 14. By Northern blot XL-piR-3, a representative piRNA, is expressed specifically in oocytes (Figure 2f) and by in situ hybridization XL-piR-3 is localized predominantly in the cytoplasm of X. laevis oocytes and it is expressed in higher levels in immature oocytes (Figure 2g).
Genetic disruption of either Drosophila PRMT5 (dPRMT5; also know as capsuleen –csul–, and dart5) or its cofactor valois, (the Drosophila homolog of MEP50/WD45), results in complete loss of sDMA modifications of Sm proteins in ovaries 5, 6. However, unlike the situation in mammals 18, 19 28, the levels or function of Sm proteins is not affected by loss of sDMAs 6 29.
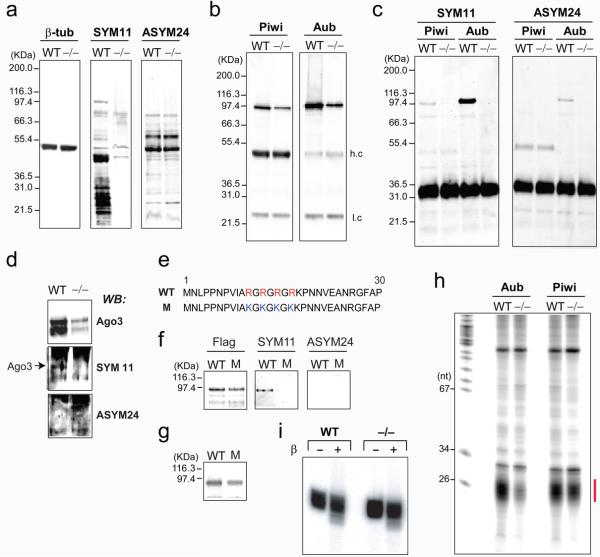
Null or hypomorphic alleles of dPRMT5 (csul, dart5) 5, 6 phenocopy aub null alleles 4 and we reasoned that dPRMT5 might be the methyltransferase that produces sDMAs in Aub, Piwi and Ago3, in vivo. To test this, we used ovaries from csulRM50/Df(2R)Jp7 females which give rise to embryos that are genetic nulls for dPRMT5 5 and w− as a wild-type control. Western blots of ovary lysates from wt and maternal null csul showed that there was near complete loss of SYM11 reactivity, indicating dramatic reduction of sDMA modified proteins in csul ovaries (Figure 3a). There was no change in ASYM24 reactivity between wt and csul, indicating that aDMA modified proteins were not affected (Figure 3a). These findings confirm that dPRMT5 (csul, dart5) activity is required specifically for sDMA modification, as previously reported 5, 6. We immunoprecipitated Piwi and Aub proteins from wt and csul mutant ovaries (Figure 3b) and probed the immunoprecipitates with SYM11 and ASYM24. As shown in Figure 3c, SYM11 reacted very strongly with Aub and also with Piwi immunopurifed from wt but not csul ovaries; ASYM24 reacted only weakly with Aub from wt ovaries. We also probed immunoprecipitates of Ago3 with SYM11 and ASYM24 and observed that only Ago3 from wt ovaries reacted with SYM11 (Figure 3d). These results indicate that, like the mouse and X. laevis Piwi family proteins, Drosophila Piwi, Aub and Ago3 contain sDMAs and that dPRMT5 is the methylase that produces sDMAs of these proteins.
Figure 3. Drosophila PRMT5 (csul, dart5) is required for arginine methylation of Aub, Piwi and Ago3 proteins in ovaries.
(a) Western blots from wild-type (WT) or csul (dPRMT5) mutant (−/−) ovary. Piwi or Aub immunoprecipitates from ovary lysates were probed on western blots with anti-Piwi and anti-Aub antibody
(b); or SYM11 and ASYM24 (c).
(d) Ago3 immunoprecipitates from WT or csul mutant (−/−) ovary lysates were probed on Western blots (WB) with indicated antibodies.
(e) Sequences of wild-type (WT) and mutant (M) Aub, where the four arginines that are substrates for methylation were substituted with lysines.
(f) Anti-Flag immunoprecipitates of S2 cells stably transfected with WT or M Flag-Aub were probed on western blots with indicated antibodies.
(g) Crosslinking of synthetic, radiolabeled piRNA to immunopurified WT or M Flag-Aub.
(h) RNA immunoprecipitation.
(i) Periodate oxidation and β-elimination of Drosophila piRNAs isolated from Piwi immunoprecipitates from wt or csul (−/−) mutant ovaries.
In Aub the four arginines that are putative substrates for symmetrical dimethylation are found in tandem very close to the amino terminus (Figure 3e). We used site-directed mutagenesis to change these arginines into lysines that are not subjected to methylation by PRMTs. We stably transfected Flag-tagged wild-type (WT) or mutant (M) Aub in Drosophila S2 cells (which express dPRMT5 6) purified the proteins by Flag immunoprecipitation and subjected them to western blot with Flag, SYM11 and ASYM24 antibodies. As shown in Figure 3f, SYM11 antibody reacted only with wild-type Aub. Next we assayed the binding of wild-type and mutant Aub to a synthetic piRNA. We incubated immunopurified, wild-type or mutant Flag-Aub with a 5′-end radiolabeled synthetic piRNA containing 4-thio-Uridine at the first position followed by crosslinking with Ultraviolet light and NuPAGE analysis. As shown in Figure 3g, there was similar binding between wild-type and mutant Aub proteins. These findings indicate that one or more of the four arginines in the amino terminus of Aub are symmetrically dimethylated and arginine methylation does not impact piRNA binding.
Next, we isolated and analyzed RNAs bound to Piwi and Aub from wt or csul ovaries. As shown in Figure 3h, piRNAs remain bound to Piwi and Aub proteins in the csul ovaries. There is mild reduction of Piwi-piRNAs and marked reduction of Aub-piRNAs in csul ovaries corresponding to concordant reduction of protein levels of Piwi and Aub (Figure 4a). The Piwi associated piRNAs were gel purified and subjected to periodate oxidation followed by β-elimination and revealed that Piwi-associated piRNAs purified from csul ovaries retain 2′-O-methylation of their 3′ termini (Figure 3i). These findings indicate that the lack of sDMA modifications of Piwi and Aub in csul ovaries does not impair the methylation of piRNAs or their binding to Piwi and Aub.
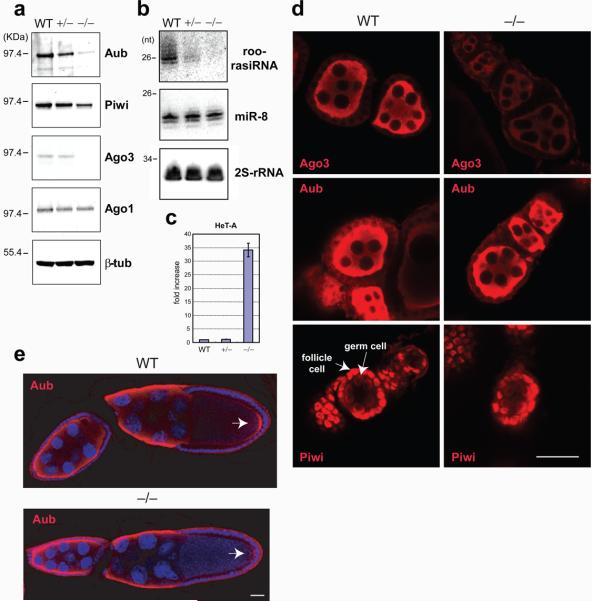
Figure 4. Reduction of Piwi proteins and piRNAs with accumulation of HeT-A retrotransposons and marked reduction of the Aub protein that localizes in the pole plasm, in the absence of dPRMT5 (csul) activity.
(a) Western blots of Drosophila ovary lysates from wild-type (wt), heterozygous (+/−) or homozygous (−/−) csul.
(b) Northern blots of total RNA from indicated ovary lysates.
(c) Fold changes of HeT-A retrotransposon transcripts in indicated ovaries assessed with qRT-PCR; n=3 and s.d. shown.
(d) Ago 3, Aub and Piwi localization in indicated early stage egg chambers; bar = 15μm
(e) Aub localization in indicated csul oocytes. Arrow indicates pole (germ) plasm; bar = 15μm
Next we compared the protein levels of Piwi family proteins between wt, heterozygous and homozygous csul ovaries. Western blot analysis showed that there was marked reduction of Aub and Ago3 protein levels and lesser reduction of Piwi levels in csul ovaries, whereas the levels of the miRNA binding protein Ago1 were not affected (Figure 4a). Since mRNA levels of Aub, Piwi and Ago3 are the same between wt and csul ovaries (Supplementary Figure 5), dPRMT5 activity might be required to stabilize the Aub, Ago3 and Piwi proteins most likely by symmetrically methylating their arginines. The level of a representative piRNA (roo-rasiRNA), was decreased in csul ovaries in accord with reduction of Piwi family proteins, while the level of a representative miRNA, miR-8, was not affected (Figure 4b). The homozygous csul ovaries showed a 30-fold increase in the levels of the HeT-A retrotransposon transcript (Figure 4c), whose expression is most sensitive to mutations that disrupt piRNA-directed silencing in the female germline 30. Collectively these results indicate that loss of dPRMT5 activity impairs the amounts of Piwi proteins and piRNAs, resulting in disruption of their function of transposon silencing.
Next we analyzed by confocal microscopy the localization of Ago3, Aub and Piwi in wt and homozygous csul early stage egg chambers. Previous studies have shown that Piwi is localized predominantly in the nuclei of follicle and germ cells 31 8 9 while Ago3 and Aub are localized in the cytoplasm of germ cells 8 9. In oocytes, Aub is concentrated in the germ (pole) plasm 4. Representative images are shown in Figure 4d and reveal that the level of Ago3 is markedly reduced in csul early stage egg chambers, while there is only a mild reduction of Aub and Piwi protein levels.
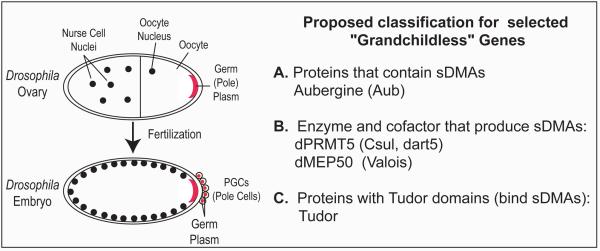
Germ cell (PGC) formation in Drosophila requires that cytoplasmic determinants are localized to the posterior pole of the embryo 32 (Figure 5, left panel). Genetic screens have identified grandchildless maternal genes that are required for PGC specification and invariably the protein or RNA products of these genes are concentrated in the pole plasm and are incorporated into the PGCs 32. Among these genes are Aub 4, dPRMT5 (csul, dart5) 5, 6, and its cofactor valois 33 and tudor 34, whose protein product contains eleven tudor domains 35. We tested the localization of Aub in csul oocytes by confocal microscopy. Representative results are presented in Figure 4e and show that the levels of Aub in the pole plasm of stage 10 egg chambers are markedly reduced. Western blotting reveals marked reduction of Aub protein levels in csul ovaries (Figure 4a) while confocal microscopy shows that Aub levels are subtly reduced in early stage egg chambers (Figure 4d) but markedly reduced in later stage egg chambers (Figure 4e), suggesting that lack of sDMAs affects Aub levels at later stages in oogenesis.
Figure 5. Proposed classification for selected Drosophila melanogaster grandchildless genes.
Our studies show that sDMA modification of Piwi family proteins is a conserved post-translational modification and we identify the methyltransferase PRMT5 (csul/dart5) as the enzyme that catalyzes sDMAs of Piwi, Ago3 and Aub in Drosophila ovaries, in vivo. Both Aub and csul/dart5 (dPRMT5) are grandchildless genes and our finding that Aub is a substrate for dPRMT5, indicates that an important function of dPRMT5 in pole plasm function and PGC specification involves methylation of Aub. Intriguingly, tudor domains bind to sDMAs 36 37 and Tudor protein is also a grandchildless gene that is required for pole plasm assembly and function. These findings suggest that pole plasm function may involve an interacting network of genes whose protein products contain sDMAs (Aub), the methylase (dPRMT5) and its cofactor (valois/dMEP50) that produce sDMAs and tudor domain (Tudor) proteins that may bind to sDMA-containing proteins (Figure 5). We note that both PRMT5 and tudor-domain-containing genes are found in all species that express Piwi family proteins and knockout of tudor domain containing 1/mouse tudor repeat 1 in mice leads to spermatogonial cell death and male sterility 38. Furthermore, we note that other Drosophila proteins whose genes are required for piRNA accumulation or function, such as Spindle-E/homeless 39 30 and Krimper 40, contain tudor domains. It will be interesting to test whether tudor domain containing proteins interact with sDMA-modified Piwi family proteins and to elucidate their function.
Methods
Antibodies
The anti-Mili monoclonal antibody 17.8 was generated by immunizing mice with recombinant GST-Mili. Other antibodies used were anti-Flag (Sigma), anti-sDMA (SYM11; Millipore), anti-aDMA (ASYM24; Millipore), anti-TMG (Santa Cruz Biotech), anti-β-tubulin (Developmental Studies Hybridoma Bank); anti-hnRNPC (4F4), anti-SMN (2B1), anti-Gemin3 (12H12), anti-Gemin5 (10G11), Y12 were gifts from G. Dreyfuss and antibodies against Drosophila Ago1, Aub, Piwi and Ago3 were gifts from MC. Siomi and H. Siomi.
Drosophila stocks
csul flies (csulRM: w−;csulRM50/CyO), were a gift from J. Anne and a deletion that uncovers csul (wa Nfa-g; Df(2R)Jp7, was obtained from Bloomington Drosophila Stock Center (Indiana University).
Xenopus laevis and piRNA sequencing
Oocytes were isolated from ovaries and defolliculated. Testis and liver tissues were procured from euthanized animals. Y12-immunopurified X. laevis piRNAs from testis and oocytes were 5′-end labeled and gel purified. Directional ligation of adapters and cDNA generation was performed using the small RNA sample prep kit (Illumina). Deep sequencing was performed on an Illumina Genome Analyzer.
Recombinant proteins and cDNA constructs
Recombinant Flag-Miwi and Flag-Mili and GST-Mili were produced in baculovirus infected Sf9 cells. The cDNAs for Flag-Miwi and Flag-Mili production and Flag-Miwi truncations mutants (BC for 68-862aa of Miwi and NP for 1-212aa) were gifts from S. Kuramochi-Miyagawa and T. Nakano.
Recombinant wild-type and mutant Flag-Aub were produced in S2 cells. The pRH-Flag-Aub expression vector was a gift from MC. Siomi and H. Siomi. To generate mutant Aub, arginine residues were changed to lysine (“WT: 10A-R-G-R-G-R-G-R-K18” changed to “mutant: 10A-K-G-K-G-K-G-K-K18”), by site-directed mutagenesis (QuickChange XL, Stratagene) using the following primers; forward: 5′-CACCAAACCCTGTAATTGCTAAAGGAAAAGGTAAAGGAAAAAAGCCCAATAATGTAGAGGC-3′ and reverse: 5′-GCCTCTACATTATTGGGCTTTTTTCCTTTACCTTTTCCTTTAGCAATTACAGGGTTTGGTG-3′. Wild-type and mutant pRH-Flag-Aub expression vectors were co-transfected with pCoBlast (Invitrogen) at a ratio of 5:1 using cellfectin at 27 °C. Twenty-four hours post-transfection, cells were grown in complete Schneider cell medium containing 15 μg/ml blasticidin. Antibiotic resistant clones were expanded and propagated in the presence of blasticidin. Recombinant Aub expression was induced by growing S2 cells seeded at 2 × 106 cells/ml in complete Schneider cell medium containing 500 μM CuSO4 for 48 hours, and the protein was purified by immunoprecipitation using anti-Flag M2 agarose (Sigma).
In situ hybridization and Northern blotting
Defolliculated oocytes were fixed with 10% neutral buffered formalin, paraffin embedded and sectioned at 5μm; in situ hybridization was performed with a Locked Nucleic Acid (LNA)-modified probe against XL-piR-3 (5′- UAAGUAGAAGAGCACCAAUGUCAUGUCC), an abundant piRNA sequenced from X. laevis oocytes. The sequence of the LNA probe was: 5′- ggAcatgaCattggtgcTcttctActta -3′-DIG (capital letters: LNA-modified nucleotides; DIG: digoxigenin). For Northern blotting, total RNA was isolated from X. laevis mixed stage oocytes and from liver and probed with 5′-end radiolabeled DNA probes antisense to XL-piR-3 (probe: 5′- GGACATGACATTGGTGCTCTTCTACTTA) and antisense to miR-16 (probe: 5′- CACCAATATTTACGTGCTGCTA).
UV crosslinking
A synthetic piRNA containing a photo-reactive residue, 4-thio-Uridine (s4U), at position 1 (5′-(s4U)GACAUGAACACAGGUGCUCAGAUAGCUUU-3′) was 5′-end labeled using [γ-32P] ATP and T4 polynucleotide kinase (T4 PNK, New England Biolabs). The 32P-labeled RNAs (∼20,000 cpm) were incubated with wild-type or mutant Flag-Aub beads at 28 °C for 60 min in a buffer containing 20 mM Tris-HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.05 % NP-40 and complete EDTA-free protease inhibitors (Roche). Crosslinking was performed on ice by irradiation for 30 min with a 365 nm hand-held lamp (EL series UV lamp, UVP). Cross-linked proteins were separated by NuPAGE and detected by storage-phosphor autoradiography.
Detailed methods are provided in the Supplement. Data and analysis for X. laevis piRNAs are found in the Supplement and in: http://cbcsrv.watson.ibm.com/piwi_modification/.
Supplementary Material
Acknowledgements
We are grateful to M.C. Siomi, H. Siomi, K. Saito, G. Dreyfuss for antibodies; to J. Anne for csul flies; to S. Kuramochi-Miyagawa and T. Nakano for Miwi and Mili cDNA constructs; to Rebecca Beerman for help with fly methodology, to Y. Kawamura for IF protocols and to members of the Mourelatos lab for discussions. Mass spectrometry was performed at the Proteomics Core Facility (University of Pennsylvania) and at the Proteomics Resource of the Keck Foundation (Yale University). Illumina sequencing was performed at the Functional Genomics Core of the University of Pennsylvania. Protein production was at the Protein Expression Core of the Wistar Institute. We apologize to colleagues whose studies were not cited because of space limitations. Supported by a Human Frontier Science Program Long Term Fellowship to YK, by NIH grant NS046573 to TAJ, GM76621 to PSK, and GM0720777, NS056070, UL1RR024134 and in part by ITMAT-PENN and URF-PENN grants to ZM.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–7. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 2.O'Donnell K,A, Boeke JD. Mighty Piwis Defend the Germline against Genome Intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartig JV, Tomari Y, Forstemann K. piRNAs--the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–13. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 4.Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–32. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 5.Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–46. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- 6.Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–89. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–27. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–48. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–50. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 12.Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–21. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582:2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 15.Houwing S, et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Krause CD, et al. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meister G, et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–4. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 20.Lerner EA, Lerner MR, Janeway CA, Jr., Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981;78:2737–41. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brahms H, et al. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–9. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- 22.Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–30. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Bowes JB, et al. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–7. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–8. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 25.Ohara T, et al. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–50. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 26.Saito K, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–8. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–72. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 28.Gonsalvez GB, et al. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol. 2007;178:733–40. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonsalvez GB, Praveen K, Hicks AJ, Tian L, Matera AG. Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster. Rna. 2008;14:878–87. doi: 10.1261/rna.940708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 31.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–14. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 32.Strome S, Lehmann R. Germ versus soma decisions: lessons from flies and worms. Science. 2007;316:392–3. doi: 10.1126/science.1140846. [DOI] [PubMed] [Google Scholar]
- 33.Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development. 2005;132:2167–77. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- 34.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 35.Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–62. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- 36.Selenko P, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 37.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–83. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 38.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103:15894–9. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–54. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–9. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.