Abstract
Members of the tristetraprolin family of tandem CCCH finger proteins can bind to AU-rich elements in the 3′-untranslated region of mRNAs, leading to their deadenylation and subsequent degradation. Partial deficiency of 1 of the 4 mouse tristetraprolin family members, Zfp36l2, resulted in complete female infertility because of early embryo death. We have now generated mice completely deficient in the ZFP36L2 protein. Homozygous Zfp36l2 knockout (KO) mice died within approximately 2 weeks of birth, apparently from intestinal or other hemorrhage. Analysis of peripheral blood from KO mice showed a decrease in red and white cells, hemoglobin, hematocrit, and platelets. Yolk sacs from embryonic day 11.5 (E11.5) Zfp36l2 KO mice and fetal livers from E14.5 KO mice gave rise to markedly reduced numbers of definitive multilineage and lineage-committed hematopoietic progenitors. Competitive reconstitution experiments demonstrated that Zfp36l2 KO fetal liver hematopoietic stem cells were unable to adequately reconstitute the hematopoietic system of lethally irradiated recipients. These data establish Zfp36l2 as a critical modulator of definitive hematopoiesis and suggest a novel regulatory pathway involving control of mRNA stability in the life cycle of hematopoietic stem and progenitor cells.
Introduction
ZFP36L2, zinc finger protein 36, C3H type–like 2 (also known as Brf2, Erf2, Tis11D), is a member of a small family of CCCH tandem zinc finger proteins capable of binding to RNA. The CCCH tandem zinc finger region in this family is characterized by 2 zinc fingers, spaced 18 amino acids apart, each containing 3 cysteines and one histidine with a strictly defined spacing, C-X8-C-X5-C-X3-H. In addition, a conserved amino-terminal sequence,(R/K)YKTEL, leads into each zinc finger.1 Tristetraprolin (TTP, ZFP36, Tis11, Nup475, Gos24) is the most extensively studied member of this protein family. Experiments conducted with bone marrow–derived macrophages from TTP knockout (KO) mice demonstrated that TTP can bind via its tandem zinc finger domain to AU-rich elements (AREs) found in the 3′-untranslated region (3′UTR) of the mRNA encoding tumor necrosis factor-α (TNF-α).2 This binding leads to the destabilization of the TNF mRNA; thus, in the absence of TTP, there is an abnormal accumulation of TNF mRNA and protein. This leads in turn to the development of a systemic inflammatory syndrome characterized by arthritis, cachexia, skin lesions, autoimmunity, and myeloid hyperplasia.3 This systemic inflammatory syndrome was almost completely alleviated by either the administration of TNF antibodies or by breeding to mice deficient in both TNF receptors.3,4 Subsequent studies have shown that TTP can also bind to and destabilize transcripts for granulocyte-macrophage colony-stimulating factor (GM-CSF) in bone marrow–derived stromal cells,5 interleukin-2β (IL-2β) in primary T lymphocytes,6 IL-10 in RAW264.7 macrophage cells,7 immediate-early response 3 (Ier3) in primary embryo fibroblasts,8 and other mRNAs.8–11
ZFP36L2, like the other 2 mammalian family members, ZFP36L1 (cMG1, Brf1, Erf1, Tis11B, Berg36) and ZFP36L3, contains a highly conserved TTP-like tandem zinc finger domain and is capable of binding to and destabilizing TNF and GM-CSF mRNAs in cell transfection and cell-free experiments.12–16 We previously generated a line of mice in which Zfp36l2 had been disrupted; this was not a complete KO but rather a first exon deletion, ΔN-ZFP36L2, leading to decreased levels of an amino-terminal truncated protein.17 This truncated protein still contained an intact, functional tandem zinc finger domain but was missing the first 29 amino acids. The ΔN-ZFP36L2 phenotype was complete female infertility. Females were able to cycle and ovulate normally, and their ova could be fertilized; however, the embryos did not progress past the 2-cell stage of development.
For the current study, we have generated mice from 2 independent clones of targeted embryonic stem (ES) cells in which the majority of the ZFP36L2 protein-coding region, including the tandem zinc finger domain, was deleted by homologous recombination. The Zfp36l2 KO mice were born at the expected Mendelian frequency, but within several weeks of birth they died rather suddenly with pallor and frequent intestinal hemorrhage. These mice exhibited pancytopenia, decreased hematopoietic progenitor cells from fetal liver and yolk sac, and ineffective hematopoietic stem cells (HSCs). These studies demonstrate that Zfp36l2 plays an important role in definitive hematopoiesis during mouse development and suggests that regulation of mRNA stability is a critical aspect of the development of the hematopoietic system.
Methods
Mice
Zfp36l2 knockout mice (B6;129-Zfp36l2tm2Pjb) were generated and genotyped as described in supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article). All results reported in this paper come from 2 independent lines of mice, derived from 2 separate ES cell clones. All mouse experiments were conducted according to the US Public Health Service policy on the humane care and use of laboratory animals. All animal procedures used in this study were approved by the National Institute of Environmental Health Sciences and the Indiana University School of Medicine Institutional Animal Care and Use Committees.
RNA isolation and Northern analysis
Mouse tissues, either from adults or embryos, were collected into RNAlater (Applied Biosystems/Ambion) according to the manufacturer's instructions. Total cellular RNA was isolated using either the RNeasy kit (QIAGEN) or the Illustra RNAspin kit (GE Healthcare). RNA was isolated directly from mouse embryo fibroblasts (MEFs) without the use of RNAlater. Mouse mammary and E7 embryo RNA was obtained from Clontech. RNA was fractionated on a 1.2% formaldehyde-agarose gel, transferred to Hybond-N+ membranes (GE Healthcare), and hybridized with random-primed, α-32P-labeled probes as previously described.18 Northern blots were hybridized with one of the following ZFP36L2 cDNA probes: a 277-bp polymerase chain reaction (PCR) fragment corresponding to nucleotides 1 to 277 of NM_001001806.2, containing the first exon (PCR primers F3 and R3 in Figure 1A); a 787-bp fragment corresponding to nucleotides 2073 to 2839 of NM_001001806.2, containing a portion of the 3′UTR (PCR primers F4 and R4 in Figure 1A); and a 703-bp EcoRV/SacII fragment excised from a mouse ZFP36L2 cDNA, p11DcDNA,19 corresponding to nucleotides 286 to 1016 of NM_001001806.2, containing the last 18 bp of exon 1 and the first 712 bp of exon 2. Some blots were also hybridized with the following expressed sequence tags obtained from the IMAGE Consortium: Cxcl1, IMAGE clone ID 1548709, GenBank accession no. BX521623.1; Mllt11, IMAGE clone ID 5688748, accession no. BM949069.1; Plekha3, IMAGE clone ID 4981486, accession no. BC031110.1; Alox12, IMAGE clone ID 40141477, accession no. BC152329.1; Mpl, IMAGE clone ID 40039147, accession no. BC146613.1; Itga2b, IMAGE clone ID 40129416, accession no. BC120493.1; Csf2rb1, IMAGE clone ID 7110372, accession no. CK483869.1; and Cd59a, IMAGE clone ID 40130366, accession no. BC132089.1. Blots were also hybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe2 or a cyclophilin cDNA probe4 to monitor gel loading. Northern blots were quantitated using a Typhoon phosphorimager (GE Healthcare).
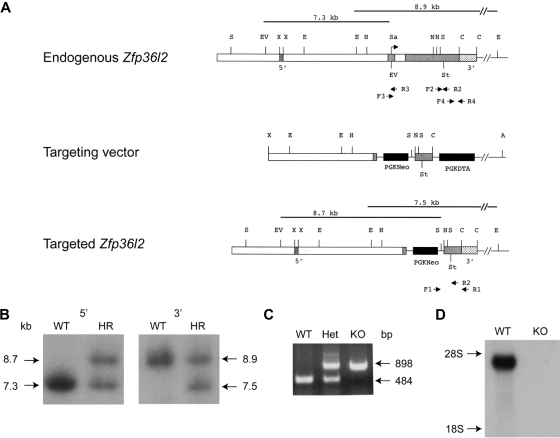
Figure 1.
Targeted disruption of Zfp36l2. (A) Schematic representation of the normal genomic locus for Zfp36l2 as well as the targeting vector and resulting disrupted allele generated by homologous recombination. The 2 exons are represented by gray boxes. The translational start site is indicated by the arrow (under Sa). Genomic 5′ and 3′ probes, located outside the targeting vector, are represented by stippled boxes. The targeting vector contained a neomycin resistance marker cassette (PGKNeo), causing the deletion of a large portion of the second exon. A diphtheria toxin resistance element was also inserted into the targeting vector (PGKDTA). The black lines above the endogenous and targeted genes represent the expected fragment length after digestion with EcoRV and SstI or EcoRI and hybridization with the 5′ probe or 3′ probe, respectively. PCR primers used to detect homologous recombination (F1, R1) as well as for genotyping offspring (F1, F2, and R2) are indicated by the arrows beneath the endogenous and targeted genes. Primers F3, R3, F4, and R4 were used to generate an exon one fragment (F3, R3) or a 3′UTR fragment (F4, R4) used as hybridization probes for Northern blots. Abbreviations for restriction enzyme sites are as follows: A indicates Asp718; C, Csp45I; E, EcoRI; EV, EcoRV; H, HindIII; N, NotI; Sa, SalI; S, SstI; St; stop codon; X, XbaI. (B) Southern blot analysis of EcoRV/SstI (5′ genomic probe) and EcoRI (3′ genomic probe)–digested genomic DNA from WT and homologously recombined (HR) ES cells. (C) PCR analysis of genomic DNA isolated from Zfp36l2 WT, heterozygous, and KO mice using PCR primers F1, F2, and R2. (D) Northern blot of total cellular RNA (15 μg) isolated from E14.5 WT and KO fetal liver and hybridized with a 32P-labeled 787-bp 3′UTR probe for Zfp36l2.
Histopathology and clinical pathology
Animals were killed at various ages, a gross examination of all organs was conducted, and carcasses and internal organs were fixed in 10% neutral-buffered formalin and used for paraffin embedding, sectioning, and staining with hematoxylin and eosin. For hematologic analysis, peripheral blood was obtained by submandibular bleeding or cardiac puncture, collected into ethylenediaminetetraacetic acid–containing Microvette collection tubes, and analyzed using a Hemavet 950 hematology analyzer (Drew Scientific). Bone marrow cells were isolated by flushing dissected femurs or tibias with 3 mL RPMI 1640 medium (Invitrogen) using a 25-gauge needle. A single-cell suspension was obtained by several passes through the needle and used to prepare cytospin slides (Shandon) that were stained with the Hemacolor staining kit (EMD Chemicals).
Isolation of MEFs and mRNA decay studies
MEFs were isolated from fetal mice at embryonic (E) day 14.5 of gestation, where E0.5 was the date of detection of the vaginal plug, and subsequent mRNA decay assays were conducted as described in the supplemental data.
In situ hybridization histochemistry
In situ hybridization was performed as described.12,20 Briefly, intact embryos within the yolk sac were isolated at E14.5 and frozen in powdered dry ice. Fifteen-micron sections were cut on a cryostat and mounted onto aminopropyltriethoxys subbed slides. Sections were dehydrated at 25°C for several minutes and then frozen at −20°C until used for hybridization. The antisense oligonucleotide probe (48 bases; Integrated DNA Technologies), 5′-GGGTGACAGAAGTGTGGTCGACATTTTTCTGGGTCCTGTAATGGTCGA-3′, was designed to be complementary to bases 229-248 of NM_001001806.2. The sense probe, 5′-TCGACCATTACAGGACCCAGAAAAATGTCGACCACACTTCTGTCACCC-3′, corresponded to the same region of the mRNA. This sequence had minimal sequence identity with the other TTP family members, Zfp36, Zfp36l1, and Zfp36l3. An oligonucleotide probe for mouse GAPDH mRNA was used as a positive control; this was complementary to bp 169 to 216 of NM_008084.1.
Probes were 3′ end-labeled with 35S-dATP (specific activity > 1000 Ci/mmol; PerkinElmer Life And Analytical Sciences) using terminal transferase (Roche Applied Science or Fermentas).
Flow cytometry
Staining of cells for flow cytometry was performed using the indicated conjugated antibodies obtained from either BD Biosciences or eBioscience. The cells were resuspended in Hanks Balanced Salt Solution (Ca2+, Mg2+ free; Invitrogen) containing 2% fetal bovine serum (FBS) [HF2] and then incubated for 30 minutes on ice with one or more of the following antibodies: Mac-1–phycoerythrin (PE; BD Biosciences), Gr-1-fluorescein isothiocyanate (FITC; BD Biosciences), B220-FITC (BD Biosciences), CD34-PE (eBioscience), CD33-PE (eBioscience), CD41-PE (BD Biosciences), Ter119-FITC (BD Biosciences), cKit-FITC (BD Biosciences PharMingen), and Sca-1-PE (BD Biosciences). To stain lineage-positive cells, a biotinylated antibody cocktail (StemCell Technologies) and streptavidin-allophycocyanin (BD Biosciences) were used. After staining, cells were washed twice with phosphate-buffered saline and then resuspended with HF2 containing 1 μg/mL propidium iodide (Sigma-Aldrich). The analyses were performed using a dual-laser FACScan with CellQuest software (BD Biosciences).
Colony-forming cell assays
Adult bone marrow and spleen hematopoietic progenitor cells.
A total of 4 × 104 bone marrow nucleated cells or splenocytes were plated onto 35-mm Petri dishes in Methocult M3434 methylcellulose medium (StemCell Technologies). Methocult M3434 is 2% methylcellulose supplemented with cytokines (50 ng/mL recombinant mouse [m] stem cell factor [SCF], 10 ng/mL rmIL-3, 10 ng/mL recombinant human [h] IL-6, and 3 U/mL rh erythropoietin [Epo]). The colony-forming cell plates were incubated at 37°C in a 5% CO2 incubator, and the number of colonies was determined 12 days after plating the cells.
Definitive fetal liver (E14.5 embryo) and yolk sac (E11.5 embryo) hematopoietic progenitor cells.
Fetal liver cells were plated at concentrations ranging from 2.0 to 30.0 × 104 mL in 1% methylcellulose culture with 30% (vol/vol) FBS (HyClone Laboratories) with 1 U/mL rhEpo, 5% (vol/vol) pokeweed mitogen mouse spleen cell conditioned medium, 50 ng/mL rmSCF, and 0.1 mM hemin (Eastman Kodak). Granulocyte-macrophage (CFU-GM), erythroid (BFU-E), multipotential (CFU-GEMM), and granulocyte-macrophage/macrophage (CFU-GM/M) colonies were scored after 7 days of incubation in a humidified atmosphere with 5% CO2 and lowered (5%) O2 as described.21,22 CFU-GM/M colonies were stimulated with 10 ng/mL rmGM-CSF, 10 ng/mL rmIL-3, 100 ng/mL rmM-CSF, and 50 ng/mL SCF. Cytokines were purchased from R&D Systems, except for Epo, which was purchased from Amgen Corp.
Primitive erythroid yolk sac (E8-8.25) progenitor cells.
Yolk sac cells were plated at 3 to 10 × 103 cells/mL in 0.9% methylcellulose-based media (StemCell Technologies) that included Iscove modified Dulbecco medium, 2 mM glutamine, 1% penicillin/streptomycin, 5% protein-free hybridoma medium-II (Invitrogen), 50 μg/mL ascorbic acid (Sigma-Aldrich), 450 μM monothioglycerol (Sigma-Aldrich), 200 μg/mL iron-saturated holotransferrin (Sigma-Aldrich), 15% plasma-derived serum (Animal Technology), and 4 U/mL rhEpo.23 Cultures were incubated at 37°C in 5% CO2, 5% O2, and colonies were counted at day 5.
In vivo long-term marrow repopulating HSCs
This assay detects functional and competitive stem cells24 and was performed as previously reported,21,22 except that donor CD45.2 fetal liver cells were used at 5 × 105 cells in combination with 5 × 105 competitor CD45.1 (B6.Boy J) adult mouse bone marrow cells for injection into CD45.1 recipients after recipient mice received a single lethal dose (950 cGy) of irradiation.
Affymetrix microarray analysis and TaqMan real-time PCR
Gene expression analysis was conducted using Affymetrix Mouse Genome 2.0 GeneChip Arrays (Mouse 430 Version 2; Affymetrix). Microarray analysis and real-time PCR were performed as outlined in the supplemental data. All microarray data have been deposited to Gene Expression Omnibus under accession no. GSE15146.
Results
Generation of Zfp36l2 KO mice
The targeting vector was designed to replace most of the protein coding region of Zfp36l2, including the tandem zinc finger domain (bases 257-1242 of NM_001001806.2), with a PGKNeo cassette (Figure 1A). Four ES cell clones were obtained that contained the correctly targeted allele as assessed by Southern analysis. Southern blots of genomic DNA from targeted ES cells digested with EcoRV and SstI (5′ end) or with EcoRI (3′ end) were hybridized with a 5′ probe and a 3′ probe, respectively (Figure 1B). The 5′ probe recognized a 7.3-kb endogenous band in the wild-type (WT or +/+) clone and a 7.3-kb endogenous band and a 8.7-kb targeted band in the homologously recombined (+/−) clone. The 3′ probe recognized an 8.9-kb endogenous band in the WT clone and an 8.9-kb endogenous plus a 7.5-kb–targeted band in the recombined clone. Two homologously recombined ES cell clones were injected into C57Bl/6J-TyrC-2J blastocysts to generate chimeric mice. Chimeric mice were bred to C57Bl/6 mice, and the resulting heterozygous offspring were intercrossed. Intercrosses of heterozygous mice resulted in offspring with an approximately Mendelian distribution of WT, heterozygous, and KO genotypes. Typical PCR genotyping results are shown in Figure 1C. To confirm the absence of Zfp36l2 mRNA expression, RNA was isolated from fetal livers from WT and KO E14.5 mice. Northern analysis demonstrated that Zfp36l2 mRNA was present in WT fetal liver, but no detectable mRNA was seen in the livers from KO mice (Figure 1D).
Zfp36l2 mRNA expression in mouse embryos and tissues
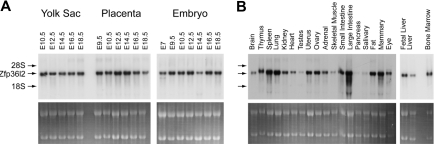
Analysis of embryonic expression of Zfp36l2 mRNA is shown in Figure 2A. Expression was readily detected in the embryo at E7, increased slightly at E9.5, and then remained relatively constant through E18.5. Expression in the yolk sac was relatively constant between E10.5 and E18.5. Placenta expression was also constant throughout development with a slight decrease observed at E18.5. Zfp36l2 mRNA expression was also examined in adult mouse tissues (Figure 2B). The highest levels of expression were seen in thymus, spleen, lung, uterus, ovary, small and large intestine, mammary gland, fat, and bone marrow; intermediate expression was found in kidney, heart, adrenal, eye, and fetal liver; and relatively low level expression was seen in brain, skeletal muscle, and adult liver. Zfp36l2 mRNA was barely detectable in testes, pancreas, and salivary glands. In some of the lanes, there was evidence of minor probe hybridization to smaller RNA species, apparently including the 18S ribosomal RNA. These minor bands may represent low levels of cross-hybridization to other mouse TTP family members because a BLAST comparison of the full-length Zfp36l2 mRNA sequence yields similarities with Zfp36l1 mRNA at a significance level of 5e-75, and with TTP mRNA of 2e-18.
Figure 2.
Developmental expression and tissue distribution of Zfp36l2 mRNA. Total cellular RNA (15 μg) was isolated from yolk sacs, placentas, and whole embryos at the indicated gestational stages (A) and from adult mouse tissues and E14.5 liver (B). Northern blots were hybridized with a 32P-labeled 787-bp 3′UTR probe for Zfp36l2. The positions of Zfp36l2 mRNA and the ribosomal RNAs are shown to the left of the blot. The bottom panels show acridine orange staining of the samples as loading controls.
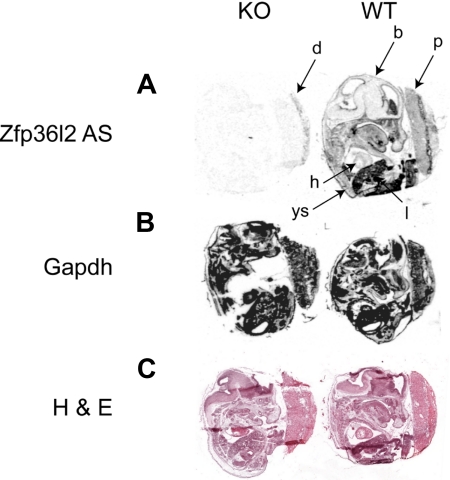
In situ hybridization with a 35S-labeled Zfp36l2 antisense probe was performed to examine the predominant sites of mRNA expression at E14.5. The highest level of expression revealed by the antisense probe was in the fetal liver (l; Figure 3A), but expression was seen in many other tissues, including yolk sac (ys). A sense probe yielded essentially no signal when hybridized to an adjacent section from the WT embryo (D.J.S., E.K.R., and P.J.B., unpublished data, 2009), whereas an antisense probe for GAPDH showed a strong signal throughout the embryo, yolk sac, and placenta (Figure 3B). There was minimal detectable signal in a KO fetus processed in parallel (Figure 3A). However, there was detectable Zfp36l2 mRNA expression in the maternal decidua (d) of the KO placenta, which should be heterozygous for Zfp36l2.
Figure 3.
In situ hybridization histochemistry of Zfp36l2 mRNA expression at E14.5. Sagittal sections of E14.5 WT and KO embryos were hybridized with Zfp36l2 (A) or GAPDH (B) antisense probes. The specificity of the Zfp36l2 antisense probe was demonstrated by the absence of signal seen with the KO embryo compared with the WT embryo. A sense probe for Zfp36l2 produced essentially no signal at this exposure time (data not shown). Note the weak hybridization seen in the KO placenta in the maternal decidua, a tissue that should be heterozygous for Zfp36l2. (C) Neighboring sections stained with hematoxylin and eosin. b indicates brain; d, decidua; h, heart; l, liver; ys, yolk sac; and p, placenta. Original magnifications ×40, obtained using the Aperio Scanscope T2 Scanner (Aperio Technologies Inc). Scanned images were imported into Aperio Imagescope, Version 6.25.0.1117.
Zfp36l2 deficiency results in pancytopenia
Zfp36l2 KO pups (from 2 independent lines of targeted ES cells) were born at approximately the expected Mendelian frequency, but most of them died within 2 weeks of birth. One female KO mouse survived 51 days, and one 32 days. Approximately half of the KO mice were smaller than their WT and heterozygous littermates, and half appeared to be of normal size. Because most KO mice died at approximately 2 weeks of age, most experimental animals were killed between 12 and 15 days. All of the KO mice necropsied had pallor of the skin, liver, and kidneys, often accompanied by overt hemorrhage, most frequently in the intestines. Living KO pups often appeared pale, and subcutaneous hemorrhages were often visible in the living mice. Heterozygous mice appeared to be healthy and fertile and showed no signs of hemorrhage or pallor.
Complete necropsies, with hematoxylin and eosin staining of most tissues, were conducted on 2 female mice (51 and 32 days of age) and one male (24 days old). Consistent findings in all 3 mice included evidence of multinodular erythropoiesis in the spleen, with evidence of maturation arrest; the primitive erythroid cells stained positive for Ter119 (data not shown). The bone marrow was hypocellular, as discussed later in this section. There were no other consistent anatomic abnormalities. Two KO and 2 WT fetal mice were also evaluated histopathologically at E18.5; there were no apparent consistent differences between the WT and KO mice at this age.
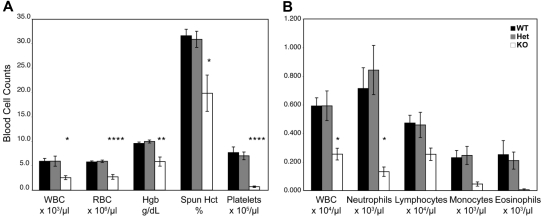
Analysis of peripheral blood, bone marrow, and spleens of Zfp36l2 KO mice revealed a range of defects. Peripheral blood counts revealed that the KO mice had markedly and significantly decreased levels of red blood cells (2.3-fold), white blood cells (2.1-fold), hemoglobin (1.6-fold), and hematocrit (1.6-fold). The most dramatic decrease was in the platelet count, where there was an 11-fold (P < .001) decrease in the number of platelets in the KO mice compared with WT and heterozygous mice (Figure 4A). The values obtained for heterozygous mice were the same as seen for WT mice. White blood cell differential counts showed that KO mice had significantly fewer neutrophils (5.4-fold) than either WT or heterozygous mice (Figure 4B). There were also marked decreases in lymphocytes, monocytes, and eosinophils in the KO animals (Figure 4B), but these changes did not achieve statistical significance, perhaps because of the relatively small sample number (n = 5-7) and the fact that comparisons were performed among multiple means.
Figure 4.
Hematologic analysis of WT, heterozygous, and KO mice. (A) Complete blood cell counts of peripheral blood from Zfp36l2 WT (n = 6), heterozygous (n = 7), and KO (n = 6) mice at 14 to 15 days of age. (B) White blood cell differential counts for peripheral blood from Zfp36l2 WT (n = 6), heterozygous (n = 7), and KO (n = 5, except WBC, n = 6) mice. Values are mean ± SEM. Statistical significance was determined using one-way analysis of variance with Tukey HSD post hoc test: *P < .05; **P < .01; ****P < .001. WBC indicates white blood cell; RBC, red blood cell; Hgb, hemoglobin; and Hct, hematocrit.
Flow-cytometric analysis of cells from postnatal spleens at 13 or 15 days of age demonstrated that there were significant decreases in several cell types in the Zfp36l2 KO mice compared with WT mice (supplemental Figure 1). Similar to the findings in the peripheral blood, myeloid (Gr-1+/Mac-1+) and megakaryocyte (CD41+) cell populations were decreased 4-fold and 14-fold, respectively (Table 1). In addition, similar to the peripheral blood findings, there was a minor decrease in the proportion of B220+ and CD3+ splenocytes that did not reach statistical significance (Table 1). Values for heterozygous mice were similar to those seen for WT mice. Bone marrow cell preparations from WT, heterozygous, and KO mice showed a 2-fold decrease in the percentage of myeloid cells in the KO bone marrow (WT 28.4%, heterozygous 31.8%, KO 15.2%; data not shown), as determined by manual counting of cytospin slides. Flow cytometric analysis of Lin−/Sca-1+/c-Kit+ (LSK) bone marrow cells, which include HSCs, showed a significant decrease in the Zfp36l2 KO mice (WT 0.09 ± 0.02, n = 7; KO 0.01% ± 0.00, n = 6; P < .005; supplemental Figure 2).
Table 1.
FACS analysis of splenocytes
| CD41+ | Ter119+ | Mac-1+ Gr-1+ | CD3+ | B220+ | |
|---|---|---|---|---|---|
| WT (n = 6) | 3.89 ± 0.58 | 40.04 ± 2.52 | 2.38 ± 0.51 | 6.23 ± 0.72 | 41.19 ± 2.81 |
| Het (n = 5) | 3.93 ± 0.49 | 39.89 ± 5.94 | 1.40 ± 0.18 | 6.96 ± 1.11 | 41.17 ± 2.59 |
| KO (n = 6) | 0.28 ± 0.07* | 52.41 ± 3.27 | 0.56 ± 0.17† | 5.74 ± 0.95 | 29.51 ± 4.92 |
Splenocytes were isolated from postnatal day 13 and 15 mice. Data represent the mean ± SEM of the percentage of cells stained with the indicated antibody.
P < .001 using one-way analysis of variance with Tukey HSD post hoc test.
P < .01 using one-way analysis of variance with Tukey HSD post hoc test.
Splenocytes from 13- or 15-day-old mice were cultured in methylcellulose supplemented with cytokines to support the proliferation and differentiation of multipotential and lineage-committed hematopoietic progenitors. Splenocytes from KO mice gave rise to significantly fewer myeloid and erythroid colonies (Table 2). Consistent with the marked pancytopenia and decreased number of LSK cells, bone marrow from KO mice (12- or 13-day-old mice) generated no colonies in methylcellulose (Table 3).
Table 2.
Spleen progenitors (colonies/104 cells)
| WT (n = 5) | Het (n = 4) | KO (n = 5) | |
|---|---|---|---|
| BFU-E | 7.2 ± 1.89 | 3.5 ± 0.88 | 0 ± 0* |
| CFU-E | 7.4 ± 2.87 | 5.0 ± 1.00 | 0.2 ± 0.14* |
| CFU-GM | 22.6 ± 6.32 | 11.0 ± 2.25 | 0.2 ± 0.14* |
A total of 40 000 splenocytes from postnatal day 13 and 15 mice were plated in duplicate in methylcellulose medium and the number of colonies determined 12 days after plating. Data represent the mean ± SEM.
P < .05 by one-way analysis of variance with Tukey HSD post hoc test.
Table 3.
Bone marrow progenitors
| WT |
KO |
|||||
|---|---|---|---|---|---|---|
| Mouse 1 | Mouse 2 | Mouse 3 | Mouse 1 | Mouse 2 | Mouse 3 | |
| BFU-E | 25 | 7 | 28 | 0 | 0 | 0 |
| CFU-GM | 42 | 17 | 31 | 0 | 0 | 0 |
| CFU-GEMM | 4 | 1 | 6 | 0 | 0 | 0 |
| Total | 71 | 25 | 65 | 0 | 0 | 0 |
A total of 40 000 bone marrow nucleated cells from postnatal day 12 and 13 mice (3 WT and 3 KO) were plated in duplicate in methylcellulose medium and the number of colonies determined 12 days after plating.
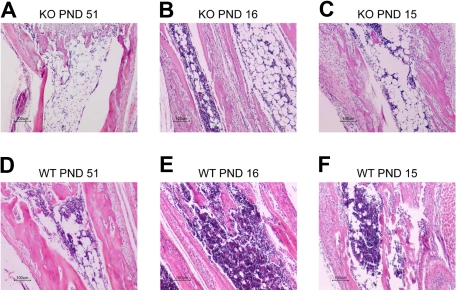
We observed hypocellular bone marrow (associated with an increase in the number of adipocytes) in postnatal day (PND) 12, 15, 16, 32, and 51 KO mice (Figure 5). However, not all KO bone marrow examined showed this hypocellularity. There was an apparent variability in the onset of severe anemia in the mice, sometimes occurring as early as PND 8 and, as mentioned, in one instance as late as PND 51. We concluded that the Zfp36l2 KO mice were most probably dying because of anemia, either as a primary cause of death or secondary to hemorrhage, possibly because of the severe thrombocytopenia. Most of the animals developed hypocellular bone marrows, especially the older ones, most probably the result of bone marrow hematopoietic failure.
Figure 5.
Histologic analysis of bone marrow in Zfp36l2 KO and WT mice. Tissue sections from the radius/ulna of PND 51 KO (A) and WT (D) mice, PND 16 KO (B) and WT (E) mice, and PND 15 KO (C) and WT (F) littermate pairs of mice were stained with hematoxylin and eosin. Note the hypocellular bone marrow along with an increased number of adipocytes in the KO samples. Original magnifications ×10, obtained with a Nikon Eclipse E600 microscope with a Nikon DXM1200 digital camera. Images were imported into the Nikon Act-1 software.
Defective definitive hematopoiesis during development in Zfp36l2 KO mice
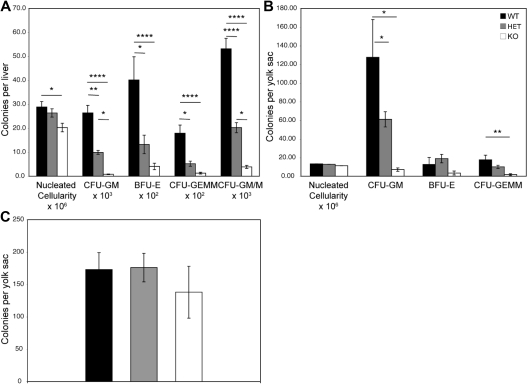
Given the hematopoietic defects in the young mice, we examined hematopoiesis during development. We initially performed hematopoietic progenitor analyses using colony-forming assays in E14.5 fetal liver cells. Fetal liver is thought to be the primary site of fetal hematopoiesis from approximately E12 until birth, after which hematopoiesis shifts to the spleen and bone marrow.25–29 Fetal liver cells were cultured in semisolid medium supplemented with various combinations of cytokines that support the proliferation and differentiation of multipotential and lineage-committed hematopoietic progenitors. We observed significant decreases in numbers of erythroid (BFU-E, 10-fold), granulocyte macrophage (CFU-GM, 32-fold), granulocyte macrophage/macrophage (CFU-GM/M, 14-fold), and multipotential (CFU-GEMM, 14-fold) progenitor cells obtained from Zfp36l2 KO fetal liver cells compared with WT cells (Figure 6A). There was a small (1.4-fold) but significant decrease in the nucleated cellularity of the KO fetal livers. We also observed smaller but significant decreases in cells from heterozygous fetal livers (Figure 6A). These results strongly support a gene dosage effect, with intermediate decreases in most cell types seen in fetal livers from the heterozygous mice.
Figure 6.
Hematopoietic progenitor colony assays from fetal liver and yolk sacs of Zfp36l2 WT, heterozygous, and KO embryos. Fetal liver cells (A) isolated from E14.5 embryos (WT, n = 15; heterozygous, n = 26, KO, n = 14) and yolk sac cells (B) isolated from E11.5 embryos (WT, n = 5; heterozygous, n = 10; KO, n = 5) were grown in methylcellulose-based medium as described in “Colony-forming cell assays.” The numbers of CFU-GM, BFU-E, and CFU-GEMM, and CFU-GM/M per fetal liver or yolk sac were determined after 7 days of culture. (C) Yolk sac cells isolated from E8 to E8.25 embryos (WT, n = 20; heterozygous, n = 31; KO, n = 10) were grown in methylcellulose-based medium. Primitive erythroid progenitor colonies, EryP-colony–forming cells (C) were determined after 5 days in culture. Values are mean ± SEM. Statistical significance was determined using one-way analysis of variance with Tukey HSD post hoc test: *P < .05; **P < .01; ****P < .001.
We next examined the effect of ZFP36L2 deficiency on hematopoietic progenitor cells in E11.5 yolk sac cells. The yolk sac is thought to be the first site of hematopoiesis in the developing embryo. Two waves of hematopoiesis occur in the yolk sac, with primitive erythroid progenitors appearing transiently between E7 and E9, whereas definitive erythroid and myeloid progenitors appear between E8.25 and E10.0.30–32 Definitive progenitors that arise in the yolk sac circulate to the liver at E9.5 to E10.5 for eventual differentiation. There were significant decreases in myeloid (CFU-GM, 18-fold) and multipotential (CFU-GEMM, 10-fold) progenitors in the KO yolk sacs (Figure 6B).
To assess whether ZFP36L2 might play a role in primitive hematopoiesis, we performed in vitro colony-forming assays on yolk sac cells derived from E8 to E8.25 embryos. No significant differences were observed in the number of primitive erythroid progenitors from WT, heterozygous, and KO E8 to E8.25 yolk sacs (Figure 6C).
Taken together, these results suggest that there is a defect in definitive hematopoiesis in Zfp36l2 KO embryos, whereas primitive hematopoiesis appears to be relatively normal.
ZFP36L2 is required for HSC reconstitution of the hematopoietic system
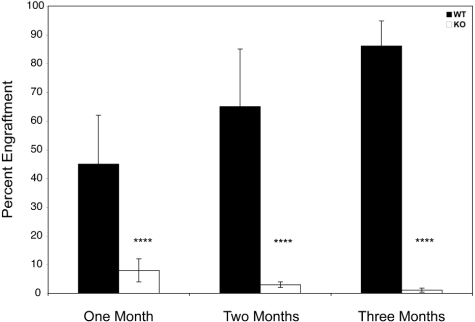
HSCs are highly enriched in the LSK population of cells. There were fewer of these phenotypically defined populations in Zfp36l2 KO compared with WT mice, as noted in “Zfp3612 deficiency results in pancytopenia.” However, phenotype does not always recapitulate function, and analysis of stem cells requires an in vivo functional assay. To this end, we used an in vivo competitive stem cell assay to assess the short (1 month) and longer (6 month) repopulating capacity of WT and KO fetal liver cells.21,22 As seen in Figure 7, both the short- and longer-term reconstituting capacities of KO fetal liver cells were greatly compromised compared with those of the WT cells. Thus, ZFP36L2 appears to be critical for functional activity of HSCs as well as progenitor cells.
Figure 7.
Competitive hematopoietic reconstitution with Zfp36l2 WT and KO fetal liver cells. A total of 5 × 105 donor-derived fetal liver cells (CD45.2+) from WT (n = 19) and KO (n = 23) E14.5 embryos were mixed with 5 × 105 adult bone marrow recipient–derived cells from B6.BoyJ (CD45.1+) mice and were injected into lethally irradiated B6.BoyJ (CD45.1+) mice. The percentage of donor CD45.2+ cells that engrafted, as determined by CD45.1/CD45.2 staining of peripheral blood, was examined at 1, 2, and 6 months after transplantation. Values are mean ± SD. Statistical significance was determined using the Student t test: ****P < .001.
Altered gene expression in Zfp36l2 KO fetal liver
Because ZFP36L2 probably functions as an ARE-containing mRNA destabilizing protein, we were interested in finding potential target transcripts that would accumulate abnormally in fetal livers from the Zfp36l2 KO mice. To do this, we performed microarray analysis on RNA derived from E14.5 WT and KO fetal livers. More than 400 transcripts were either increased or decreased at a significance level of P < .001. Of the 239 transcripts significantly upregulated in KO fetal livers, only 3 were found to contain promising AREs of the TTP-binding type in their 3′ UTRs (Table 4): Cxcl1, Plekha3, and Mllt11. The Cxcl1 mRNA has an ARE that is a good potential target for TTP family member proteins and has recently been shown to be a target of TTP in peritoneal macrophages.11 Using real-time PCR to verify the microarray results, we confirmed that Cxcl1 mRNA levels were increased 4-fold in Zfp36l2 KO fetal livers compared with WT (P < .002). We then performed actinomycin D time courses with primary MEFs isolated from Zfp36l2 WT and KO embryos. Serum-deprived fibroblasts were treated with 10% FBS for 90 minutes, at which time actinomycin D was added and cells were harvested at various times for RNA purification. Under these conditions, there appeared to be no differences in the decay rates for Cxcl1 mRNA in serum-stimulated WT and KO fibroblasts (data not shown). We also confirmed the increased mRNA levels for Plekha3 and Mllt11 by Northern analysis (data not shown). Actinomycin D time-course experiments demonstrated that there were no differences in the decay rates for Mllt11 mRNA comparing serum-stimulated WT and KO fibroblasts (data not shown). Plekha3 mRNA, unlike both Cxcl1 and Mllt11, did not decay detectably in response to actinomycin D treatment, either in WT or KO serum-stimulated fibroblasts, in time courses up to 120 minutes (data not shown).
Table 4.
Genes differentially expressed in fetal liver cells of Zfp36l2 knockout mice
| Gene name | Gene ID | Function | Microarray KO vs WT | Northern/real-time PCR KO vs WT |
|---|---|---|---|---|
| 3′ UTR ARE-containing | ||||
| Cxcl1 (KC) | NM_008176 | Chemokine (C-X-C motif) ligand 1 | 5.9, 3.7, 2.2 | 4,* 7† |
| Plekha3 (FAPP1) | BB780848 | Pleckstrin homology domain-containing, family A (phosphoinositide-binding specific member 3 | 1.7 | 1.6† |
| Mllt11 (Af1q) | NM_019914 | Myeloid/lymphoid or mixed lineage leukemia (trithorax homolog, Dros) translocated to 11 | 1.6 | 2.2† |
| Hematopoiesis | ||||
| Il6st (gp130, CD130) | AK017211 | Interleukin 6 signal transducer | 1.79 | |
| Fga | BC005467 | Fibrinogen, α-polypeptide | 1.54 | |
| Cxcl4 (Pf4) | NM_019932 | Chemokine (C-X-C motif), ligand 4 (platelet factor 4) | −2.6 | |
| Cxcl7 (Ppbp) | NM_023785 | Chemokine (C-X-C motif), ligand 7 (pro-platelet basic protein) | −2.57 | |
| Gp5 | NM_008148 | Glycoprotein 5 (platelet) | −2.45 | |
| Gp1bb | NM_010327 | Glycoprotein Ib, β-polypeptide | −2.44 | |
| Gp1ba | NM_010326 | Glycoprotein Ib, α-polypeptide | −2.35 | |
| Itgb3 (CD61, GP3A) | AV352983 | Integrin β3 (platelet glycoprotein IIIa) | −2.29 | |
| Treml1 | AK017256 | Triggering receptor expressed on myeloid cells-like 1 | −2.24 | |
| Rab27b | BB121269 | Rab27b, member of RAS oncogene family | −2.11 | |
| Alox12 | BB554189 | Arachidonate 12-lipoxygenase | −2.05 | −1.97† |
| Mpl (TPO-R, CD110) | NM_010823 | Myeloproliferative leukemia virus oncogene (thrombopoietin receptor) | −1.92 | −2.5† |
| Itga2b (CD41, GpIIb) | NM_010575 | Integrin α2b (platelet glycoprotein IIb) | −1.78 | −2.0† |
| Thbs1 | AI385532 | Thrombospondin 1 | −1.66, −1.61 | |
| CD59a | AK005507 | CD59a antigen | −1.42 | −1.77† |
| Other | ||||
| Csf2rb | BB769628 | Colony stimulating factor 2, β, low-affinity (granulocyte-macrophage) | 2.34 | 2.0† |
| Ccr1l1 (MIP-1 alphaRL1) | NM_007718 | Chemokine (C-C motif) receptor 1-like 1 (macrophage inflammatory protein-1 alpha receptor-like 1) | −2.4 | |
| HSCs | ||||
| Vwf | BB667216 | von Willebrand factor homolog | −1.92‡ | |
| Slamf1 (Slam, CD150) | BB132695 | Signaling lymphocytic activation molecule family member 1 | −1.83 | |
| Pbx1 | BG073018 | Pre-B-cell leukemia transcription factor | −1.62§ |
Partial list of genes differentially expressed in Zfp36l2 KO fetal liver at a significance level of P < .001. Total RNA was isolated from 5 KO and 5 WT E14.5 fetal livers and used for gene expression analysis using Affymetrix Mouse Genome 2.0 GeneChip Arrays (Mouse 430, Version 2). Values represent the fold changes (positive numbers indicate increased expression and negative numbers indicate decreased expression) in gene expression in KO fetal liver compared with WT fetal liver.
Microarray mRNA changes confirmed by real-time PCR.
Microarray mRNA changes confirmed by Northern analysis.
P = .003.
P = .02.
Of the transcripts found to be significantly downregulated in KO fetal livers, many have been associated in some way with hematopoiesis or platelet function. For example, Thbs1, the CXC chemokines Cxcl4 (Pf4) and Cxcl7 (Ppbp), and Treml1, Itga2b (CD41), Itgb3 (CD61), Rab27b, and Alox12 are all expressed in platelets (Table 4).
Discussion
In this study, we demonstrated that Zfp36l2 plays an essential role in definitive hematopoiesis during mouse development. Zfp36l2 KO mice exhibited a range of hematopoietic defects. Peripheral blood analysis indicated that KO mice had significantly decreased levels of red and white blood cells, hemoglobin, hematocrit, and platelets. KO mice generally died within approximately 2 weeks of birth with anemia and thrombocytopenia, and often had intestinal and other hemorrhages. Analysis of postnatal spleens and bone marrow demonstrated that there were decreased myeloid and megakaryocyte cell populations as well as decreased hematopoietic progenitor cells and HSCs. Examination of embryonic hematopoiesis showed that the lack of ZFP36L2 expression led to a decrease in hematopoietic progenitor cells in E10.5 yolk sacs and E14.5 fetal livers. Primitive erythroid progenitor cell number, as measured in E8 to E8.25 yolk sacs, did not appear to be affected by ZFP36L2 deficiency. Competitive reconstitution studies revealed that Zfp36l2 KO fetal liver cells were unable to effectively reconstitute the hematopoietic system in lethally irradiated recipients, suggesting that ZFP36L2 might be required for self-renewal and/or maintenance of adult HSCs.
TTP, the most widely studied member of this family of CCCH tandem zinc finger proteins, has been shown to bind to AREs located in the 3′UTR of several mRNAs.33 TTP binding leads to the deadenylation and destabilization of these mRNAs.14,15 ZFP36L2 has also been shown in cell transfection and cell-free experiments to bind to some of the identified TTP mRNA targets, resulting in the deadenylation and subsequent destabilization of the bound mRNAs.12–16 Based on the observation that TNF and GM-CSF, known physiologic targets of TTP,2,5 are abnormally stabilized in TTP knockout mice, leading to the development of a systemic inflammatory syndrome, it seems probable that the absence of ZFP36L2 protein expression may lead to the abnormal stabilization of one or more mRNAs whose protein product may directly or indirectly affect HSC function.
To begin to address this possibility, we isolated RNA from E14.5 WT and KO fetal livers and examined differentially expressed mRNAs using microarray analysis. Of the 239 transcripts upregulated in KO fetal liver, only 3 were found to contain AREs in their 3′UTR that contained more than 3 UAUUUAU sequences, of which at least 2 were overlapping, and were conserved between mouse and human. These 3 transcripts represent possible ZFP36L2 targets, but verification of their identity as true targets will require mRNA stability measurements in cells derived from WT and KO mice. Of these 3 transcripts (Cxcl1, Mllt11, and Plekha3), Cxcl1 mRNA stability has recently been shown to be regulated by TTP in peritoneal macrophages isolated from TTP WT and KO mice.11 Cxcl1 (also known as KC, growth-related oncogene α, Gro-α, and melanoma growth stimulatory activity) is a member of the CXC subfamily of chemokines. It acts as a functional homolog of human IL-8 in the mouse.34 Chemokines are mediators of inflammation and immunity that are thought to be responsible for the recruitment of leukocytes to injured tissues. Some chemokines play a role in stem cell survival, development and homeostasis, and angiogenesis.34–36 At least 25 chemokines in the CC and CXC subfamilies have been shown to have suppressive effects on the proliferation of myeloid progenitor cells.37 Cxcl1 was found to have no suppressive effect on human bone marrow progenitor cells,37,38 but it was capable of suppressing IL-3–dependent proliferation of the murine myeloid progenitor cell line 32D.39 However, at least in our studies in MEFs, we were not able to demonstrate an effect of ZFP36L2 deficiency on Cxcl1 mRNA stability. Obviously, these fibroblasts may not be an appropriate cell type in which to do this analysis, given the profound effects of ZFP36L2 deficiency on hematopoiesis. In addition, it is possible that ZFP36L2 deficiency may cause the secondary abnormal accumulation of one or more transcripts but in the absence of direct mRNA stabilization. Finally, it has recently been shown that another member of the TTP family of CCCH tandem zinc finger proteins, ZFP36L1, has been implicated in the expression of vascular endothelial growth factor-A at the translational level,40 rather than at the level of mRNA stability. Further studies will be necessary to determine whether or not the increased expression of Cxcl1 mRNA and the other potential target transcripts that were seen in Zfp36l2 KO fetal livers are the direct result of changes in mRNA stability affected by ZFP36L2 binding and, in turn, whether these changes in transcript levels are directly related to the observed hematopoietic phenotype.
We stress that this initial microarray survey involved intact fetal liver and that there are probable changes in hematopoietic cell number and type in this tissue in the KO mice. For example, if HSCs were decreased in number in the KO mice, per fetal liver, we would be attempting to detect increased accumulation of transcripts in decreased numbers of cells, clearly an unsatisfactory situation. Alternatively, certain subpopulations of hematopoietic cells could be increased in the fetal livers from the KO mice, resulting in false-positive apparent increases in transcript accumulation. In future experiments, we will attempt to analyze gene expression in subsets of cells from fetal liver, particularly populations enriched in HSCs. At this point, we cannot completely distinguish between decreased intrinsic proliferative ability of HSCs and progenitors, versus sharply decreased numbers of stem cells; it is even possible that abnormalities exist in both the numbers and functions of these cells.
Hematopoiesis is a complex hierarchical process, which involves the expansion and differentiation of a limited number of HSCs into multipotential and lineage-restricted progenitors, leading to the production of mature and functional blood cells. During this process, HSCs that arise in distinct hematopoietic tissues during development31,41–45 must migrate to the liver, where expansion takes place, and then subsequently migrate to the spleen and the bone marrow. In the bone marrow, HSCs must maintain the proper balance between proliferation and differentiation. Deregulation of hematopoiesis can lead to a number of hematologic abnormalities. Regulation of hematopoiesis can occur at a number of steps along the pathway from the embryonic generation of HSCs and progenitor cells, to the migration, homing, and self-renewal and/or differentiation of HSCs. A number of cytokines and transcription factors are known to be involved in the regulation and modulation of hematopoiesis.37,46–50 Our competitive reconstitution studies demonstrate that Zfp36l2 KO fetal liver cells are defective in maintaining hematopoiesis in irradiated recipient mice. Additional studies will be needed to determine what step(s) along the hierarchical pathway of hematopoiesis are influenced by ZFP36L2. It is interesting to speculate that ZFP36L2 might be influencing the abundance of one or more known or unknown protein factors involved in HSC development, function, or renewal.
In conclusion, we have demonstrated that Zfp36l2 plays an important role in definitive hematopoiesis during mouse development. Definitive hematopoiesis can occur, although suboptimally, but ZFP36L2-deficient HSCs are apparently unable to maintain adequate hematopoiesis for survival in the postnatal animal. Further studies will be required to address the mechanism by which ZFP36L2 is regulating hematopoiesis and to determine whether or not regulating the stability of an mRNA encoding one or more critical regulatory proteins is involved.
Supplementary Material
Acknowledgments
The authors thank Dori Germolec and Anton Jetten for reviewing the manuscript, Sandra Ward for analysis of the bone marrow cytospin slides, the National Institute of Environmental Health Sciences Microarray facility for performing and analyzing the microarray experiments, Ruchir Shah of the Costella Group for analysis of the microarray data, and Dee Wenzel for animal husbandry and breeding support.
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, National Cancer Institute, and National Institutes of Health (grants R01 HL56416 and R01 HL67384, H.E.B.; grant R01 HL63169, M.C.Y.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.J.S. performed and analyzed the research and wrote the paper; H.E.B. supervised and analyzed the fetal liver definitive progenitor assays and in vivo transplantation assays; T.W. helped with animal work; S.C. performed and analyzed the fetal liver progenitor assays; G.H. performed and analyzed the transplantation assays; P.D.A. supervised and analyzed the spleen and bone marrow progenitor FACS and progenitor assays; Y.J.C. performed and analyzed the spleen and bone marrow FACS and progenitor assays; M.C.Y. supervised and analyzed the yolk sac primitive progenitor assays; W.C.S. designed and performed the yolk sac primitive progenitor assays; E.K.R. performed the in situ hybridization analysis; M.K.R. made the knockout mice; and P.J.B. supervised the entire study and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Perry J. Blackshear, A2-05 National Institute of Environmental Health Sciences, 111 Alexander Dr, Research Triangle Park, NC 27709; e-mail: black009@niehs.nih.gov.
References
- 1.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265(27):16556–16563. [PubMed] [Google Scholar]
- 2.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281(5379):1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 3.Taylor GA, Carballo E, Lee DM, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 4.Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98(8):2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- 5.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95(6):1891–1899. [PubMed] [Google Scholar]
- 6.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174(2):953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 7.Stoecklin G, Tenenbaum SA, Mayo T, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283(17):11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26(24):9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmons J, Townley-Tilson WH, Deleault KM, et al. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA. 2008;14(5):888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frasca D, Landin AM, Alvarez JP, Blackshear PJ, Riley RL, Blomberg BB. Tristetraprolin, a negative regulator of mRNA stability, is increased in old B cells and is involved in the degradation of E47 mRNA. J Immunol. 2007;179(2):918–927. doi: 10.4049/jimmunol.179.2.918. [DOI] [PubMed] [Google Scholar]
- 11.Datta S, Biswas R, Novotny M, et al. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180(4):2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 12.Blackshear PJ, Phillips RS, Ghosh S, Ramos SB, Richfield EK, Lai WS. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod. 2005;73(2):297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 13.Lai WS, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J Biol Chem. 2001;276(25):23144–23154. doi: 10.1074/jbc.M100680200. [DOI] [PubMed] [Google Scholar]
- 14.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275(23):17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 15.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol. 2003;23(11):3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res Ther. 2004;6(6):248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos SB, Stumpo DJ, Kennington EA, et al. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131(19):4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 18.Stumpo DJ, Graff JM, Albert KA, Greengard P, Blackshear PJ. Molecular cloning, characterization, and expression of a cDNA encoding the “80- to 87-kDa” myristoylated alanine-rich C kinase substrate: a major cellular substrate for protein kinase C. Proc Natl Acad Sci U S A. 1989;86(11):4012–4016. doi: 10.1073/pnas.86.11.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips RS, Ramos SB, Blackshear PJ. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J Biol Chem. 2002;277(13):11606–11613. doi: 10.1074/jbc.M111457200. [DOI] [PubMed] [Google Scholar]
- 20.McCoy L, Cox C, Richfield EK. Antipsychotic drug regulation of AMPA receptor affinity states and GluR1, GluR2 splice variant expression. Synapse. 1998;28(3):195–207. doi: 10.1002/(SICI)1098-2396(199803)28:3<195::AID-SYN2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Broxmeyer HE, Orschell CM, Clapp DW, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201(8):1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopherson KW, 2nd, Hangoc G, Mantel CR, Broxmeyer HE. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305(5686):1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 23.Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood. 2008;111(7):3435–3438. doi: 10.1182/blood-2007-08-107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood. 1980;55(1):77–81. [PubMed] [Google Scholar]
- 25.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1(1):57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand JY, Giroux S, Golub R, et al. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc Natl Acad Sci U S A. 2005;102(1):134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi K. The hemangioblast: a common progenitor of hematopoietic and endothelial cells. J Hematother Stem Cell Res. 2002;11(1):91–101. doi: 10.1089/152581602753448568. [DOI] [PubMed] [Google Scholar]
- 28.Dzierzak E, Medvinsky A. Mouse embryonic hematopoiesis. Trends Genet. 1995;11(9):359–366. doi: 10.1016/s0168-9525(00)89107-6. [DOI] [PubMed] [Google Scholar]
- 29.Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101(2):508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 30.McGrath KE, Palis J. Hematopoiesis in the yolk sac: more than meets the eye. Exp Hematol. 2005;33(9):1021–1028. doi: 10.1016/j.exphem.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133(19):3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 32.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126(22):5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 33.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30(6):945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 34.Romagnani P, Lasagni L, Annunziato F, Serio M, Romagnani S. CXC chemokines: the regulatory link between inflammation and angiogenesis. Trends Immunol. 2004;25(4):201–209. doi: 10.1016/j.it.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Baggiolini M. Chemokines in pathology and medicine. J Intern Med. 2001;250(2):91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 36.Broxmeyer HE. Chemokines in hematopoiesis. Curr Opin Hematol. 2008;15(1):49–58. doi: 10.1097/MOH.0b013e3282f29012. [DOI] [PubMed] [Google Scholar]
- 37.Youn BS, Mantel C, Broxmeyer HE. Chemokines, chemokine receptors and hematopoiesis. Immunol Rev. 2000;177:150–174. doi: 10.1034/j.1600-065x.2000.17701.x. [DOI] [PubMed] [Google Scholar]
- 38.Broxmeyer HE, Kim CH. Regulation of hematopoiesis in a sea of chemokine family members with a plethora of redundant activities. Exp Hematol. 1999;27(7):1113–1123. doi: 10.1016/s0301-472x(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez X, Suetomi K, Cousins-Hodges B, Horton JK, Navarro J. CXC chemokines suppress proliferation of myeloid progenitor cells by activation of the CXC chemokine receptor 2. J Immunol. 1998;160(2):906–910. [PubMed] [Google Scholar]
- 40.Bell SE, Sanchez MJ, Spasic-Boskovic O, et al. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn. 2006;235(11):3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 41.Cumano A, Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol. 2007;25:745–785. doi: 10.1146/annurev.immunol.25.022106.141538. [DOI] [PubMed] [Google Scholar]
- 42.Dzierzak E. The emergence of definitive hematopoietic stem cells in the mammal. Curr Opin Hematol. 2005;12(3):197–202. doi: 10.1097/01.moh.0000160736.44726.0e. [DOI] [PubMed] [Google Scholar]
- 43.Mikkola HK, Gekas C, Orkin SH, Dieterlen-Lievre F. Placenta as a site for hematopoietic stem cell development. Exp Hematol. 2005;33(9):1048–1054. doi: 10.1016/j.exphem.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8(3):377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26(47):6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 47.Loose M, Swiers G, Patient R. Transcriptional networks regulating hematopoietic cell fate decisions. Curr Opin Hematol. 2007;14(4):307–314. doi: 10.1097/MOH.0b013e3281900eee. [DOI] [PubMed] [Google Scholar]
- 48.Pina C, Enver T. Differential contributions of haematopoietic stem cells to foetal and adult haematopoiesis: insights from functional analysis of transcriptional regulators. Oncogene. 2007;26(47):6750–6765. doi: 10.1038/sj.onc.1210759. [DOI] [PubMed] [Google Scholar]
- 49.Robb L. Cytokine receptors and hematopoietic differentiation. Oncogene. 2007;26(47):6715–6723. doi: 10.1038/sj.onc.1210756. [DOI] [PubMed] [Google Scholar]
- 50.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453(7193):306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.