Abstract
Human CMV is the predominant infectious cause of congenital birth defects and an opportunistic pathogen in immunosuppressed individuals, including AIDS patients. Most individuals are infected early during their life followed by life-long latent infection. During this latent phase, frequent reactivation and antigen production continue to stimulate the immune system. While the immune response is able to control the virus, it is unable to eradicate it. Moreover, super-infection by different CMV strains has been observed despite a strong immune response. Long-term immune stimulation by CMV has also been implicated in immune senescence and chronic conditions such as atherosclerosis. CMVs are highly species-specific and the relatedness of CMV genomes exactly mirrors the relatedness of their hosts. Thus, each CMV species is highly adapted to its respective host species, but is unable to infect other, even closely related hosts. While fascinating from an evolutionary perspective, this host restriction prevents studying HCMV in experimental animals. Exceptions are severely immunocompromised mice, e.g. SCID mice, or SCID/NOD mice, which might allow partial reconstitution of CMV infection in rodents. More practical however, is to study CMVs in their natural host, e.g. murine, rat or guinea pig CMVs. However, while these small animal models have many advantages, such as the availability of inbred animals as well as lower cost, the limited homology of the viral genomes with HCMV limits the functional analysis of homologous gene products. The closest relative to HCMV is chimpanzee CMV (CCMV), but this is not a practical animal model since chimps are a protected species, extremely expensive and of very limited availability. In contrast, rhesus macaques are a more widely used experimental animal species and, while more distant than CCMV, rhesus CMV (RhCMV) contains most of the HCMV gene families thus allowing the study of their role in acute and latent CMV infection. In this review we will discuss the current state of developing RhCMV as a model for HCMV.
Keywords: Cytomegalovirus, CMV, Rhesus cytomegalovirus, Rhesus CMV, Cytomegalovirus animal models
Introduction
Human cytomegalovirus (HCMV), a β-herpesvirus, is a successful and widespread pathogen [1]. Immunological and PCR-based assays revealed that 60–100% of the adult population is infected with the virus, with frequency of infection highest in urban areas. Infection is generally asymptomatic in healthy individuals, but the virus is a major cause of morbidity and mortality in immunocompromised hosts, who are less able to control primary infection or reactivation of latent virus. Indeed, HCMV is an opportunistic infection during AIDS and represents a common, life-threatening post-transplant complication in allograft recipients [2]. Congenital infection is also a serious problem with HCMV being the largest infectious cause of birth defects in the United States [3]. Total health care costs related to HCMV disease in the US alone exceed $4.4 billion annually with costs for CMV-induced sensorineural hearing loss and mental retardation alone exceeding $1 billion [4]. After initial infection, CMV persists in the host indefinitely and viral shedding in multiple body fluids (saliva, tears, urine, genital secretions, breast milk) can occur years after initial exposure. Like other herpesviruses, CMV can manifest latent infection of its target cells [5]. However, the frequency of shedding [6], kinetics of reactivation after transplantation [7], and the high CMV-specific T cell response [8] suggest that active CMV replication, or at least in some cases gene expression [9], occurs frequently within the infected host. This persistent, life-long infection together with the continuous stimulation of the immune system has also been implicated in chronic diseases, such as atherosclerosis, restenosis and transplant vascular sclerosis (reviewed in [10]).
One of the limitations of studying CMV is the fact that HCMV does not infect animals. This species-specificity is a characteristic trait of β-herpesviruses in general, and therefore, viral evolution has accompanied mammalian speciation such that each species has their own uniquely adapted CMV, and CMV relatedness generally parallels species relatedness [11]. Thus, the genomes of primate CMVs (human [12], chimp [13], rhesus [14]) are much more closely related to each other than rodent CMVs (mouse [15], rat [16], guinea pig [17]). Consequently, animal models for CMV depend on studying the CMVs of the respective mammal species. The most widely used model is murine CMV (MCMV), which can address many fundamental questions for biology, pathogenesis and immunology of cytomegalovirus [18, 19]. However, the genomes of MCMV and HCMV are quite different and many “non-essential genes”, i.e. genes that are not required for growth in vitro, are also non-conserved in MCMV or other rodent viruses.
The closest relative of HCMV that infects primates is chimpanzee CMV (CCMV) [13]. However, chimpanzees are a protected species, costly and only available in very small numbers, which limits their use as a model for HCMV. In contrast, rhesus macaques (RM) are a widely used and available species for non-human primate (NHP) studies. Like all mammals, RM harbor their own species of CMV, RhCMV [20]. In this review we will summarize the current knowledge of RhCMV pathogenesis, gene functions and its usefulness as animal model for HCMV.
Pathogenesis of RhCMV
Similar to human CMV, the majority of monkeys within a rhesus population are positive for CMV, usually measured by serology. For instance, 95% of RM at “monkey temples” in India tested positive for RhCMV [21]. Similarly, close to 100% of RM in primate centers are CMV positive [22]. Most monkeys seroconvert during the first year of their life [23]. Once infected, NHP continue shedding virus for the rest of their lives in urine and saliva [6, 20]. Thus, to study primary CMV infection in RM, CMV-free animals need to be kept separate from the rest of the colony, which adds considerable cost and logistical challenges to this animal model. Experimental infection of naïve animals (oral or i.v.) was shown to result in initial viremia in the blood and virus could be detected in various organs, particularly the spleen [24]. All infections were asymptomatic, similar to primary CMV infection of adult humans. To facilitate detection of CMV both in vivo and in vitro, Peter Barry and colleagues recently developed a recombinant CMV expressing GFP [25] as well as RhCMV strain 68.1 in a bacterial artificial chromosome, which greatly facilitates generating recombinant viruses [26]. Importantly, both recombinant viruses retained their ability to infect RM suggesting that molecular manipulations do not reduce RhCMV pathogenicity. Taken together, these data suggest that the pathogenesis, prevalence, and transmission of RhCMV are highly similar to that of HCMV.
RhCMV as a model for CMV-mediated congenital defects
Primary infection of seronegative pregnant women is a well-known cause of birth defects, particularly sensorineural defects [3]. In addition, recent observations suggest that hearing loss in children born to latently infected mothers occurred at the same frequency, albeit with slower progression, as in infants from primary infected mothers [27]. The close developmental, immunological, anatomical, and biochemical similarities between RM and humans render RhCMV particularly suited for the development of models for congenital CMV infection (recently reviewed in [28]). While natural congenital infection is difficult to detect in a monkey colony, intrauterine inoculation is being used to study the effect of CMV infection on the developing fetus. In one study it was shown that intra-amniotic inoculation with RhCMV leads to CNS lesions in the newborn [29]. However, more severe disease was observed upon intra-cranial inoculation of the fetus [30]. Similar to humans, severity of neurological defects was much higher when infections occurred during the first trimester of infection [28]. Thus, intrauterine infection with RhCMV clearly resembles some of the features of congenital HCMV infection. However, there is also a need to develop models that permit studying the impact of vertical transfer of RhCMV from pregnant mothers to infants, particularly during primary infection with RhCMV. The generation of CMV-free animal colonies at various primate centers should facilitate the development of such models.
RhCMV as model to test CMV anti-viral drugs
Rhesus macaques are routinely used at late stages of preclinical testing of vaccines as well as drugs in many areas of research. The main reason to use primates for these studies is that bio-availability and toxicity profiles obtained in primates are likely more predictive for humans than those obtained in small animal models. Primary and long-term RhCMV infected RM are thus ideally suited to examine the impact of novel drugs as well as anti-CMV vaccines. For instance, the pharmacokinetics of Ganciclovir, a well-established anti-CMV drug, was tested in blood and CNS in RM and was found to be similar to humans [31]. Moreover, RhCMV was found to be equally susceptible to Ganciclovir as HCMV and the activity of the Ganciclovir-kinase, UL97, was found to be similar [32]. RhCMV also showed comparable sensitivity to HCMV for benzimidazole nucleosides, a promising new class of anti-CMV drugs [33]. Thus, RhCMV will be an excellent model to test new anti-CMV drugs in vivo.
Rhesus macaques were also used to test the effect of anti-CD40 and anti-CD28 antibodies to prolong renal allograft survival. Interestingly, there was a significant reactivation of RhCMV in anti-CD40/CD28-treated animals with severe systemic manifestations of disseminated CMV disease [34]. This observation suggests that monitoring RhCMV will be an important aspect during the pre-clinical testing of immune suppressive treatments.
Opportunistic infection and AIDS
Human CMV is a well-known opportunistic infection in AIDS patients. A common manifestation is CMV retinitis, which has become rare thanks to the availability of highly active antiretroviral therapy (HAART). Ocular CMV-infection was also noticed in SIV-infected monkeys [35]. However, a more common occurrence is disseminated CMV disease in animals developing spontaneous SAIDS [36] or upon experimental infection with SIV [37]. Affected organs include the gastro-intestinal tract [38, 39]. The occurrence of disseminated CMV in SIV-infected animals is thus a unique and highly relevant model to study the interaction of these two viruses. The reactivation of CMV from latency and the development of disseminated CMV correlated with the deterioration of CMV-specific cellular immune responses [40]. In a progressive study, a clear correlation was observed in the magnitude and rate of decline of CMV-specific antibodies and CD4+ and CD8+ T lymphocytes and the degree by which macaques progressed to CMV disease [41]. These data indicate that both humoral and cellular immune responses seem to protect from developing CMV disease and it is the combined decay that results in AIDS-opportunistic infection. The rhesus model also allows the study of the development of RhCMV and SIV infections upon co-infection with both viruses, thus mimicking situations where primary infection with each virus occurs almost simultaneously [42]. These experiments suggested that con-current primary infection with CMV could augment the development of AIDS.
RhCMV immunology
The immune response to CMVs and the components necessary for disease suppression have been well characterized using the MCMV model in mice as well as data from HCMV in humans. Thus far, RhCMV has largely been used as a tool by immunologists to help provide insight into the macaque immune response that provides critical information for the development of the RM model in general. This includes the characterization of the CD4+ CD8+ T cell population [43], the characterization of the cytolytic properties of CD8+ T cells [44], defining of the memory T cell population [45], and the description of useful techniques such as polychromatic flow cytometry analysis of immune cells during infection [46].
Another focus of immunological study using RhCMV has been characterizing the response to RhCMV during SIV infection. This helps to establish the model for CMV pathogenesis during AIDS (see previous section). CTL responses to RhCMV were similar in both SIV-negative animals and those infected with pathogenic and attenuated SIV strains [47]. However, the frequency of CMV-specific CD4+ T cells was found to be reduced in SIV-positive macaques [40]. Furthermore, the regulatory T cell response to RhCMV is delayed compared to SIV [48].
Similar to HCMV in humans, RhCMV gB appears to be a strong target of the humoral immune response. gB antibodies are detected in the serum of infected animals and there is a strong correlation between neutralizing and gB-specific antibody levels [49].
Genomics
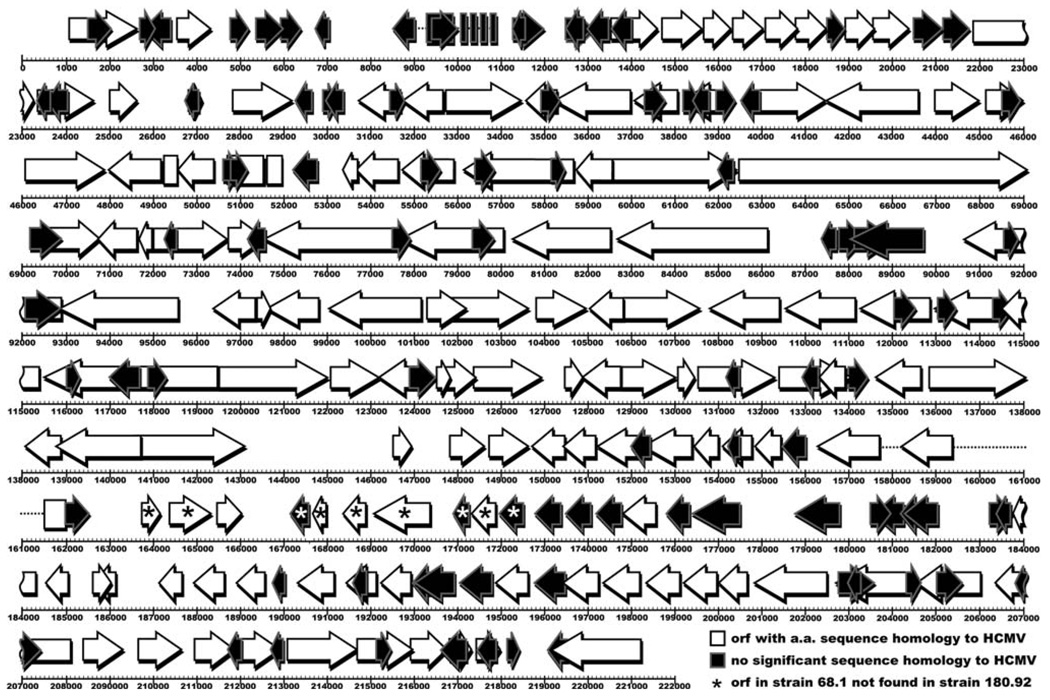
The RhCMV genome (strain 68.1) is 221,459 bp in length, slightly smaller than HCMVs 229,354 bp [14]. The genomes are colinear and share a similar structure, although unlike HCMV, RhCMV does not appear to isomerize [14]. Initially 230 open reading frames (ORFs) were predicted within strain 68.1, although this number was later proposed to be modified to 260 [50]. Of these, 135 are homologous to known HCMV proteins (Fig. 1). These homologs include members of the RL11, UL25, UL82, US1, US6, US12, US22, and seven-transmembrane protein families. Large loci of RhCMV-specific genes are located at the ends of the RhCMV genome and in the rh165–180 ORFs, a region at the same position as the UL/b′ region of HCMV, which is deleted or truncated in laboratory strains [51]. There are currently two full-length RhCMV strains sequenced [14, 50]. The two genomes are 97% identical at the nucleotide level, although strain 180.92 is 5,781 nucleotides shorter than strain 68.1, is missing 10 ORFs found in strain 68.1, and contains 8 ORFs not identified in strain 68.1 [50]. This indicates variability between RhCMV strains not unlike differences seen between strains of HCMV. Along with the identification of orthologs in CCMV, the overall similarity between RhCMV and HCMV has helped redefine the coding potential for HCMV [13, 52]. And while there is a large amount of coding potential seemingly unique to RhCMV, many RhCMV proteins have maintained functional homology to HCMV despite limited sequence homology [53].
Fig. 1.
RhCMV open reading frame (ORF) homology to HCMV. This diagram details the predicted ORFs in the RhCMV genome (strain 68.1). ORFs in white have significant amino acid (a.a.) sequence homology to HCMV proteins, while ORFs in black have no significant homology to HCMV. Asterisks indicate ORFs in strain 68.1 not found in strain 180.92. Rectangles indicate known exons. Dashed lines indicate known introns. For a more detailed analysis see Refs. [14] and [50]
Functional characterizations of RhCMV proteins
Multiple RhCMV genes and gene products have already been characterized. Early studies on the RhCMV immediate early 1 and 2 (IE1/IE2) gene and promoter region showed a conservation of gene structure, transcription, and protein sequence with HCMV IE1/IE2 [54, 55]. gB, a major target of anti-HCMV antibodies, is also well conserved in RhCMV. The cloned RhCMV gB is proteolytically processed similarly to HCMV gB, portions of RhCMV gB cross-react with anti-HCMV gB antibodies, and RhCMV gB antibodies can be detected during RhCMV infection [56, 57]. Phosphoprotein 65 (pp65) is another dominant target for the immune response to HCMV. RhCMV encodes two pp65 homologs. Much like HCMV pp65, Rh-pp65–2 localizes to the nucleus, is contained within the virion, and is the target of both humoral and cellular immunity [58]. The conservation of the immune system targets IE1/IE2, gB and pp65 makes RhCMV a viable model for studying potential CMV vaccines.
The ability to study HCMV-homologous immunomodulatory proteins in vivo also makes RhCMV attractive. HCMV inhibitors of apoptosis are conserved in RhCMV. Both the UL36 (viral inhibitor of caspase-8-induced apoptosis, vICA) and UL37 (mitochondria inhibitor of apoptosis, vMIA) homologs in RhCMV were able to prevent Fas-mediated apoptosis in HeLa cells, whereas the MCMV UL37 homolog M37 was not [59]. The IL-10 homolog of CMVs was first identified in RhCMV and was shown to be expressed in vivo, targeted by the humoral immune response, and to have immunosuppressive properties [60, 61]. RhCMV also contains homologs of HCMV US2, US3, US6, and US11 [53]. This gene family inhibits expression of major histocompatibility complex class I (MHC I) molecules to avoid CD8+ T-cell detection, and while MHC I modulators are found in rodent CMVs, the US2-US11 gene family is not. RhCMV contains five homologs to the HCMV chemokine receptor US28, a unique seven-transmembrane domain gene family not found in rodent CMVs. One of these, RhUS28.5, has been shown to have a similar ligand binding profile as HCMV US28 [62]. The US12 gene family, another seven-transmembrane domain gene family found only in primate CMVs, is also well conserved in RhCMV [63].
Among the RhCMV-specific genes is a cyclooxygenase-2 (COX-2) homolog that was shown to be a critical factor in viral growth in endothelial cells [64]. HCMV, while not having a COX-2 homolog, does upregulate cellular COX-2 upon infection, and COX-2 inhibitors prevent normal viral replication [65]. Lastly, RhCMV is able to block expression of interferon-stimulated genes (ISGs) by preventing activation of interferon regulatory factor-3 (IRF3) [66]. The failure to induce ISGs by an enveloped virus is thus far unique to RhCMV.
Concluding remarks
The development of RhCMV as an animal model for Human CMV has clearly come of age. Many useful tools have been developed in recent years that facilitate handling and manipulating the RhCMV genome such as bacterial artificial chromosomes [26] and telomerized rhesus fibroblasts [67]. In addition, immunological markers of rhesus immune cells have been developed [45] and several models of RhCMV pathogenesis, primary infection and re-infection are now available [24]. Due to cost and the unavailability of inbred and knockout animals, RM will not replace mice for many questions in CMV biology and immunology. However, for many important scientific questions, including vaccine development, drug-development, latent CMV infection and immune senescence, RhCMV is rapidly becoming the model of choice.
References
- 1.Mocarski ES, Shenk T, Pass RF. Cytomegalovirus. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roiz-man B, Straus SE, editors. Fields virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- 2.Nichols WG, Boeckh M. Recent advances in the therapy and prevention of CMV infections. J Clin Virol. 2000;16:25–40. doi: 10.1016/s1386-6532(99)00065-7. [DOI] [PubMed] [Google Scholar]
- 3.Alford CA, Stagno S, Pass RF, Britt WJ. Congenital and perinatal cutomegalovirus infection. Rev Infect Dis. 1990;12:745–753. doi: 10.1093/clinids/12.supplement_7.s745. [DOI] [PubMed] [Google Scholar]
- 4.Stratton KR, Durch JS, Lawrence RS, editors. Washington, DC: National Academy Press; 2000. Vaccines for the 21st century. [PubMed] [Google Scholar]
- 5.Söderberg-Naucler C, Nelson JY. Human cytomegalovirus latency and reactivation—a delicate balance between the virus and its host’s immune system. Intervirology. 1999;42:314–321. doi: 10.1159/000053966. [DOI] [PubMed] [Google Scholar]
- 6.Huff JL, Eberle R, Capitanio J, Zhou SS, Barry PA. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J Gen Virol. 2003;84:83–92. doi: 10.1099/vir.0.18808-0. [DOI] [PubMed] [Google Scholar]
- 7.Reinke P, Prosch S, Kern F, Volk HD. Mechanisms of human cytomegalovirus (HCMV) (re)activation and its impact on organ transplant patients. Transpl Infect Dis. 1999;1:157–164. doi: 10.1034/j.1399-3062.1999.010304.x. [DOI] [PubMed] [Google Scholar]
- 8.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon CO, Holtappels R, Tervo HM, Bohm V, Daubner T, Oehr-lein-Karpi SA, Kuhnapfel B, Renzaho A, Strand D, Podlech J, Reddehase MJ, Grzimek NK. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J Virol. 2006;80:10436–10456. doi: 10.1128/JVI.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streblow DN, Orloff SL, Nelson JA. Do pathogens accelerate atherosclerosis? J Nutr. 2001;131:2798S–2804S. doi: 10.1093/jn/131.10.2798S. [DOI] [PubMed] [Google Scholar]
- 11.McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 12.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, 3rd, Kouzarides T, Martignetti JA, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomega-lovirus genome. J Gen Virol. 2003;84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 14.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cyto-megalovirus. J Virol. 2003;77:6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vink C, Beuken E, Bruggeman CA. Complete DNA sequence of the rat cytomegalovirus genome. J Virol. 2000;74:7656–7665. doi: 10.1128/jvi.74.16.7656-7665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGregor A, Liu F, Schleiss MR. Identification of essential and non-essential genes of the guinea pig cytomegalovirus (GPC-MV) genome via transposome mutagenesis of an infectious BAC clone. Virus Res. 2004;101:101–108. doi: 10.1016/j.virusres.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Hudson JB. The murine cytomegalovirus as a model for the study of viral pathogenesis and persistent infections. Arch Virol. 1979;62:1–29. doi: 10.1007/BF01314900. [DOI] [PubMed] [Google Scholar]
- 19.Reddehase MJ, Podlech J, Grzimek NK. Mouse models of cytomegalovirus latency: overview. J Clin Virol. 2002;25 suppl 2:S23–S36. doi: 10.1016/s1386-6532(02)00087-2. [DOI] [PubMed] [Google Scholar]
- 20.Asher DM, Gibbs CJ, Jr, Lang DJ, Gajdusek DC, Chanock RM. Persistent shedding of cytomegalovirus in the urine of healthy Rhesus monkeys. Proc Soc Exp Biol Med. 1974;145:794–801. doi: 10.3181/00379727-145-37897. [DOI] [PubMed] [Google Scholar]
- 21.Jones-Engel L, Engel GA, Heidrich J, Chalise M, Poudel N, Viscidi R, Barry PA, Allan JS, Grant R, Kyes R. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerg Infect Dis. 2006;12:900–906. doi: 10.3201/eid1206.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swack NS, Hsiung GD. Natural and experimental simian cytomegalovirus infections at a primate center. J Med Primatol. 1982;11:169–177. [PubMed] [Google Scholar]
- 23.Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 1994;44:25–30. [PubMed] [Google Scholar]
- 24.Lockridge KM, Sequar G, Zhou SS, Yue Y, Mandell CP, Barry PA. Pathogenesis of experimental rhesus cytomegalovirus infection. J Virol. 1999;73:9576–9583. doi: 10.1128/jvi.73.11.9576-9583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang WL, Tarantal AF, Zhou SS, Borowsky AD, Barry PA. A recombinant rhesus cytomegalovirus expressing enhanced green fluorescent protein retains the wild-type phenotype and pathogenicity in fetal macaques. J Virol. 2002;76:9493–9504. doi: 10.1128/JVI.76.18.9493-9504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang WL, Barry PA. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J Virol. 2003;77:5073–5083. doi: 10.1128/JVI.77.9.5073-5083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross SA, Fowler KB, Ashrith G, Stagno S, Britt WJ, Pass RF, Boppana SB. Hearing loss in children with congenital cytomegalovirus infection born to mothers with preexisting immunity. J Pediatr. 2006;148:332–336. doi: 10.1016/j.jpeds.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Barry PA, Lockridge KM, Salamat S, Tinling SP, Yue Y, Zhou SS, Gospe SM, Jr, Britt WJ, Tarantal AF. Nonhuman primate models of intrauterine cytomegalovirus infection. Ilar J. 2006;47:49–64. doi: 10.1093/ilar.47.1.49. [DOI] [PubMed] [Google Scholar]
- 29.London WT, Martinez AJ, Houff SA, Wallen WC, Curfman BL, Traub RG, Sever JL. Experimental congenital disease with simian cytomegalovirus in rhesus monkeys. Teratology. 1986;33:323–331. doi: 10.1002/tera.1420330311. [DOI] [PubMed] [Google Scholar]
- 30.Tarantal AF, Salamat MS, Britt WJ, Luciw PA, Hendrickx AG, Barry PA. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta) J Infect Dis. 1998;177:446–450. doi: 10.1086/514206. [DOI] [PubMed] [Google Scholar]
- 31.Serabe BM, Murry DJ, Dauser R, Nuchtern J, Durfee J, McGuffey L, Berg S, Blaney SM. Plasma and CSF pharmacokinetics of ganciclovir in nonhuman primates. Cancer Chemother Pharmacol. 1999;43:415–418. doi: 10.1007/s002800050916. [DOI] [PubMed] [Google Scholar]
- 32.Swanson R, Bergquam E, Wong SW. Characterization of rhesus cytomegalovirus genes associated with anti-viral susceptibility. Virology. 1998;240:338–348. doi: 10.1006/viro.1997.8935. [DOI] [PubMed] [Google Scholar]
- 33.North TW, Sequar G, Townsend LB, Drach JC, Barry PA. Rhesus cytomegalovirus is similar to human cytomegalovirus in susceptibility to benzimidazole nucleosides. Antimicrob Agents Chemother. 2004;48:2760–2765. doi: 10.1128/AAC.48.7.2760-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson TC, Trambley J, Odom K, Anderson DC, Cowan S, Bray R, Lin A, Hollenbaugh D, Aruffo A, Siadak AW, Strobert E, Hennigar R, Larsen CP. Anti-CD40 therapy extends renal allograft survival in rhesus macaques. Transplantation. 2002;74:933–940. doi: 10.1097/00007890-200210150-00006. [DOI] [PubMed] [Google Scholar]
- 35.Conway MD, Didier P, Fairburn B, Soike KF, Martin L, Murphey-Corb M, Meiners N, Insler MS. Ocular manifestation of simian immunodeficiency syndrome (SAIDS) Curr Eye Res. 1990;9:759–770. doi: 10.3109/02713689008999571. [DOI] [PubMed] [Google Scholar]
- 36.Henrickson RV, Maul DH, Osborn KG, Sever JL, Madden DL, Ellingsworth LR, Anderson JH, Lowenstine LJ, Gardner MB. Epidemic of acquired immunodeficiency in rhesus monkeys. Lancet. 1983;1:388–390. doi: 10.1016/s0140-6736(83)91503-9. [DOI] [PubMed] [Google Scholar]
- 37.Baskin GB. Disseminated cytomegalovirus infection in immunodeficient rhesus monkeys. Am J Pathol. 1987;129:345–352. [PMC free article] [PubMed] [Google Scholar]
- 38.Kaup F, Matz-Rensing K, Kuhn E, Hunerbein P, Stahl-Hennig C, Hunsmann G. Gastrointestinal pathology in rhesus monkeys with experimental SIV infection. Pathobiology. 1998;66:159–164. doi: 10.1159/000028015. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn EM, Stolte N, Matz-Rensing K, Mach M, Stahl-Henning C, Hunsmann G, Kaup FJ. Immunohistochemical studies of productive rhesus cytomegalovirus infection in rhesus monkeys (Macaca mulatta) infected with simian immunodeficiency virus. Vet Pathol. 1999;36:51–56. doi: 10.1354/vp.36-1-51. [DOI] [PubMed] [Google Scholar]
- 40.Kaur A, Hale CL, Noren B, Kassis N, Simon MA, Johnson RP. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J Virol. 2002;76:3646–3658. doi: 10.1128/JVI.76.8.3646-3658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaur A, Kassis N, Hale CL, Simon M, Elliott M, Gomez-Yafal A, Lifson JD, Desrosiers RC, Wang F, Barry P, Mach M, Johnson RP. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J Virol. 2003;77:5749–5758. doi: 10.1128/JVI.77.10.5749-5758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sequar G, Britt WJ, Lakeman FD, Lockridge KM, Tarara RP, Canfield DR, Zhou SS, Gardner MB, Barry PA. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J Virol. 2002;76:7661–7671. doi: 10.1128/JVI.76.15.7661-7671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macchia I, Gauduin MC, Kaur A, Johnson RP. Expression of CD8alpha identifies a distinct subset of effector memory CD4+ T lymphocytes. Immunology. 2006;119:232–242. doi: 10.1111/j.1365-2567.2006.02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan KS, Kaur A. Flow cytometric detection of degranulation reveals phenotypic heterogeneity of degranulating CMV-specific CD8+ T lymphocytes in rhesus macaques. J Immunol Methods. 2007;325:20–34. doi: 10.1016/j.jim.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 46.Walker JM, Maecker HT, Maino VC, Picker LJ. Multicolor flow cytometric analysis in SIV-infected rhesus macaque. Methods Cell Biol. 2004;75:535–557. doi: 10.1016/s0091-679x(04)75022-0. [DOI] [PubMed] [Google Scholar]
- 47.Kaur A, Daniel MD, Hempel D, Lee-Parritz D, Hirsch MS, Johnson RP. Cytotoxic T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J Virol. 1996;70:7725–7733. doi: 10.1128/jvi.70.11.7725-7733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estes JD, Li Q, Reynolds MR, Wietgrefe S, Duan L, Schacker T, Picker LJ, Watkins DI, Lifson JD, Reilly C, Carlis J, Haase AT. Premature induction of an immunosuppressive regulatory T cell response during acute simian immunodeficiency virus infection. J Infect Dis. 2006;193:703–712. doi: 10.1086/500368. [DOI] [PubMed] [Google Scholar]
- 49.Yue Y, Zhou SS, Barry PA. Antibody responses to rhesus cytomegalovirus glycoprotein B in naturally infected rhesus macaques. J Gen Virol. 2003;84:3371–3379. doi: 10.1099/vir.0.19508-0. [DOI] [PubMed] [Google Scholar]
- 50.Rivailler P, Kaur A, Johnson RP, Wang F. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J Virol. 2006;80:4179–4182. doi: 10.1128/JVI.80.8.4179-4182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci USA. 2003;100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pande NT, Powers C, Ahn K, Fruh K. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol. 2005;79:5786–5798. doi: 10.1128/JVI.79.9.5786-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alcendor DJ, Barry PA, Pratt-Lowe E, Luciw PA. Analysis of the rhesus cytomegalovirus immediate-early gene promoter. Virology. 1993;194:815–821. doi: 10.1006/viro.1993.1323. [DOI] [PubMed] [Google Scholar]
- 55.Barry PA, Alcendor DJ, Power MD, Kerr H, Luciw PA. Nucleotide sequence and molecular analysis of the rhesus cyto-megalovirus immediate-early gene and the UL121–117 open reading frames. Virology. 1996;215:61–72. doi: 10.1006/viro.1996.0007. [DOI] [PubMed] [Google Scholar]
- 56.Kravitz RH, Sciabica KS, Cho K, Luciw PA, Barry PA. Cloning and characterization of rhesus cytomegalovirus glycoprotein B. J Gen Virol. 1997;78(Pt 8):2009–2013. doi: 10.1099/0022-1317-78-8-2009. [DOI] [PubMed] [Google Scholar]
- 57.Kropff B, Mach M. Identification of the gene coding for rhesus cytomegalovirus glycoprotein B and immunological analysis of the protein. J Gen Virol. 1997;78(Pt 8):1999–2007. doi: 10.1099/0022-1317-78-8-1999. [DOI] [PubMed] [Google Scholar]
- 58.Yue Y, Kaur A, Zhou SS, Barry PA. Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65) J Gen Virol. 2006;87:777–787. doi: 10.1099/vir.0.81516-0. [DOI] [PubMed] [Google Scholar]
- 59.McCormick AL, Skaletskaya A, Barry PA, Mocarski ES, Goldmacher VS. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology. 2003;316:221–233. doi: 10.1016/j.virol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Lockridge KM, Zhou SS, Kravitz RH, Johnson JL, Sawai ET, Blewett EL, Barry PA. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology. 2000;268:272–280. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- 61.Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol. 2002;76:1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Penfold ME, Schmidt TL, Dairaghi DJ, Barry PA, Schall TJ. Characterization of the rhesus cytomegalovirus US28 locus. J Virol. 2003;77:10404–10413. doi: 10.1128/JVI.77.19.10404-10413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesniewski M, Das S, Skomorovska-Prokvolit Y, Wang FZ, Pellett PE. Primate cytomegalovirus US12 gene family: a distinct and diverse clade of seven-transmembrane proteins. Virology. 2006;354:286–298. doi: 10.1016/j.virol.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 64.Rue CA, Jarvis MA, Knoche AJ, Meyers HL, DeFilippis VR, Hansen SG, Wagner M, Fruh K, Anders DG, Wong SW, Barry PA, Nelson JA. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tro-pism. J Virol. 2004;78:12529–12536. doi: 10.1128/JVI.78.22.12529-12536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci USA. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeFilippis V, Fruh K. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J Virol. 2005;79:6419–6431. doi: 10.1128/JVI.79.10.6419-6431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang WL, Kirchoff V, Pari GS, Barry PA. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods. 2002;104:135–146. doi: 10.1016/s0166-0934(02)00060-5. [DOI] [PubMed] [Google Scholar]