Abstract
OMP decarboxylase (ODCase) generates a very large rate enhancement without the assistance of metals or other cofactors. The uncatalyzed decarboxylation of 1-methylorotate in water is shown to involve the monoanion, although uncharged 1-methylorotic acid is decarboxylated at a similar rate. To measure the extent to which the rate of the nonenzymatic decarboxylation of orotate derivatives might be enhanced by their removal from solvent water, the 1-phosphoribosyl moiety of OMP was replaced by 1-substituents that would allow it to enter less polar solvents. When the tetrabuytlyammonium salt of 1-cyclohexylorotate was transferred from water to a series of dipolar aprotic solvents, its rate of decarboxylation increased markedly, varying with the relative ability of each solvent to release the substrate in the ground state from stabilization by solvent water acting as a proton donor. These findings are consistent with the view that separation of the substrate from solvent water may contribute, at least to a limited extent, to the rate enhancement produced by ODCase. This enzyme's active site, like that of another cofactorless enzyme recently shown to produce a rate enhancement of similar magnitude (uroporphyrinogen decarboxylase), is equipped with an ammonium group positioned in such a way as to balance the electrostatic charge of the carboxylate group of the substrate and later supply a proton to the incipient carbanion in a relatively waterless environment.
Compared with the sluggish pace of the uncatalyzed reaction, orotidine 5'-phosphate decarboxylase (ODCase; EC 4.1.1.23) generates one of the largest rate enhancements known to be produced by any enzyme, and it achieves this feat as a pure protein catalyst, without the assistance of metal ions or other cofactors (1). Of the 8 invariant amino acid residues that are conserved in ODCase from all species that have been examined, 7 make direct contact with the substrate, and mutation of any one of those residues to alanine leads to a drastic loss of activity (2). But only 1 of those 8 residues (Lys-93 in the yeast sequence, approaches the pyrimidine ring at a position near the site of CO2 elimination, as indicated by the structure of the enzyme complex with the transition state analogue 6-hydroxyuridine 5’-phosphate (3). The other 7 residues appear to play a less direct role in the action of ODCase, bracing the substrate in position for effective catalysis and assisting the folding of loops that enclose the substrate during the central chemical events in catalysis (3, 4).
An enzyme's tendency to bind the substrate while the enzyme is in an open configuration, and then enclose the altered substrate in the transition state (S‡), has been observed in transition state analogue complexes formed by triosephosphate isomerase (5, 6), adenosine deaminase (7), cytidine deaminase (8) and ODCase (9). There appear to be several possible reasons why that tendency might promote effective catalysis. First, closure of an initially open active site would be expected to allow maximization of the solid angle of contact between the enzyme (a large molecule) and the altered substrate in the transition state (usually a small molecule), and hence maximization of their forces of mutual attraction (10). Second, closure of an initially open active site tends to involve removal of the substrate from solvent water, which would, in itself, be expected to enhance reactivity, at least in some cases. Earlier work has shown, for example, that the nonenzymatic elimination of CO2 from 2-keto acids in the presence of thiamine (11), and the uncatalyzed decarboxylation of 3-carboxy-6-benzisoxazole (CBI) (12), are greatly accelerated when these reactions are conducted in nonpolar solvents.
In the present work, we set out to determine the extent to which removal of orotate derivatives from solvent water, by transfer into nonpolar solvents, might enhance the spontaneous rate of decarboxylation of orotate derivatives.
Earlier, the uncatalyzed decarboxylation of orotate derivatives was found to proceed so slowly in water that it was necessary to conduct experiments at elevated temperatures if rate constants were to be determined within a reasonable period of time. Because the 1-glycosidic bonds of pyrimidine nucleosides undergo hydrolysis much more rapidly than the carboxylate group is eliminated from orotate, those experiments on nonenzymatic decarboxylation were conducted using the relatively stable 1-methyl derivative of orotic acid and related molecules (13 - 22). In the present experiments, we sought to replace the phosphoribosyl moiety of OMP by nonpolar substituents that would allow orotate to enter relatively nonpolar solvents in the presence of alkylammonium counter ions. The 1-cyclohexyl substituent proved suitable for that purpose.
Materials and Methods
Orotic acid, 1-methyluracil (1-MeU), 1-cyclohexyluracil (1-ChxU) and uracil-6-acetic acid (U6AA) were purchased from Sigma-Aldrich Corp. 1-Methylorotic acid (1-MeO) and 1-cyclohexylorotic acid (1-ChxO) were prepared from the corresponding uracil derivatives as described by Landesman (23). In both cases, the 1-alkyluracil in dry DMF solution was treated with bromine (in CCl4) to generate the 5-bromo derivative, followed by reaction with potassium cyanide in the presence of 18-crown-6 to produce the 6-cyano derivative, and finally by heating of the 6-cyano derivative in 1 N NaOH to yield the corresponding 1-alkylorotic acid. 3-Methylorotic acid (3-MeO) and 1,3-dimethyl orotic acid (1,3-Me2O) were prepared as described by Curran and Angier (24). The identity of each product was confirmed by 1H NMR and UV spectrometry in acidic, neutral and basic aqueous solutions (25), and their purities, as measured by 1H-NMR, were judged to be >95%.
To measure the effect of changing pH on the rate of decarboxylation, reaction mixtures containing 1-MeO (0.01 M) were prepared in HCl, and in buffers (sodium acetate, potassium phosphate, sodium borate, sodium carbonate, 0.1 M), and KOH distributed over the Ho-pH range between ∼ −1 and 11 at 25 °C. The heats of ionization of these anionic buffers are relatively small (26), as would also be expected to be true of the −COOH groups of orotic acid derivatives. Thus, the state of ionization of these orotate derivatives in buffered solution is not expected to change much with increasing temperature in the acid range (pKa1 for 1-MeO (pKa1 ∼0.6 at 25 °C) (25). Because the heats of ionization of the N3 proton of uracil derivatives (pKa2 for 1-MeO (∼9.8 at 25 °C)) (26) do not appear to have been reported, we used a pH meter to determined the temperature dependence of pKa2 for 1-MeO as described in Results.
1H NMR spectra were recorded using with a Varian Unity 500 MHz Spectrometer, equipped with a high-sensitivity cryoprobe operated by Solaris 9 software. To obtain a signal-to-noise ratio suitable for accurate measurement of the integrated intensities of the resonances arising from both the starting material and product in the kinetic experiments described below, we employed 4 to 16 transients with a 60 second pulse delay.
For rate measurements in aqueous solution, samples (0.1 mL) of orotate derivatives (0.025 M) in potassium phosphate buffer (0.1 M, pH 7.0) were sealed under vacuum in quartz tubes and heated in Thermolyne 47900 ovens for times sufficient to achieve 15 to 85% reaction. For analysis by proton NMR, samples (0.1 mL) were then diluted with D2O (0.5 mL, containing 0.01 M pyrazine (4H, δ = 8.60 ppm) as an integration standard). For rate measurements in nonpolar solvents, the tetrabutylammonium salt of 1-cyclohexylorotic acid (1-ChxO-TBA) was prepared by titration of 1-ChxO with tetrabutylammonium hydroxide to pH 7, lyophilized to remove water, dried over magnesium perchlorate, and dissolved in the nonpolar solvent. To minimize residual 1H signals arising from the solvent during analysis by 1H NMR, reaction mixtures were prepared in DMSO-d6, acetone-d6, dioxane-d8, chloroform-d1, dimethylformamide-d7, dimethylacetamide-d8, or tetrahydrofuran-d6 purchased from Sigma-Aldrich Corp.; in formamide-d3 purchased from CDN Isotopes, Inc.; in methyl formamide-d4 purchased from Medical Isotopes, Inc.; or in acetonitrile-d3 purchased from Cambridge Isotopes Laboratories. Samples dissolved in nonpolar solvents were frozen by immersion in dry ice-acetone and sealed in quartz tubes under vacuum. After heating, samples were prepared for analysis by 1H NMR as described above, except that DMSO-d6 was used as the solvent instead of D2O in some cases. D2O and DMSO-d6 were found to be devoid of NMR signals in the region used for observing signals arising from the pyrimidine protons and from the C1′ proton of the 1-cyclohexyl substituent. 1H NMR spectra were acquired as described above.
To follow the progress of reactions conducted in aqueous solution, the integrated intensities of the 1-alkyl protons and the C-H protons of the pyrimidine ring were used to determine the concentrations of the reactant and product (Table 1). For reactions conducted in nonpolar solvents, the integrated intensities of the 1-ChxO resonances at ∼5.58 ppm (C5H, singlet) and ∼3.72 ppm (C1′H, multiplet) and the 1-ChxU resonances at ∼7.69 ppm, (C6H, doublet), ∼5.79 ppm (C5H, doublet), and ∼4.27 ppm (C1′H, multiplet) were used instead. Under all conditions examined, decarboxylation followed simple first-order kinetics to completion, and the resulting rate constants yielded linear Arrhenius plots when they were plotted as a logarithmic function of 1000/T (Kelvin).
Table 1.
1H NMR chemical shifts of derivatives of orotic acid and uracil in D2O.
| C5H | 1-Me | 3-Me | C1′Ha | CH2b | C6H | C5H | 1-Me | 3-Me | C1′Ha | CH3b | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Orotate | 6.14 | U | 7.48 | 5.75 | ||||||||
| 1-MeO | 5.74 | 3.29 | 1-MeU | 7.55 | 5.74 | 3.32 | ||||||
| 3-MeO | 6.20 | 3.23 | 3-MeU | 7.44 | 5.81 | 3.22 | ||||||
| 1,3-Me2O | 5.78 | 3.34 | 3.25 | 1,3-Me2U | 7.53 | 5.83 | 3.36 | 3.26 | ||||
| 1-ChxO | 5.58 | 3.72 | 1-chxU | 7.69 | 5.79 | 4.27 | ||||||
| U6AA | 5.63 | 3.36 | 6-MeU | 5.59 | 2.14 |
Multiplet arising from the cyclohexyl C1'-H group linked to N1 of the pyrimidine ring.
The CH2 singlet of U6AA and the CH3 singlet of 6-MeU were also used in the calculations.
Results
Effects of pH on the rate of decarboxylation of 1-MeO in water
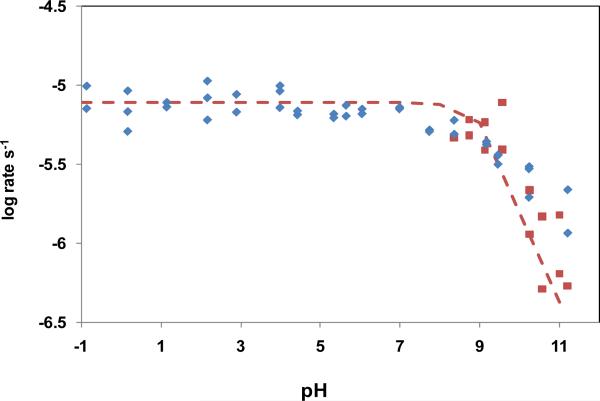
At 160 °C, the rate of decarboxylation of 1-MeO remained essentially constant in the acidic and neutral range (Figure 1), but decreased at higher pH values, presumably in response to the ionization of the N3 proton. Since hydroxide ion displaces silicate from quartz in alkaline solution at elevated temperatures (27), experiments above pH 8 were also conducted in Teflon-lined bombs. The heat of ionization of the N3 proton of 1-MeO was also determined by monitoring the pH of a solution of mM 1-MeO (0.07 M, pH 9.8 at 22°C) half-titrated with KOH, as a function of increasing temperature. A van't Hoff plot of those data (not shown) yielded a heat of ionization of 6.1 kcal/mol.
Figure 1.
 or Teflon cups in steel bombs
or Teflon cups in steel bombs  . The solid line is calculated for the equation:
. The solid line is calculated for the equation: The pKa value of the −COOH group of 1-MeO is 0.7 (25). Because carboxylic acids tend to undergo decarboxylation in their anionic (−COO−) forms (28), the absence of any significant effect of changing pH on the rate of decarboxylation of 1-MeO between Ho −1 and pH 3 is somewhat surprising. As in certain other cases in which formally uncharged carboxylic acids have been found to be unexpectedly reactive (29), the actual reactant in the range between Ho −1 and pH 3 seems likely to be a rare zwitterionic form of the reactant (1-MeO±) in which the carboxylate group remains negatively charged and the reactant is protonated at another position (N1, O2 or O4). 2 Such a zwitterion would be expected to be much more rapidly decarboxylated than 1-MeO−. As a result, the reactivity of the major species (1-MeO−) might (by coincidence) appear to remain unchanged by protonation.
Effects of 1-substituents on the rate of decarboxylation of orotate derivatives in water
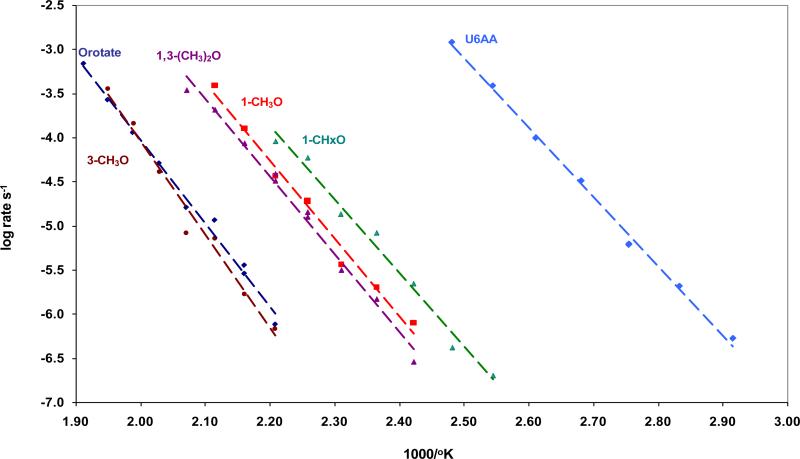
In potassium phosphate buffer (0.1 M, pH 7), the decarboxylation of orotate derivatives followed simple first order kinetics, yielding linear Arrhenius plots with similar slopes (Figure 2) that were extrapolated to estimate the rates of decarboxylation of these molecules at ordinary temperatures.
Figure 2.
Rate constants (log k, s−1) for the decarboxylation of orotic acid derivatives in potassium phosphate buffer (0.1 M, pH 7.0), plotted as a function of the reciprocal of absolute temperature.
Of the reactants examined here, the monoanions of orotate and 3-MeO underwent decarboxylation most reluctantly, with extrapolated rate constants of 1.3 × 10−17 s−1 and 3.9 × 10−17 s−1, respectively, at 25 °C. These very slow decarboxylations were accompanied by a concurrent reaction that led to opening of the pyrimidine ring and ultimately to the release of acetaldehyde (see below). 1,3-Me2O and 1-MeO reacted considerably more rapidly, with rate constants of 2.4 × 10−15 s−1 and 3.4 × 10−15 s−1 at 25 °C, and no ring opening accompanied decarboxylation. 1-Cyclohexylation of orotate resulted in a further increase in the rate constant for decarboxylation, to 4.9 × 10−14 s−1 at 25 °C, with no concurrent ring opening. Heats and entropies of activation for each of these reactions, obtained from Arrhenius plots are shown in Table 2.3
Table 2.
Thermodynamics of activation for decarboxylation of orotic acid derivatives in potassium phosphate buffer (0.1M, pH 7.0)
| Temp Range | k25°C s−1 | log k25°C | ΔH‡ kcal | ΔG‡ kcal | TΔS‡ kcal | |
|---|---|---|---|---|---|---|
| Orotate | 180 – 250°C | 1.3 × 10−17 | −16.89 | 42.8 | 40.4 | 2.5 |
| 3-MeO | 180 – 250°C | 3.9 × 10−17 | −16.41 | 47.8 | 42.4 | 5.4 |
| 1,3-Me2O | 140 – 210°C | 2.4 × 10−15 | −14.62 | 39.7 | 37.3 | 2.4 |
| 1-MeO | 140 – 200°C | 3.4 × 10−15 | −14.47 | 39.8 | 37.1 | 2.8 |
| 1-chxO | 120 – 180°C | 4.9 × 10−14 | −13.31 | 37.3 | 35.5 | 1.8 |
| U6AA | 70 – 130°C | 1.7 × 10−10 | − 9.77 | 35.1 | 30.7 | 4.4 |
For the decarboxylation of 1-MeO at 25 °C (3.4 × 10−15 s−1), the rate constant obtained in the present experiments was somewhat larger than a value (2.8 × 10−16 s−1) reported earlier by one of us (1), based on UV spectrophotometric analysis of the progress of decarboxylation. We attribute that difference to superiority of the present 1H-NMR method, which allowed independent identification and more precise quantitation of the reactant and product at each stage of the reaction.
Ring opening of orotate and 3-methylorotate in water
During the very slow decarboxylation of aqueous solutions of orotate and 3-MeO at temperatures between 180 and 250 °C, other products were formed in significant amounts. Two of those products were identified as acetaldehyde and the corresponding gem-diol, by the characteristic aldehyde proton (∼9.62 ppm, quartet, J =3Hz) and the methine proton of the covalent hydrate (∼5.2 ppm, quartet, J = 5.2 Hz) and the doublets for the respective methyl groups (∼2.19 ppm aldehyde, 1.28 ppm hydrate). Those products would be expected to arise from the opening of the pyrimidine ring between N3 and C4 to generate the corresponding derivative of ureidosuccinaldehyde, followed by elimination of acetaldehyde. Additional minor peaks appeared in the aliphatic region of the 1H NMR spectra, presumably representing other products of ring opening. Decomposition by this alternative route proceeded at ∼57% of the rate of decarboxylation of orotate, and at ∼32% of the rate of decarboxylation of 3-MeO. No significant ring opening was observed at any temperature for 1-MeO, 1,3-Me2O, 1-ChxO or U6AA, whose rate of decarboxylation was considerably more rapid than that of orotate or 3-MeO. Rate constants for the decarboxylation of orotate and 3-MeO were obtained by applying the observed product ratio (as reflected in the appearance of acetaldehyde and its hydrate compared with the product of decarboxylation (U or 3-MeU) to the first order rate constant observed for the disappearance of the substrate. Because that correction was substantial, particularly for orotate, the rate constants for orotate and 3-MeO in Table 2 should be considered approximate.
Decarboxylation of 1-cyclohexylorotate in nonpolar solvents
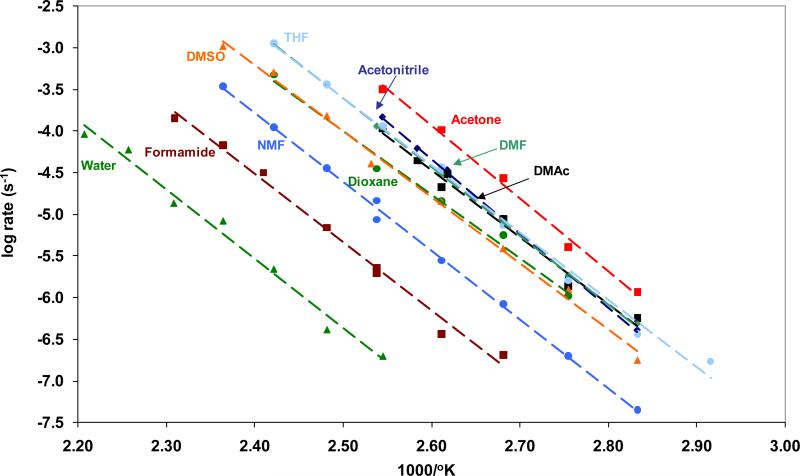
To permit the ionized form of 1-ChxO to enter organic solvents at concentrations sufficient to monitor its decomposition, we converted the acid to its tetrabutylammonium salt (1-ChxO - TBA). TBA was selected to both enhance solubility of 1-ChxO in organic solvents and to provide a counter ion that did not contain a proton, which could hydrogen bond to the orotate carboxylate and potentially reduce its reactivity toward decarboxylation. In a series of solvents ranging from formamide to acetone, we observed quantitative conversion of 1-ChxO to 1-ChxU. The logarithm of the first order rate constants of these reactions, plotted as a function of absolute temperature, yielded linear Arrhenius plots as shown in Figure 3.
Figure 3.
Rate constants (log k, s−1) for the decarboxylation of 1-ChxO - TBA salt in various solvents, compared with 1-ChxO in potassium phosphate buffer (0.1 M, pH 7), plotted as a function of the reciprocal of absolute temperature.
Extrapolation of these Arrhenius plots to 25 °C showed that decarboxylation in formamide, N-methylformamide (NMF), and dimethylformamide (DMF) proceeded ∼10 fold, ∼100 fold and ∼1000 fold more rapidly than decarboxylation in water. Acetonitrile and dimethyl sulfoxide produced rates of decarboxylation that were intermediate between those observed in NMF and DMF. Decarboxylation proceeded most rapidly (∼3 × 10−11 s−1) in dimethylacetamide, tetrahydrofuran, dioxane and acetone. The thermodynamic activation parameters observed in these solvents are shown in Table 3.
Table 3.
Thermodynamics of activation for decarboxylation of the TBA Salt of 1-ChxO in nonpolar solvents.
| Temp Range | k25°C s−1 | log k25°C | ΔH‡ kcal | ΔG‡ kcal | TΔS‡ kcal | |
|---|---|---|---|---|---|---|
| Water | 120 – 180°C | 4.9 × 10−14 | −13.31 | 37.3 | 35.5 | 1.8 |
| Formamide | 110 – 160°C | 4.3 × 10−13 | −12.37 | 37.0 | 34.2 | 2.8 |
| NMF | 80 – 150°C | 2.1 × 10−12 | −11.68 | 37.2 | 33.3 | 3.9 |
| Acetonitrile | 80 – 120°C | 8.3 × 10−12 | −11.08 | 40.3 | 32.5 | 7.8 |
| DMSO | 80 – 150°C | 1.6 × 10−11 | −10.80 | 35.7 | 32.1 | 3.6 |
| DMF | 80 – 140°C | 2.2 × 10−11 | −10.66 | 37.1 | 31.9 | 5.1 |
| Me2-Acetamide | 80 – 120°C | 2.9 × 10−11 | −10.54 | 36.2 | 31.7 | 4.5 |
| Tetrahydrofuran | 70 – 140°C | 3.0 × 10−11 | −10.52 | 36.5 | 31.7 | 4.7 |
| Dioxane | 90 – 140°C | 3.0 × 10−11 | −10.52 | 34.3 | 31.7 | 2.6 |
| Acetone | 80 – 120°C | 3.0 × 10−11 | −10.52 | 39.3 | 31.7 | 7.6 |
6-Deuteration during decarboxylation in nonpolar solvents
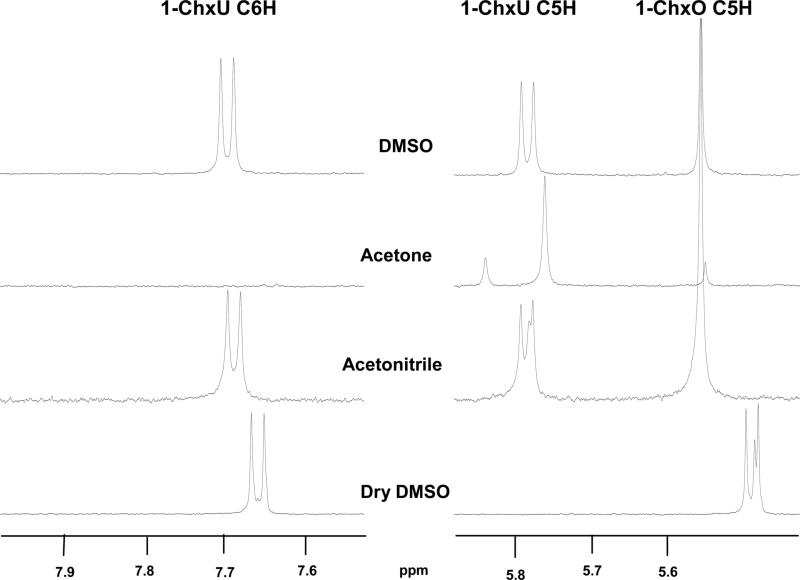
In the present experiments, deuterated solvents were used to reduce interference by proton signals arising from the solvent. To generate uracil derivatives in these solvents, it would be necessary to abstract a proton or deuteron from the solvent unless traces of water were present in the solvent. In some cases, deuteration of C6 was in fact observed in the 1H NMR spectra, with a singlet for the 5-proton of 1-ChxU resulting from the loss of the 6-proton coupling due to deuteration. (Figure 4).
Figure 4.
1H NMR spectra of the C6 and C5 protons of 1-ChxU arising from the decarboxylation of 1-ChxO conducted in three solvents. The top three samples were diluted in D2O, while the fourth was diluted in DMSO-d6.When traces of H2O were present in these reactions, two doublets (J = 7.9 Hz) were produced when the C6 carbanion extracted a proton from water. In acetone-d6, the C6-carbanion extracted a deuteron from the solvent, leaving only the C5 proton resonance as a singlet. Acetonitrile-d3 and “dry” DMSO showed evidence of both reactions. In DMSO-d6, the resonances were shifted upfield from their positions in D2O.
The extent of 6-deuteration varied in different solvents. Thus, the product of 1-ChxO decarboxylation in DMSO-d6 showed two doublets arising from 6-protonation by traces of H2O (top spectrum of Figure 4). But in acetone-d6, only the C5 proton resonance of the product appeared as a singlet, implying deuteration at C6. In solvents in which limited amounts of water were present, a mixture of protonated and deuterated products was observed, as illustrated by the two lower spectra of Figure 4. Similarly, Beak and Siegel (14) reported deuteration at C6 when 1,3-Me2O was heated in sulfolane-d4 at 206 °C. In these cases, the observed deuteration at C6 implies an ability of the carbanion to extract deuterium even from solvents of very weak acidity. That property is not unexpected, in view of the estimated pKa values of acetone, 20; acetonitrile, 25; Me2-acetamide, 25; and DMSO, 31; compared with an estimated pKa value of ∼34 for the 6C-H group of UMP (30).
Discussion
Table 2 shows that the rate of decarboxylation of orotate derivatives in water is markedly affected by the nature of substituents at N1. In earlier experimental work on the nonenzymatic reactivity of orotic acid derivatives in water and other solvents, 1-methylated derivatives were used to circumvent complications arising from the instability of the 1-phosphoribofuranosyl substituent at the elevated temperatures that were needed to observe decarboxylation (1, 13-14). In the present work, the slowest rates of decarboxylation were observed for unsubstituted orotate and 3-MeO, which also underwent slow but significant ring opening with release of acetaldehyde at elevated temperatures. But 1-MeO and 1,3-Me2O undergo decarboxylation ∼100-fold more rapidly than does orotate, and a further 10-fold increase in rate was produced by the presence of a 1-cyclohexyl substituent, introduced for the purpose of enhancing solubility in non-polar solvent. Model building discloses a steric conflict between the 1-substituent and the 6-carboxylate group if the latter is oriented in the same plane as the pyrimidine ring. That steric conflict, present in 1-substituted forms of orotate but not in orotate or in 3-MeO, is presumably relieved in carbanionic intermediates approaching the transition state for decarboxylation, accounting for their differing rates of reaction.
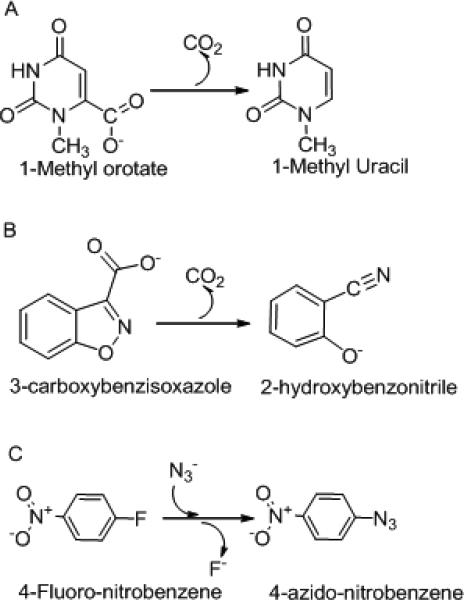
Substantial increases in the rate of decarboxylation of 1-ChxO were observed in nonaqueous solvents. Table 4 shows the logarithms of the rate constants for decarboxylation of 1-ChxO (obtained by extrapolation to 25°C) in various solvents, and also some values recorded in the literature for two well characterized, quite different reactions that involve delocalization of negative charge as the substrates proceed from the ground state to the transition state: the reaction of azide with 4-fluoro-nitrobenzene (FNB) at 100°C (31) and the decomposition of 3-carboxybenzisoxazole (CBI) at 30 °C (32-34).4, Despite their differences in mechanism, (see Figure 5) all three of these reactions can be seen to be very closely related to each other in the relative ordering of their kinetic responses to the various solvents (as indicated by the R2 values for linear regression at the foot of each column of Table 4) the span of relative reactivities is greater for azide attack on FNB and for decomposition of CBI than for decarboxylation of 1-ChxO (as indicated by the slopes in Table 4).
Table 4.
Solvent effects on rate constants (log k, s−1) reported for for 1-ChxO decarboxylation (this work), CBI decarboxylation (ref. 12) and azide attack on 1-fluoro-4-nitrobenzene (ref. 31).
| ChxO | CBIa | azide + FNBb | acityc | basityc | acity + basity | |
|---|---|---|---|---|---|---|
| Water | −13.3 | −5.13 | 0.0 | 1.00 | 1.00 | 2.00 |
| Formamide | −12.4 | −3.13 | 0.8 | 0.66 | 0.99 | 1.65 |
| NMF | −11.7 | −2.09 | — | — | — | — |
| Acetonitrile | −11.1 | 0.60 | 3.9 | 0.37 | 0.86 | 1.22 |
| DMSO | −10.8 | 1.00 | 3.9 | 0.34 | 1.08 | 1.41 |
| DMF | −10.7 | 1.56 | 4.5 | 0.30 | 0.93 | 1.23 |
| Acetone | −10.5 | 1.38 | 4.9 | 0.25 | 0.81 | 1.06 |
| Dioxane | −10.5 | −1.39 | — | 0.19 | 0.67 | 0.86 |
| DMAc |
−10.5 |
2.20 |
5.0 |
— |
— |
— |
| linear regressiond | ||||||
| n | 9 | 7 | 7 | 7 | 7 | |
| R2 | 0.81 | 0.98 | 0.98 | 0.25 | 0.87 | |
| slope | 0.36 | 0.53 | (−3.8) | (−3.9) | (−2.7) | |
log k (s−1) for decarboxylation of CBI (ref. 12).
log k (s−1) for azide attack on FNB (ref. 31).
Swain's scales of acity (anion solvating tendency) and basity (cation solvating tendency), based on a survey of solvent effects on the rates of 77 reactions (ref. 34).
The number of data points (n), squared regression coefficient (R2) and slope are shown for linear regression of log k (s−1) observed for decarboxylation of chxO in the present work (column 2), against the logarithms of rate constants recorded for CBI decarboxylation (column 3) and for azide attack on FNB (column 4), and against Swain's scales of acity (anion solvating tendency, column 5), basity (cation solvating tendency, column 6)34 and overall polarity (acity + basity, column 7).
Figure 5.
The decarboxylation of 1-methylorotate , the decarboxylation of 3-carboxybenzisoxazole and the reaction of NaN3 with 4-fluoro-nitrobenzene.
These reactions are characterized by delocalization of charge in the transition state.
Such reactions, in the words of Parker, “are faster in dipolar aprotic solvents because the reactant anion is much more solvated by protic than by dipolar (aprotic) solvents and this outweighs any effects due to transition state anion...solvation.”(35). Also shown are Swain's scales of “acity” (anion solvating tendency) and “basity” (cation solvating tendency), based on a survey of the rates of 77 reactions in various solvents. (36 In keeping with Parker's analysis, the rate of 1-ChxO decarboxylation shows a strong negative correlation (slope = −3.8) with Swain's acity scale and no significant correlation with his basity scale. One is led to infer that orotate derivatives are markedly stabilized in their ground states (relative to their transition states) by H-bonding to solvent water, and that this effect tends to disappear in solvents of increasingly aprotic character.
The present findings are consistent with the possibility that extraction of the substrate from solvent water enhances the value of kcat and contributes to the rate enhancement (kcat/knon) produced by OMP decarboxylase. It should be recognized that simple desolvation of a hydrophilic substrate would not, by itself, expected to have much effect on the value of kcat/Km, because desolvation is expected to increase Km to the same extent that it enhances kcat (37, 38). Nevertheless, if a substrate is equipped with substituents distant from the site of bond making and breaking, whose interaction with the enzyme can be used to draw the scissile portion of the substrate into a nonaqueous environment that enhances the reaction rate, then those substituents can be considered to contribute indirectly to catalysis by paying the cost of desolvating the scissile part of the substrate. In the case of ODCase, the phosphoribosyl group (39)—and more specifically the phosphoryl group (40)—of substrate OMP make very large contributions (11-12 kcal./mol in free energy) to stabilizing the altered substrate in the transition state by interacting with basic groups at the active site, without themselves playing any direct role in bond breaking (37). In a somewhat similar way, quaternary ammonium and phosphonium ions form ion pairs with the thiophenoxide ion, enabling it to enter the organic layer of a 2-phase system where it reacts rapidly with 1-bromooctane, in the first well-characterized example of phase transfer catalysis (41).
The present solvent effects on the decarboxylation of 1-ChxO are less pronounced than those observed for the reaction of azide with FNB or the decomposition of CBI (see slopes in Table 4), suggesting that simple desolvation of the substrate is unlikely to account for more than a limited part of the catalytic power of ODCase. Moreover, the effects of structural modification make it clear that much of this enzyme's catalytic effect depends on specific interactions between the substrate and polar residues of this enzyme's active site (2, 42). In the present model experiments, a positively charged ammonium group was necessary to assist removal of the carboxylate group from water. In ODCase, Lys-93 may play a comparable role in assisting the extraction of the substrate from solvent water during formation of the enzyme-substrate complex. Thus, the affinity of ODCase for OMP is substantially reduced when Lys-93 is converted to alanine, or when OMP is converted to UMP (3). Kinetic isotope effects indicate that, after substrate binding, the enzyme reaction proceeds in stepwise fashion, with carbanion formation preceding incorporation of the C6 proton into product UMP (42–47). The ammonium group of Lys-93 presumably stabilizes the carbanion generated by CO2 elimination and furnishes the proton that takes its place (11).
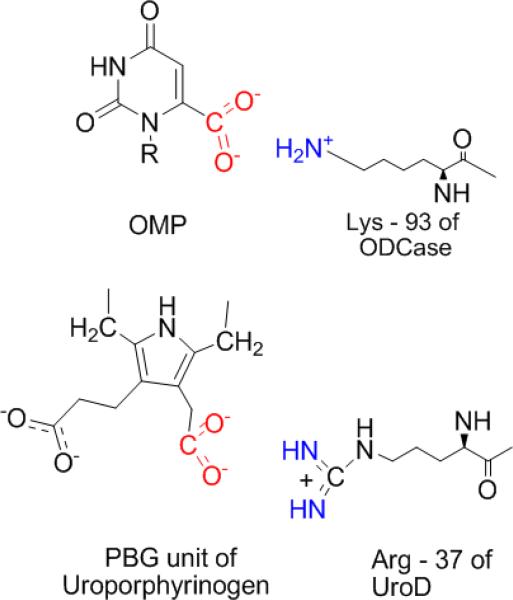
The likelihood of that scenario receives unexpected support from the recent finding that another enzyme reaction, involving a very different substrate (uroporphyrinogen), proceeds with a t1/2 of 2.3 × 109 years in water in the absence of the enzyme (48). Like ODCase, uroporphyrinogen decarboxylase (UroD, EC 4.1.1.37) catalyzes a difficult decarboxylation reaction without the assistance of metals or other cofactors. The only structural feature shared by these enzymes is the presence at the active site of a single basic side chain (Lys-93 in the case of yeast ODCase; Arg-37 in the case of UroD) that is indispensable for catalysis (Figure 6). During the evolution of their active sites, these enzymes appear to have converged on a common strategy in which cationic side-chains are in position to assist in the extraction of the substrate from solvent water into a relatively waterless non-polar cavity, stabilize carbanions that approach the transition state in structure, and furnish a proton to generate the decarboxylated product. Other active site side-chains appear to play important – but indirect – roles in maintaining the structural surroundings in which these events occur.
Figure 6.
Proposed similarity of interaction of the ammonium groups of Lys-93 in ODCase and the Arg-37 of Urogen III Decarboxylase with the carboxylate groups of their respective OMP and urogen III substrates.
Abbreviations
- OMP
orotidine 5′-monophosphate
- ODCase
orotidine 5′-monophosphate decarboxylase
- 1-MeU
1-methyluracil
- 1-ChxU
1-cyclohexyluracil
- U6AA
uracil-6-acetic acid
- 1-MeO
1-methylorotic acid
- 1-ChxO
1-cyclohexylorotic acid, 3-MeO, 3-methylorotic acid
- 1,3-Me2O
1,3-dimethylorotic acid
- TBA
tetrabutylammonium
- 1-ChxO-TBA
1-cyclohexylorotate – tetrabutylammonium salt
- 3-MeU
3-methyluracil
- 1,3-Me2U
1,3-dimethyluracil
- 6-MeU
6-methyluracil
- DMAc
N,N-Dimethylacetamide
- CBI
3-carboxy-6-benzisoxazole
- FNB
4-fluoro-nitrobenzene
- UroD
uroporphyrinogen decarboxylase
Footnotes
This work was supported by National Institutes of Health Grant GM-18325
However, the C5 proton of 1-MeO showed an increase in chemical shift below pH 2 confirming protonation of 1-MeO, to yield an acid with a pKa value of ∼0.7 (25) (Data not shown).
We also tested the effect of inserting a methylene group between C6 of orotic acid and its carboxylate group, by examining the behavior of U6AA as a reactant. The insertion of that methylene group enhanced the rate constant for decarboxylation by almost 7 orders of magnitude, to 1.7 × 10−10 s−1 at 25 °C. In contrast, the acetate ion has been shown to undergo no detectable decarboxylation after 14 days at 360 °C (29). The reasons for this unusual reactivity remain to be established, but the decarboxylation of U6AA presumably generates a 6-methyl anion in which negative might be delocalized to either of the oxygen atoms at C2 or C4.
Although the decomposition of CBI involves a decarboxylation, that decarboxylation occurs in concert with the opening of the isoxazole ring and does not involve formation of a discrete carbanion intermediate as indicated by the lack of solvent tritium incorporation into the product (Kemp 1970). Benzisoxazole (BI), which lacks a 3-carboxylate group, decomposes by an E2 elimination reaction in the presence of acetate as a catalyst, shows kinetic solvent effects (exemplified by a 7 orders of magnitude rate increase from water to hexamethylphosphoramide) closely comparable in magnitude with those observed for CBI. But when BI reacts in the presence of triethylamine, the kinetic solvent effect was reduced to less than a single order of magnitude. Hilvert et al. (1993) suggested that large solvent effects might be associated with the presence of a carboxylate functionality in the reactant or the catalyst, rather than solvent interactions with the polarizable benzisoxazole.
References
- 1.Radzicka A, Wolfenden R. A Proficient Enzyme. Science. 1995;267:90–94. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Traut TW, Temple BRS. The Chemistry of the Reaction Determines the Invariant Amino Acids during the Evolution and Divergence of Orotidine 5′-Monophosphate Decarboxylase. J. Biol.Chem. 2000;275:28675–28681. doi: 10.1074/jbc.M003468200. [DOI] [PubMed] [Google Scholar]
- 3.Miller BG, Snider MJ, Wolfenden R, Short S. Dissecting a Charged Network at the Active Site of Orotidine-5′-Phosphate Decarboxylase. J. Biol. Chem. 2001;276:15174–15176. doi: 10.1074/jbc.M011429200. [DOI] [PubMed] [Google Scholar]
- 4.Barnett SA, Amyes TL, Wood BM, Richard JP. Dissecting the Total Transition State Stabilization Provided by Amino Acid Side Chains at Orotidine 5′- Monophosphate Decarboxylase: a Two-Part Substrate Approach. Biochemistry. 2008;47:7785–7787. doi: 10.1021/bi800939k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson LN, Wolfenden R. Changes in Absorption Spectrum and Crystal Structure of Triose Phosphate Isomerase Brought about by 2-Phosphoglycollate, a Potential Transition State Analogue. J. Mol. Biol. 1970;47:93–100. doi: 10.1016/0022-2836(70)90404-3. [DOI] [PubMed] [Google Scholar]
- 6.Lolis E, Petsko GA. Crystallographic Analysis of the Complex Between Triosephosphate Isomerase and 2-Phosphoglycollate at 2.5-Å Resolution: Implications for Catalysis. Biochemistry. 1990;29:6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DK, Rudolph FB, Quiocho FA. Atomic Structure of Adenosine Deaminase with a Transition-State Analog: Understanding Catalysis and Immunodeficiency Mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 8.Betts L, Xiang S, Wolfenden R, Carter CW., Jr. Cytidine Deaminase. The 2.3 Å Crystal Structure of and Enzyme: Transition State Analog Complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 9.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a Proficient Enzyme: the Structure of Orotidine 5′-Monophosphate Decarboxylase in the Presence and Absence of a Potential Transition State Analog. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfenden R. Enzyme Catalysis: Conflicting Requirements of Substrate Access and Transition State Affinity. Mol. Cell Biochem. 1974;3:207–211. doi: 10.1007/BF01686645. [DOI] [PubMed] [Google Scholar]
- 11.Crosby J, Lienhard GE. Mechanisims of Thiamine-Catalyzed Reactions. A Kinetic Analysis of the Decarboxylation of Pyruvate by 3,4-Dimethylthiazolium Ion in Water and Ethanol. J. Am Chem. Soc. 1970;92:5707–5716. doi: 10.1021/ja00722a027. [DOI] [PubMed] [Google Scholar]
- 12.Kemp DS, Paul KG. The Physical Organic Chemistry of Benzisoxazoles. III. The Mechanism and the Effects of Solvents on Rates of Decarboxylation of Benzisoxazole-3-carboxylic Acids. J. Am. Chem. Soc. 1975;97:7305–7311. [Google Scholar]
- 13.Beak P, Siegel B. Mechanism of Decarboxylation of 1,3-Dimethylorotic Acid: A Possible Role for Orotate Decarboxylase. J. Amer. Chem. Soc. 1973;95:7919–7920. doi: 10.1021/ja00804a088. [DOI] [PubMed] [Google Scholar]
- 14.Beak P, Siegel B. Mechanism of Decarboxylation of 1,3-Dimethylorotic Acid. A Model for Orotidine 5′-Phosphate Decarboxylase. J. Amer. Chem. Soc. 1976;98:3601–3606. doi: 10.1021/ja00428a035. [DOI] [PubMed] [Google Scholar]
- 15.Wu W, Ley-Han AF, Wong M, Austin TJ, Miller S. Decarboxylation of 1,3-Dimethylorotic Acid Revisited: Determining the Role of N-1. Bioorg. & Med. Chem. Letters. 1997;7:2623–2628. [Google Scholar]
- 16.Kakanishi MP, Wu W. Mechanism of Decarboxylation of 1,3-Dimethylorotic Acid Revisited: Trapping of the Reaction Intermediate. Tet. Letters. 1998;39:6271–6272. [Google Scholar]
- 17.Singleton DA, Merrigan SR, Kim BJ, Beak P, Phillips LM, Lee JK. 13C Kinetic Isotope Effects and the Mechanism of the Uncatalyzed Decaroxylation of Orotic Acid. J. Amer. Chem. Soc. 2000;122:3296–3300. [Google Scholar]
- 18.Feng WY, Austin TJ, Chew F, Gronert S, Wu W. The Mechanism of Orotidne 5′-Monophosphate Decarboxylase: Catalysis by Destabilization of the Substrate. Biochemistry. 2000;39:1778–1783. doi: 10.1021/bi992553w. [DOI] [PubMed] [Google Scholar]
- 19.Tran NL, Colvin ME, Gronert S, Wu W. Catalysis of Decarboxylation by an Adjacent Negative Charge: a Theoretical Approach. Bioorg. Chem. 2003;31:271–277. doi: 10.1016/s0045-2068(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 20.Shem DL, Gronert S, Wu W. Modest Catalysis of the Decarboxylation of Orotate by Hydrogen Bonding: a Theoretical Model for Orotidine-5′-Monophosphate Decarboxylase. Bioorg. Chem. 2004;32:76–81. doi: 10.1016/j.bioorg.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Wong FM, Wu W. Accelerated Decarboxylation of 1,3-Dimethylorotic Acid in Ionic Liquid. Bioorg. Chem. 2006;34:99–104. doi: 10.1016/j.bioorg.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Yeoh FY, Cuasito RR, Capule CC, Wong FM, Wu W. Carbanions from Decarboxylation of Orotate Analogues: Stability and Mechanistic Implications. Biorg. Chem. 2007;35:338–343. doi: 10.1016/j.bioorg.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landesman PW. Ph. D. Dissertation. State University of New York; Buffalo: 1982. Design, Synthesis and Evaluation of Potential Inhibitors of Pyrimidine Biosynthesis: A Mechanistic Approach. [Google Scholar]
- 24.Curran WV, Angier RB. The Synthesis of Orotidine and its Isomer, 3-β-D-Ribofuranosylorotic Acid, and the Methylation of Orotic Acid. J. Org. Chem. 1966;31:201–205. [Google Scholar]
- 25.Fox JJ, Young N, Wempen I. Spectrophotometric Studies of Nucleic Acid Derivatives and Related Compounds as a Function of pH. IV. On the Structure of Orotidines. A Study of N-Methylated Orotic Acids. Biochim. et Biophys. Acta. 1957;23:295–305. doi: 10.1016/0006-3002(57)90331-1. [DOI] [PubMed] [Google Scholar]
- 26.Edsall JT, Wyman J. J. Biophysical Chemistry. Vol. 1. Academic Press; New York: 1958. pp. 452–453. [Google Scholar]
- 27.Schroeder G, Lad C, Wyman P, Williams NH, Wolfenden R. Time Required for Water Attack at the Phosphorus Atom of Simple Phosphodiesters and of DNA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown BR. The Mechanism of Thermal Decarboxylation. Quart. Rev. Chem. Soc. 1951;5:131–146. [Google Scholar]
- 29.Fairclough J. The Kinetics of Decarboxylation of Certain Organic Acids. J. Chem. Soc. 1938:1186–1190. [Google Scholar]
- 30.Sievers A, Wolfenden R. Equilibrium Formation of the 6-Carbanion of UMP, a Potential Intermediate in the Action of OMP Decarboxylase. J. Amer. Chem. Soc. 2002;124:13986–13987. doi: 10.1021/ja021073g. [DOI] [PubMed] [Google Scholar]
- 31.Miller J, Parker AJ. Dipolar Aprotic Solvents in Bimolecular Substitution Reactions. J. Amer. Chem. Soc. 1961;83:117–123. [Google Scholar]
- 32.Kemp DS, Paul K. Decarboxylation of Benzisoxazole-3-carboxylic Acids. Catalysis by Extraction of Possible Relevance to the Problem of Enzymatic Mechanism. J. Amer. Chem. Soc. 1970;92:2553–2554. [Google Scholar]
- 33.Kemp DS, Cox DD, Paul KG. The Physical Organic Chemistry of Benzioxazoles. IV. The origins and Catalytic Nature of the Solvent Rate Acceleration for Decarboxylation of 3-Carboxybenzisoxazoles. J. Amer. Chem. Soc. 1975;97:7312–7318. [Google Scholar]
- 34.Grate JW, McGill RA, Hilvert D. Analysis of Solvent Effects on the Decarboxylation of Benzisoxazole-3-carboxylate Ions Using Linear Solvation Energy Relationships: Relevance to Catalysis in an Antibody Binding Site. J. Amer. Chem. Soc. 1993;115:8577–8584. [Google Scholar]
- 35.Parker AJ. Protic-Dipolar Aprotic Solvent Effects on Rates of Bimolecular Reactions. Chem. Rev. 1969;69:1–32. [Google Scholar]
- 36.Swain CG, Swain MS, Powell AL, Alunni S. Solvent Effects on Chemical Reactivity. Evaluation of Anion and Cation Solvation Components. J. Amer. Chem. Soc. 1983;105:502–513. [Google Scholar]
- 37.Miller BG. Insight into the Catalytic Mechanism of Orotidine 5′-Phosphate Decarboxylase from Crystallography and Mutagenesis. Top Curr. Chem. 2004;238:43–62. [Google Scholar]
- 38.Warshel A, Florián J. Computer Simulations of Enzyme Catalysis: Finding out What has been Optimized by Evolution. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5950–5957. doi: 10.1073/pnas.95.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller BG, Snider MJ, Short SA, Wolfenden R. Contribution of Enzyme-Phosphoribosyl Contacts to Catalysis by Orotidine 5′- Phosphate Decarboxylase. Biochemistry. 2000;39:8113–8118. doi: 10.1021/bi000818x. [DOI] [PubMed] [Google Scholar]
- 40.Sievers A, Wolfenden R. The Effective Molarity of the Substrate Phosphoryl Group in the Transition State for Yeast OMP decarboxylase. Bioorg. Chem. 2005;33:45–52. doi: 10.1016/j.bioorg.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Herriott AW, Picker D. Phase Transfer Catalysis. An Evaluation of Catalysts. J. Am. Chem. Soc. 1975;97:2345–2349. [Google Scholar]
- 42.Lee JK, Tantillo DJ, editors. Topics in Current Chemistry 238: Orotidine Monophosphate Decarboxylase: A Mechanistic Dialogue. Springer-Verlag; New York: 2004. [Google Scholar]
- 43.Richavy MA, Cleland WW. Determination of the Mechanism of Orotidine 5′-Monophosphate Decarboxylase by Isotope Effects. Biochemistry. 2000;39:4569–4574. doi: 10.1021/bi000376p. [DOI] [PubMed] [Google Scholar]
- 44.van Vleet JL, Reinhardt LA, Miller BG, Sievers A, Cleland WW. Carbon Isotope Effect Study on Orotidine 5′-Monophosphate Decarboxylase: Support for an Anionic Intermediate. Biochemistry. 2008;47:798–803. doi: 10.1021/bi701664n. [DOI] [PubMed] [Google Scholar]
- 45.Toth K, Amyes TL, Wood B, Chan K, Gerlt JA, Richard JP. Product Deuterium Isotope Effect for Orotidine 5′-Monophosphate Decarboxylase: Evidence for a Short-Lived Carbanion Intermediate. J. Amer. Chem. Soc. 2007;129:12946–12947. doi: 10.1021/ja076222f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. Formation and Stability of a Vinyl Carbanion at the Active Site of Orotidine 5′-Monophosphate Decarboxylase: pK of the C-6 Proton of Enzyme-Bound UMP. J. Amer. Chem. Soc. 2008;130:1574–1575. doi: 10.1021/ja710384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KK, Wood BM, Federov A, Federov EV, Imker HJ, Amyes TL, Richard JP, Almo SC, Gerlt JA. Mechanism of the Orotidine 5′-Monophosphate Decarboxylase-Catalyzed recation: Evidence for Substrate Destabilzation. Biochemistry. 2009;48:5518–5531. doi: 10.1021/bi900623r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis CA, Jr., Wolfenden R. Uroporphyrinogen Decarboxylation as a Benchmark for the Catalytic Proficiency of Enzymes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17328–17333. doi: 10.1073/pnas.0809838105. [DOI] [PMC free article] [PubMed] [Google Scholar]