Abstract
In humans, NM23-H1 is a metastasis suppressor whose expression is reduced in metastatic melanoma and breast carcinoma cells, and which possesses the ability to inhibit metastatic growth without significant impact on the transformed phenotype. NM23-H1 exhibits three enzymatic activities in vitro, each with potential to maintain genomic stability, a 3′–5′exonuclease and two kinases, nucleoside diphosphate kinase (NDPK), and protein histidine kinase. Herein we have investigated the potential contributions of NM23 proteins to DNA repair in the yeast, Saccharomyces cerevisiae, which contains a single NM23 homolog, YNK1. Ablation of YNK1 delayed repair of UV-and etoposide-induced nuclear DNA damage by 3–6 hrs. However, YNK1 had no impact upon the kinetics of MMS-induced DNA repair. Furthermore, YNK1 was not required for repair of mitochondrial DNA damage. To determine whether the nuclear DNA repair deficit manifested as an increase in mutation frequency, the CAN1 forward assay was employed. An YNK1 deletion was associated with increased mutation rates following treatment with either UV (2.6x) or MMS (1.6x). Mutation spectral analysis further revealed significantly increased rates of base substitution and frameshift mutations following UV treatment in the ynk1Δ strain. This study indicates a novel role for YNK1 in DNA repair in yeast, and suggests an anti-mutator function that may contribute to the metastasis suppressor function of NM23-H1 in humans.
1. Introduction
NM23-H1 was first identified by virtue of its reduced expression in highly metastatic melanoma and breast carcinoma cells, and the ability of forced NM23-H1 expression to inhibit metastatic potential without significant impact on the transformed phenotype [1]. The metastatic process requires the accumulation of mutations and high levels of genomic instability to permit tumor cells to overcome the barriers to metastatic growth [2–4]. Despite the fact that NM23-H1 has been recognized to play a pivotal role in the development of metastasis, the underlying mechanisms by which NM23-H1 exhibits its anti-metastastic effect remains unknown.
Consistent with a role in DNA repair the NM23 molecule possesses at least three distinct enzymatic activities that could participate in genomic maintenance and antimutator activity [5]. NM23-H1 possess significant 3′–5′ exonuclease (3′–5′ EXO) activity [6,7] and these DNA cleaving molecules are predominantly involved with maintaining genomic fidelity during DNA synthesis and repair [8]. Accordingly, deficiencies in 3′–5′ EXO activity have been shown to be associated with the mutator phenotype [8–11]. NM23-H1 also exhibits a nucleoside diphosphate kinase (NDPK) activity that maintains homeostasis of nuclear nucleotide pools which may limit pro-mutagenic mismatches during DNA repair [12,13]. Furthermore, a protein histidine kinase (hisK) activity has been described for NM23-H1, implicated as an inhibitor of signaling pathways underlying cell motility [14], but which could also initiate signaling to DNA repair pathways. Moreover, despite the repair-relevant enzymatic activities of NM23-H1 in vitro, its contribution to maintenance of genomic integrity in vivo is poorly understood.
Previous studies strongly suggest that NM23 proteins exhibit functions consistent with DNA repair. DNA damage has been reported to induce nuclear localization of NM23-H1, consistent with a role in the DNA damage response [7]. In addition, co-incubation of NM23-H1 with the base excision repair (BER) enzyme uracil-DNA glycosylase (UDG) results in enhanced 3′–5′ EXO activity against single-stranded oligodeoxynucleotide substrates in vitro, suggesting the potential for functional cooperativity between these proteins [7]. A function in genomic stability was also suggested earlier by the marked mutator phenotype of ndk-null E. coli, which exhibit elevated rates of base substitutions and frameshifts [15]. While a recent study suggested that NDK possesses an intrinsic UDG function, subsequent studies show that NDK exhibits little if any intrinsic UDG activity [16–18], but does enhance that of the prototypical UDG, UNG, upon physical association between the proteins [17]. Consistent with this function, a very recent study has attributed the mutator phenotype of the ndk-null strain of E. coli to excess misincorporation of uracil, as well as a defect in the uracil base excision pathway [19].
To explore the potential function of NM23 proteins in maintenance of genomic integrity, we have employed the yeast S. cerevisiae, which harbors a single NM23 homologue, YNK1. Despite of its phylogenetic distance from the eight human NM23 isoforms, ynk1p shares approximately 60% amino acid sequence identity and structural similarities with human NM23-H1 and NM23-H2 [20], including conservation of glutamic acid-5 and lysine-12 residues which are critical for the 3′–5′ EXO function [6], histidine-118, which is essential for NDPK activity, and proline-96, which has been implicated in the histidine kinase function [21]. Our results demonstrate that an YNK1-null strain exhibits significantly reduced kinetics of nuclear DNA repair in response to damage induced by UV irradiation, and etoposide, as well as increased rates of UV-induced mutations.
2. Materials and methods
2.1 Saccharomyces cerevisiae strains and media
S. cerevisiae strains harboring single genetic lesions were obtained commercially (Open Biosystems) and were derived from BY4741 wild-type strains and listed in Table 1 (Supplemental Table 1). Open reading frames for the gene of interest were replaced with a KanMX marker by a PCR-based strategy. Yeast strains were grown in standard media consisting of yeast extract/peptone/dextrose (YPD) medium (Fisher Scientific).
Table 1.
The percentage of specific mutation types in wild-type and ynk1Δ strains
| −UV Exposure |
+UV Exposure |
|||||||
|---|---|---|---|---|---|---|---|---|
| Base substitution | Frameshift | Complex | No mutation | Base substitution | Frameshift | Complex | No mutation | |
| Wild-type | 59 | 0 | 26 | 15 | 53 | 0 | 21 | 26 |
| ynk1Δ | 70 | 0 | 12 | 18 | 70 | 15 | 10 | 5 |
Data is expressed as the percentage of mutation type i.e. base substitution, frameshift, complex or no mutation of the total number of mutational events in wild-type and ynk1Δ strains.
2.2 MMS, etoposide and UV treatment
S. cerevisiae strains (1 × 107 cells/ml) were treated with MMS (0.1%; Sigma) or etoposide (1 mM; Sigma) for 1 h at 30°C shaking at 250 rpm, followed by centrifugation at 5,000 × g for 5 min. The pellet was washed in 50 mM potassium phosphate, pH 7.0, aspirated, centrifuged at 5,000 × g for 5 min, and YPD added. The repair time course was 0.5, 1, 3 and 6 h for MMS and 0.5, 3, 6, 24 and 48 h for etoposide at 30°C. For UVB exposure (Model XX-15M, UVP Products), S. cerevisiae were grown on YPD plates for 48 h at 30°C, exposed to UV (192 J/m2) and maintained at 30°C and for a repair time course of 0.5, 1, 3 and 6 h.
2.3 Quantitative extended-length PCR (QXL-PCR)
QXL-PCR measures the average lesion frequency and works on the premise that damage on the DNA template will block a thermostable polymerase, resulting in reduced amplification of the DNA fragment. Thus, only DNA templates devoid of polymerase blocking lesions will be amplified. DNA lesion frequencies were calculated as the amplification of damaged (treated) samples (Ad) relative to the amplification of non-damaged fragment controls (A0) resulting in the ratio (Ad/A0). To determine average lesion frequency, a random distribution of lesions was assumed, and the following equation was used, λ = −ln Ad/A0 [22,23]. The DNA lesion frequencies were used to determine percentage repair of the initial DNA damage caused by the DNA damaging agent. The PCR conditions and primer sequences used are shown in supplemental information.
2.4 CAN1 forward mutation assay and sequence analysis
The standard CAN1 forward mutation assay was performed as previously described [24,25]. Independent CAN1 colonies were isolated and the CAN1 gene sequenced at University of Kentucky Genetic Technologies Center.
2.5 Statistical analyses
A two-tailed t test was used for comparison between two treatments and for comparison between three or more experimental groups, one way ANOVA with the Bonferroni post hoc test was used. Values of p<0.05 were considered statistically significant.
3. Results
3.1 A ynk1Δ strain exhibits attenuated repair of etoposide- and UV radiation--induced nuclear DNA damage
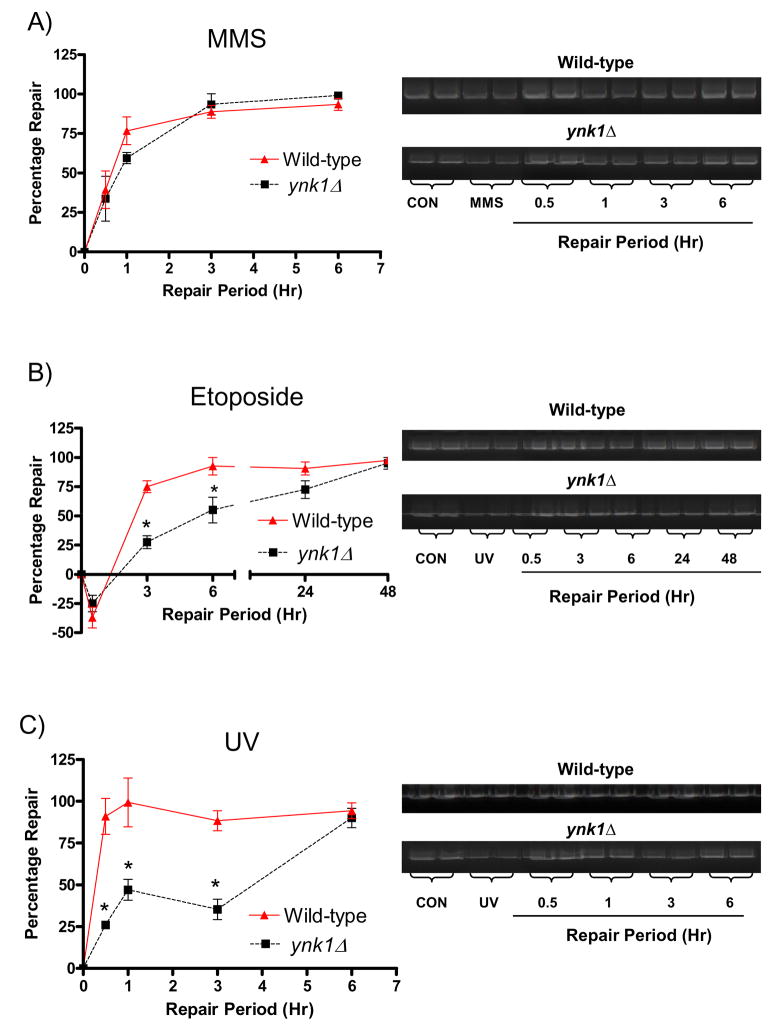
To determine whether YNK1 has a functional role in DNA repair, we compared the repair rates of ynk1Δ versus wild-type cells following MMS (0.1%), etoposide (1 mM) and UV (192 J/m2) treatment (Figure 1). Repair of MMS-induced nDNA damage did not significantly differ throughout the repair period between ynk1Δ and wild-type strains. In contrast, ynk1Δ mutants demonstrated a significantly reduced capacity to repair etoposide- and UV-induced nuclear DNA damage compared to wild-type up to 6 hr and 3 hr post-treatment, respectively (p < 0.05).
Figure 1. ynk1Δ attenuates nDNA repair of etoposide-and UV-induced damage but not MMS.
Cells were harvested at the indicated time points, DNA extracted and QXL-PCR performed. The lesion frequencies were determined as described in Materials and Methods and data expressed as percentage repair of the initial DNA damage. A) MMS-induced DNA damage and repair at 0.5, 1, 3, and 6 h post-treatment; B) etoposide-induced DNA damage and repair at 0.5, 3, 6, 24 and 48 h post-treatment and C) UV-induced DNA damage and repair at 0.5, 1, 3 and 6 h post-treatment. Bars represent mean ± SEM. *P < 0.05, ynk1Δ vs. wild-type, one way ANOVA, n = 3 per group.
3.2 YNK1 does not have a role in the repair of MMS-, etoposide-, or UV-induced mitochondrial DNA damage
The mitochondrial genome is frequently challenged by DNA damaging agents, and mitochondrial genomic instability is associated with impaired nucleotide metabolism and development of the mutator phenotype [26,27]. Intriguingly, a fraction of the total cellular ynk1p has been localized to the mitochondrion [28]. However, its function within this compartment is not fully understood. Therefore, we aimed to examine whether YNK1 has a functional role in mitochondrial DNA repair following DNA damage caused by MMS (0.1%), etoposide (1mM) and UV (192 J/m2) (Supplemental Figure 1). The repair of MMS-, etoposide-and UV-induced mtDNA damage did not significantly differ between the ynk1Δ and wild-type strains, throughout the respective repair periods. Furthermore, only ~25% of the initial lesions were repaired at 6hr post-treatment for all DNA damaging agents.
3.3 The ynk1Δ strain exhibits a mutator phenotype following MMS and UV exposure
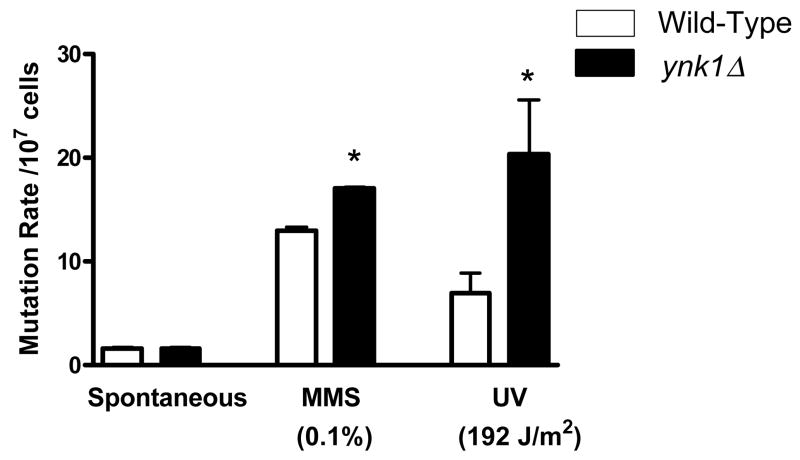
The ynk1Δ strain displayed a significantly slower repair of DNA damage induced by UV irradiation. To determine whether this impairment was manifested as an increase in mutation frequency, the CAN1 forward mutation assay was employed under spontaneous conditions (no treatment), and following exposure to MMS and UV irradiation (Figure 2). Under spontaneous conditions, no significant difference in mutation rate between ynk1Δ and wild-type strains occurred. In contrast, treatment with UV (192J/m2) and MMS (0.1%) generated 2.6-fold and 1.6-fold increases in the mutation rate of ynk1Δ compared to wild-type, respectively (p < 0.05).
Figure 2. ynk1Δ causes a mutator phenotype following MMS and UV exposure.
The CAN1 forward mutation rate was measured in ynk1Δ and wild-type strains under spontaneous conditions or following treatment with either MMS (0.1%) or UV (192 J/m2). The CAN1 mutation rate was determined from the median in a fluctuation test of more than 10 independent cultures. *P < 0.05, ynk1Δ vs. wild-type.
3.4 YNK1 mutants display a greater rate of base substitutions and frameshift mutations
Mutation spectra for ynk1Δ and wild-type strains were obtained following UV exposure (192 J/m2) by sequencing the CAN1 gene (Table 1). The ynk1Δ strain displayed a greater rate of base substitutions compared to the wild-type strain (70% vs. 59%), and a lower rate of complex alterations (12% vs. 26%). Following UV irradiation, there was no significant change in base substitutions or more complex alterations in either the ynk1Δ or wild-type strains. However, only the ynk1Δ mutants underwent frameshift events following UV-induced damage (15% vs. 0%), with mutations found at higher frequencies in simple repeat tracts of thymines in ynk1Δ mutants (Supplemental Figure 2).
4. Discussion
This investigation provides novel information regarding the role of YNK1 in the maintenance of genomic integrity. Firstly, YNK1 is required for the repair of UV- and etoposide-induced damage of nuclear DNA. However, YNK1 has no measurable impact upon the kinetics of repair of nuclear DNA damage caused by MMS. Secondly, YNK1 is not required for the repair of mitochondrial DNA damage. Thirdly, YNK1 ablation promotes a mutator phenotype following UV and MMS exposure. This is the first study to our knowledge demonstrating a direct involvement of YNK1 in DNA repair and in providing an anti-mutator function in vivo.
To test the hypothesis that YNK1 has a role in DNA repair, we used QXL-PCR to study DNA repair kinetics [22,23]. This approach demonstrated that ablation of YNK1 attenuated the repair of a 9.3-kb fragment in the nuclear phosphofructokinase-2 gene (a key enzyme involved in glycolysis) following UV and etoposide treatment. In the case of UV-induced DNA damage, which is repaired primarily by nucleotide excision repair (NER) [29], YNK1 could be involved in the NER pathway by functioning as a redundant 3′–5′ EXO. Furthermore, this function is also a plausible candidate for repair of etoposide-induced topoisomerase II-mediated DNA strand breaks [30]. In contrast, YNK1 did not appear to facilitate the repair of DNA damage caused by MMS, as both wild-type and ynk1Δ displayed similar repair rates. The DNA damage caused by MMS is largely repaired by the BER [31]. Thus, this suggests YNK1 does not have a functional involvement in BER, at least not against MMS-induced lesions (e.g., 7-methylguanine, O6-methylguanine and, 3-methyladenine) [32,33].
The mitochondrial DNA comprises approximately 15% of the DNA content of S. cerevisiae, and its stability is crucial for cell viability [34]. A fraction of cellular ynk1p is localized to the mitochondria’s intermembrane [28], yet its function within this compartment in yeast is not fully understood. YNK1 exhibited no beneficial effect in the repair of MMS, etoposide or UV-induced mtDNA lesions in a 6.9-kb fragment of the mitochondrial COX1 gene. However, this data has revealed important differences with respect to the repair of DNA damage in the nucleus and mitochondria in S. cerevisiae, and is consistent with the general notion, that mtDNA in higher eukaryotes is highly susceptible to genomic injury with limited repair potential [35].
To determine whether the deficit in DNA repair manifests to an increased incidence of mutations in S. cerevisiae, mutation rates were measured and sequenced following DNA damage. Indeed, mutation rates significantly increased in the ynk1Δ strain following treatment with UV and MMS. Intriguingly, NM23-H1 is an autonomous 3′–5′ EXO [5,6,8], an enzymatic function often implicated in enhancing the proofreading capacities of error-prone DNA polymerases during translesion repair (TLS). Thus, it is possible NM23 may have a direct proofreading function during TLS or as a facilitory role in protein complexes associated with DNA during TLS [8,36,37]. Mutation spectral analysis of the CAN1 gene revealed increased frameshift mutations in ynk1Δ after UV treatment. In particular, homonucleotide tracts of thymines were hot spots for these UV-induced mutations, which is consistent with the primary lesions generated by UV exposure (i.e. bulky lesions such as pyrimidine dimers and 6-4 photoproducts) [38]. These observed mutational events further support a role of YNK1 in mutation avoidance, and possibly a function in TLS, as this mutation spectra is consistent with both 3′-5 EXO deficiency and impaired TLS following UV damage [36,38–40].
While DNA repair is an obvious potential function of YNK1, an alternative and complementary role in maintaining genomic stability must also be considered, via its NDPK activity [41]. NDPK has a central role in the maintenance of dNTP pools with perturbations in the dNTP balance associated with increased base substitutions, frameshift mutations as well as strong mutator phenotypes [42–46]. A previous study has provided evidence that YNK1 possesses NDPK activity [28], and E. coli deficient in NDK suffer chronic perturbations in dNTP pools and increased mutagenesis [47,48]. These mutator phenotypes have been attributed to an imbalance in nucleotide pool sizes as a consequence of NDPK deficiencies, of particular note elevation in intracellular concentrations of dCTP [47]. Furthermore, of relevance to the present study, alterations in the intracellular dNTP pool may also compromise DNA replicative synthesis, and enhance fragile sites where chromosomes are susceptible to breakage and inhibit DNA repair [26,49–51].
Together, these observations suggest novel mechanisms underlying the metastasis suppressor activity of NM23-H1. We postulate that YNK1 is an important factor in the mutation avoidance machinery by facilitating efficient DNA repair and limiting the generation of DNA mutations, primarily by virtue of its 3′–5′ EXO and/or NDPK activity.
Supplementary Material
Supplemental Figure 1. ynk1Δ does not have a role in the repair of mtDNA damage caused by MMS, etoposide or UV. Cells were harvested at the indicated time points, DNA extracted and QXL-PCR performed. The lesion frequencies were determined as described in Materials and Methods and data expressed as percentage repair of the initial DNA damage. A) MMS-induced DNA damage and repair at 0.5, 1, 3, and 6 h post-treatment; B) etoposide-induced DNA damage and repair at 0.5, 3, 6, 24 and 48 h post-treatment and C) UV-induced DNA damage and repair at 0.5, 1, 3 and 6 h post-treatment. Bars represent mean ± SEM. *P < 0.05, ynk1Δ vs. wild-type, one way ANOVA, n = 3 per group.
Supplemental Figure 2. Thymine tracts are vulnerable to alterations in ynk1Δ following UV exposure. The mutation spectra for ynk1Δ and wild-type strains were determined by sequencing the CAN1 gene. A T tract alteration was classified as a homonucleotide run of at least 4 Ts which after UV treatment underwent at least 1 incorrect base alteration. To determine independence of each reversion event, only one colony from each culture was used for mutation spectrum analysis from 20 independent CAN1 isolates.
Acknowledgments
This work was supported by NIH/NCI; CA83237-7 (DMK).
References
- 1.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 2.Callaghan KA, Becker TE, Ellsworth DL, Hooke JA, Ellsworth RE, Shriver CD. Genomic instability and the development of metastatic lymph node tumors. Ann Surg Oncol. 2007;14:3125–3132. doi: 10.1245/s10434-007-9504-7. [DOI] [PubMed] [Google Scholar]
- 3.Fukino K, Shen L, Patocs A, Mutter GL, Eng C. Genomic instability within tumor stroma and clinicopathological characteristics of sporadic primary invasive breast carcinoma. Jama. 2007;297:2103–2111. doi: 10.1001/jama.297.19.2103. [DOI] [PubMed] [Google Scholar]
- 4.Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68:2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 5.Kaetzel DM, Zhang Q, Yang M, McCorkle JR, Ma D, Craven RJ. Potential roles of 3′–5′ exonuclease activity of NM23-H1 in DNA repair and malignant progression. J Bioenerg Biomembr. 2006;38:163–167. doi: 10.1007/s10863-006-9040-3. [DOI] [PubMed] [Google Scholar]
- 6.Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3′–5′ exonuclease activity. J Biol Chem. 2004;279:18073–18084. doi: 10.1074/jbc.M400185200. [DOI] [PubMed] [Google Scholar]
- 7.Yoon JH, Singh P, Lee DH, Qiu J, Cai S, O’Connor TR, Chen Y, Shen B, Pfeifer GP. Characterization of the 3′--> 5′ exonuclease activity found in human nucleoside diphosphate kinase 1 (NDK1) and several of its homologues. Biochemistry. 2005;44:15774–15786. doi: 10.1021/bi0515974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevelev IV, Hubscher U. The 3′ 5′ exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 9.Lehtinen DA, Perrino FW. Dysfunctional proofreading in the Escherichia coli DNA polymerase III core. Biochem J. 2004;384:337–348. doi: 10.1042/BJ20040660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison A, Sugino A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 11.Foury F, Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. Embo J. 1992;11:2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal RP, Robison B, Parks RE., Jr Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol. 1978;51:376–386. doi: 10.1016/s0076-6879(78)51051-3. [DOI] [PubMed] [Google Scholar]
- 13.Postel EH. NM23-NDP kinase. Int J Biochem Cell Biol. 1998;30:1291–1295. doi: 10.1016/s1357-2725(98)00087-9. [DOI] [PubMed] [Google Scholar]
- 14.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 15.Miller JH, Funchain P, Clendenin W, Huang T, Nguyen A, Wolff E, Yeung A, Chiang JH, Garibyan L, Slupska MM, Yang H. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics. 2002;162:5–13. doi: 10.1093/genetics/162.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett SE, Chen CY, Mosbaugh DW. Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease. Proc Natl Acad Sci U S A. 2004;101:6391–6396. doi: 10.1073/pnas.0401031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goswami SC, Yoon JH, Abramczyk BM, Pfeifer GP, Postel EH. Molecular and functional interactions between Escherichia coli nucleoside-diphosphate kinase and the uracil-DNA glycosylase Ung. J Biol Chem. 2006;281:32131–32139. doi: 10.1074/jbc.M604937200. [DOI] [PubMed] [Google Scholar]
- 18.Postel EH, Abramczyk BM. Escherichia coli nucleoside diphosphate kinase is a uracil-processing DNA repair nuclease. Proc Natl Acad Sci U S A. 2003;100:13247–13252. doi: 10.1073/pnas.2333230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordman J, Wright A. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc Natl Acad Sci U S A. 2008;105:10197–10202. doi: 10.1073/pnas.0802816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Bao R, Jiang C, Yang Z, Zhou CZ, Chen Y. Structure of Ynk1 from the yeast Saccharomyces cerevisiae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:572–576. doi: 10.1107/S1744309108015212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem. 1996;271:25107–25116. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 22.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 23.Stuart GR, Santos JH, Strand MK, Van Houten B, Copeland WC. Mitochondrial and nuclear DNA defects in Saccharomyces cerevisiae with mutations in DNA polymerase gamma associated with progressive external ophthalmoplegia. Hum Mol Genet. 2006;15:363–374. doi: 10.1093/hmg/ddi454. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi G, Daigaku Y, Nagata Y, Ihara M, Yamamoto K. Saccharomyces cerevisiae RAD27 complements its Escherichia coli homolog in damage repair but not mutation avoidance. Genes Genet Syst. 2004;79:183–187. doi: 10.1266/ggs.79.183. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi G, Endo K, Doi A, Fujita A, Daigaku Y, Nunoshiba T, Yamamoto K. Spontaneous mutagenesis in haploid and diploid Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;325:928–933. doi: 10.1016/j.bbrc.2004.10.120. [DOI] [PubMed] [Google Scholar]
- 26.Desler C, Munch-Petersen B, Rasmussen LJ. The role of mitochondrial dNTP levels in cells with reduced TK2 activity. Nucleosides Nucleotides Nucleic Acids. 2006;25:1171–1175. doi: 10.1080/15257770600894501. [DOI] [PubMed] [Google Scholar]
- 27.Mathews CK, Song S. Maintaining precursor pools for mitochondrial DNA replication. FASEB J. 2007;21:2294–2303. doi: 10.1096/fj.06-7977rev. [DOI] [PubMed] [Google Scholar]
- 28.Amutha B, Pain D. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem J. 2003;370:805–815. doi: 10.1042/BJ20021415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rass K, Reichrath J. UV damage and DNA repair in malignant melanoma and nonmelanoma skin cancer. Adv Exp Med Biol. 2008;624:162–178. doi: 10.1007/978-0-387-77574-6_13. [DOI] [PubMed] [Google Scholar]
- 30.Sabourin M, Nitiss JL, Nitiss KC, Tatebayashi K, Ikeda H, Osheroff N. Yeast recombination pathways triggered by topoisomerase II-mediated DNA breaks. Nucleic Acids Res. 2003;31:4373–4384. doi: 10.1093/nar/gkg497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaina B, Ochs K, Grosch S, Fritz G, Lips J, Tomicic M, Dunkern T, Christmann M. BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis. Prog Nucleic Acid Res Mol Biol. 2001;68:41–54. doi: 10.1016/s0079-6603(01)68088-7. [DOI] [PubMed] [Google Scholar]
- 32.Calleja F, Jansen JG, Vrieling H, Laval F, van Zeeland AA. Modulation of the toxic and mutagenic effects induced by methyl methanesulfonate in Chinese hamster ovary cells by overexpression of the rat N-alkylpurine-DNA glycosylase. Mutat Res. 1999;425:185–194. doi: 10.1016/s0027-5107(99)00034-2. [DOI] [PubMed] [Google Scholar]
- 33.Elder RH, Jansen JG, Weeks RJ, Willington MA, Deans B, Watson AJ, Mynett KJ, Bailey JA, Cooper DP, Rafferty JA, Heeran MC, Wijnhoven SW, van Zeeland AA, Margison GP. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol Cell Biol. 1998;18:5828–5837. doi: 10.1128/mcb.18.10.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuin A, Gabrielli N, Calvo IA, Garcia-Santamarina S, Hoe KL, Kim DU, Park HO, Hayles J, Ayte J, Hidalgo E. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS ONE. 2008;3:e2842. doi: 10.1371/journal.pone.0002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 36.Abdulovic AL, Minesinger BK, Jinks-Robertson S. The effect of sequence context on spontaneous Polzeta-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:2082–2093. doi: 10.1093/nar/gkn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beliakova NV, Kravetskaia TP, Legina OK, Ronzhina NL, Shevelev IV, Krutiakov VM. Complex of repair DNA polymerase beta with autonomous 3′-->5′-exonuclease shows increased accuracy of DNA synthesis. Izv Akad Nauk Ser Biol. 2007:517–523. [PubMed] [Google Scholar]
- 38.Ikehata H, Ono T, Tanaka K, Todo T. A model for triplet mutation formation based on error-prone translesional DNA synthesis opposite UV photolesions. DNA Repair (Amst) 2007;6:658–668. doi: 10.1016/j.dnarep.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Tran HT, Gordenin DA, Resnick MA. The 3′-->5′ exonucleases of DNA polymerases delta and epsilon and the 5′-->3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 42.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proc Natl Acad Sci U S A. 1990;87:4946–4950. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J Biol Chem. 1992;267:3589–3596. [PubMed] [Google Scholar]
- 44.Mattano SS, Palella TD, Mitchell BS. Mutations induced at the hypoxanthine-guanine phosphoribosyltransferase locus of human T-lymphoblasts by perturbations of purine deoxyribonucleoside triphosphate pools. Cancer Res. 1990;50:4566–4571. [PubMed] [Google Scholar]
- 45.Meuth M. Sensitivity of a mutator gene in Chinese hamster ovary cell to deoxynucleoside triphosphate pool alterations. Mol Cell Biol. 1981;1:652–660. doi: 10.1128/mcb.1.7.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meuth M, L’Heureux-Huard N, Trudel M. Characterization of a mutator gene in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1979;76:6505–6509. doi: 10.1073/pnas.76.12.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 48.Shen R, Wheeler LJ, Mathews CK. Molecular interactions involving Escherichia coli nucleoside diphosphate kinase. J Bioenerg Biomembr. 2006;38:255–259. doi: 10.1007/s10863-006-9041-2. [DOI] [PubMed] [Google Scholar]
- 49.Cohen A, Thompson E. DNA repair in nondividing human lymphocytes: inhibition by deoxyadenosine. Cancer Res. 1986;46:1585–1588. [PubMed] [Google Scholar]
- 50.Desler C, Munch-Petersen B, Stevnsner T, Matsui S, Kulawiec M, Singh KK, Rasmussen LJ. Mitochondria as determinant of nucleotide pools and chromosomal stability. Mutat Res. 2007;625:112–124. doi: 10.1016/j.mrfmmm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Ke PY, Kuo YY, Hu CM, Chang ZF. Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev. 2005;19:1920–1933. doi: 10.1101/gad.1322905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. ynk1Δ does not have a role in the repair of mtDNA damage caused by MMS, etoposide or UV. Cells were harvested at the indicated time points, DNA extracted and QXL-PCR performed. The lesion frequencies were determined as described in Materials and Methods and data expressed as percentage repair of the initial DNA damage. A) MMS-induced DNA damage and repair at 0.5, 1, 3, and 6 h post-treatment; B) etoposide-induced DNA damage and repair at 0.5, 3, 6, 24 and 48 h post-treatment and C) UV-induced DNA damage and repair at 0.5, 1, 3 and 6 h post-treatment. Bars represent mean ± SEM. *P < 0.05, ynk1Δ vs. wild-type, one way ANOVA, n = 3 per group.
Supplemental Figure 2. Thymine tracts are vulnerable to alterations in ynk1Δ following UV exposure. The mutation spectra for ynk1Δ and wild-type strains were determined by sequencing the CAN1 gene. A T tract alteration was classified as a homonucleotide run of at least 4 Ts which after UV treatment underwent at least 1 incorrect base alteration. To determine independence of each reversion event, only one colony from each culture was used for mutation spectrum analysis from 20 independent CAN1 isolates.