Summary
Mitotic chromosome segregation requires that kinetochores attach to microtubule polymers and harness microtubule dynamics to drive chromosome movement. In budding yeast, the Dam1 complex couples kinetochores with microtubule depolymerization. However, a metazoan homologue of the Dam1 complex has not been identified. To identify proteins that play a corresponding role at the vertebrate kinetochore-microtubule interface, we isolated a 3-subunit human Ska1 complex, including the previously uncharacterized protein Rama1, that localizes to the outer kinetochore and spindle microtubules. Depletion of Ska1 complex subunits severely compromises proper chromosome segregation. Reconstituted Ska1 complex possesses two separable biochemical activities; direct microtubule-binding through the Ska1 subunit, and microtubule-stimulated oligomerization imparted by the Rama1 subunit. The full Ska1 complex forms assemblies on microtubules that can facilitate the processive movement of microspheres along depolymerizing microtubules. In total, these results demonstrate a critical role for the Ska1 complex in interacting with dynamic microtubules at the outer kinetochore.
Keywords: Kinetochore, mitosis, spindle, chromosome segregation
Introduction
Chromosome segregation during mitosis requires kinetochores to generate physical connections between chromosomes and spindle microtubule polymers. The microtubule interface represents a particular challenge since the kinetochore must not only form contacts with microtubules, but must also remain attached as the microtubules polymerize and depolymerize, as well as couple this growth and shrinkage to chromosome movement. Previous studies have identified a number of components of the kinetochore that form direct connections with microtubules (reviewed in (Cheeseman and Desai, 2008; Tanaka and Desai, 2008). The majority of these proteins, such as the plus end tracking proteins Bim1/EB1, Bik1/CLIP-170, Stu2/XMAP215, the kinesin-related protein kinesin-8/Kip3, and the multi-subunit KNL1/Mis12 complex/Ndc80 complex (KMN) network, are conserved throughout eukaryotes where they play similar mitotic functions. The combination of these proteins provides the ability to generate stable attachments to microtubule polymers and control the polymerization state of kinetochore-bound microtubules.
In addition to the activities of these conserved outer kinetochore proteins, it is also critical for kinetochores to maintain connections with dynamic microtubule polymers and couple microtubule disassembly to chromosome movement. In budding yeast, this function appears to be provided by the Dam1 complex (also called DASH). The Dam1 complex is a 10-subunit microtubule binding complex (Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002) that can form rings (Miranda et al., 2005; Westermann et al., 2005) that encircle and slide along microtubule polymers (Asbury et al., 2006; Grishchuk et al., 2008b; Westermann et al., 2006). However, vertebrate homologues of the Dam1 complex have not been identified. One possibility to explain the absence of a Dam1 homologue is that, in organisms where multiple microtubules form a dynamic kinetochore bundle, the energy of the microtubule depolymerization may be harnessed using an alternate mechanism (for example, see McIntosh et al., 2008). Alternately, a distinct protein or complex may function to couple kinetochore-microtubule dynamics with chromosome segregation.
To understand the formation of kinetochore-microtubule interactions in vertebrates and the way that these interactions are coupled to microtubule depolymerization, we sought to identify additional components of the human kinetochore-microtubule interface. Recent work identified Ska1 as a component of the human kinetochore (Hanisch et al., 2006). However, the specific role of Ska1 in mediating chromosome segregation was unclear. Here, we demonstrate that the 3-subunit Ska1 complex is a direct component of the kinetochore-microtubule interface. Importantly, the Ska1 complex displays several key properties that suggest an important role in coupling chromosome movement to microtubule depolymerization including dual localization to kinetochores and to spindle microtubules during mitosis, a critical role in proper chromosome segregation, a direct microtubule binding activity, the formation of cooperative oligomeric assemblies on microtubules, and the ability to couple a cargo to the depolymerization of a microtubule polymer.
Results
Purification of a multi-subunit outer kinetochore Ska1 complex
Recent work identified the human kinetochore protein Ska1 (Hanisch et al., 2006). Based on the localization of Ska1 to both kinetochores and microtubules, we hypothesized that it might function at the kinetochore-microtubule interface. To test this, we began by generating stable clonal human cell lines expressing either GFPLAP-tagged Ska1 or Ska2, which has been shown to associate with Ska1 by two-hybrid analysis (Hanisch et al., 2006). Consistent with this previous work, GFPLAP-Ska1 and GFPLAP-Ska2 both localized to kinetochores and spindle microtubules from prometaphase through telophase (Fig. 1A). Purification of LAP-tagged Ska1 and Ska2 from these cells isolated a complex containing Ska1, Ska2, and the previously uncharacterized protein Rama1 (also referred to as C13orf3, LOC221150, and Q8IX90.2) (Fig. 1B). In Ska1 and Ska2 purifications, we also identified the vesicle trafficking protein SNAP29. However, SNAP29 did not exhibit a similar localization or depletion phenotype to Ska1 (data not shown; Steegmaier et al., 1998), and thus we excluded this protein from further analysis. Less stringent one step purifications (IPs) with Ska1, but not multi-step LAP purifications, additionally isolated small amounts of the spindle checkpoint proteins Bub1 and Bub3 (Fig. 1B).
Figure 1. A 3 subunit human Ska1 complex localizes to outer kinetochore and microtubules.
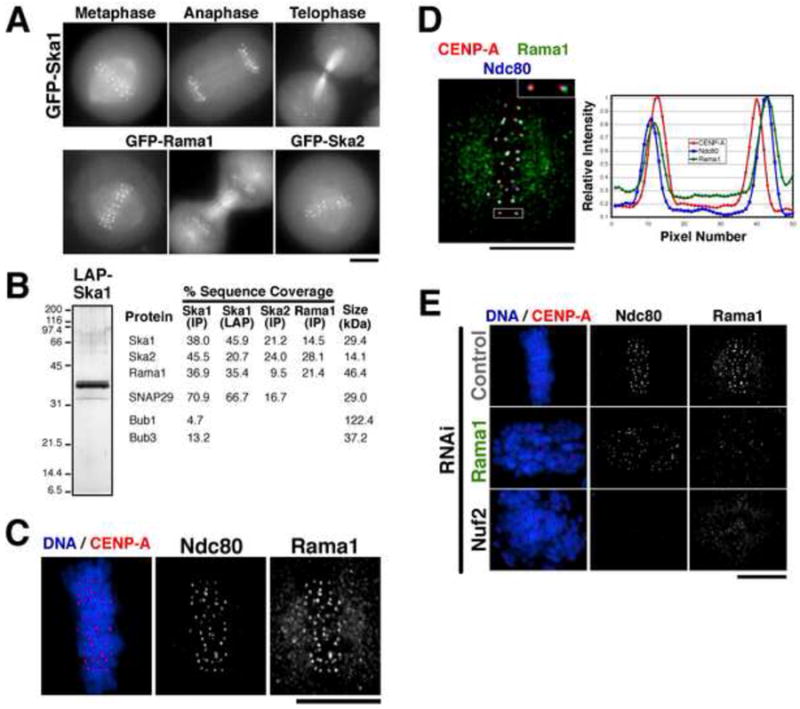
(A) Images of mitotic cells from clonal human cell lines stably expressing moderate amounts of Ska1, Ska2, and Rama1 as GFPLAP fusions. Each fusion protein localizes to kinetochores and microtubules throughout mitosis. (B) Purification of a 3-subunit Ska1 complex. GFPLAP tagged fusions were used to isolate Ska1, Ska2, or Rama1 from the stable cell lines shown in (A) using either one step IPs, or tandem affinity (LAP) purifications. Left, silver stained gel of the Ska1 LAP purification. Right, percent sequence coverage from the mass spectrometric analysis of these samples of proteins identified in the Ska1 complex purifications, but not unrelated controls. (C) Localization of endogenous Rama1. Immunofluorescence using anti-Rama1, anti-HEC1, and anti-GFP (against GFP-CENP-A) antibodies. Rama1 shows a punctuate cellular background staining throughout the cell cycle, but shows pronounced mitotic localization to kinetochores following after nuclear envelope breakdown. (D) Left, merged image showing co-localization of GFP-CENP-A, Rama1, and HEC1. Right, graph showing a linescan of the fluorescent intensity of the highlighted kinetochore. Rama1 co-localizes with Hec1 at the outer kinetochore, and localizes peripherally to CENP-A. (E) Rama1 kinetochore localization depends on the Ndc80 complex, but the Ndc80 complex localizes to kinetochores independently of Rama1. Immunofluorescence of control cells, Rama1 depleted cells, and Nuf2 depleted cells showing localization of GFP-CENP-A, Ndc80/HEC1, and Rama1. Scale bars, 10 μm.
To confirm that Rama1 is a subunit of the Ska1 complex, we generated a cell line stably expressing GFPLAP-Rama1. Similarly to Ska1 and Ska2, GFPLAP-Rama1 localized to kinetochores and microtubules during mitosis, but not during interphase or prophase (Fig. 1A). Purification of GFPLAP-tagged Rama1 isolated Ska1 and Ska2, confirming this interaction (Fig. 1B). Immunofluorescence against using affinity-purified polyclonal antibodies against endogenous Rama1 also revealed kinetochore localization (Fig. 1C), but under the examined fixation conditions localization to microtubules was not clear. In addition to the kinetochore-associated signal, anti-Rama1 antibodies detected a weak punctuate background staining throughout the cell that is not eliminated by RNAi.
To determine the localization of Rama1 within the kinetochore, we compared the spatial localization of Rama1 relative to the inner kinetochore protein CENP-A and outer kinetochore protein Ndc80 (Fig. 1D). Rama1 precisely co-localized with Ndc80 at the outer kinetochore, and both proteins localized peripherally to CENP-A. Consistent with this localization to the outer kinetochore and prior work on the localization of Ska1 (Hanisch et al., 2006), depletion of the Ndc80 complex subunit Nuf2 abolished the kinetochore localization of Rama1 (Fig. 1E). In contrast, depletion of Rama1 eliminated its kinetochore localization, but did not affect localization of hNdc80 (Hec1) to kinetochores (Fig. 1E). In total, these results indicate that a 3 subunit complex containing Ska1, Ska2, and Rama1 localizes to the outer kinetochore and spindle microtubule during mitosis in an Ndc80 complex-dependent manner.
The Ska1 complex is required for mitotic chromosome segregation and microtubule organization
To determine the functional roles of the Ska1 complex, we conducted both overexpression and depletion studies. HeLa cells expressing low or moderate amounts of Ska1, Ska2, or Rama1 GFP fusions, either as stable clonal cell lines or using transiently expressed proteins, displayed the mitotic spindle and kinetochore localization described above (Fig. 1A). However, in cells transiently overexpressing GFP-Ska1 (Fig. 2A), but not Ska2 or Rama1 (not shown), the fusion protein localized to microtubules in interphase cells. In these cases, the microtubules were bundled suggesting that Ska1 is a microtubule-associated protein that may play a role in microtubule organization.
Figure 2. The Ska1 complex is required for kinetochore-microtubule attachments.
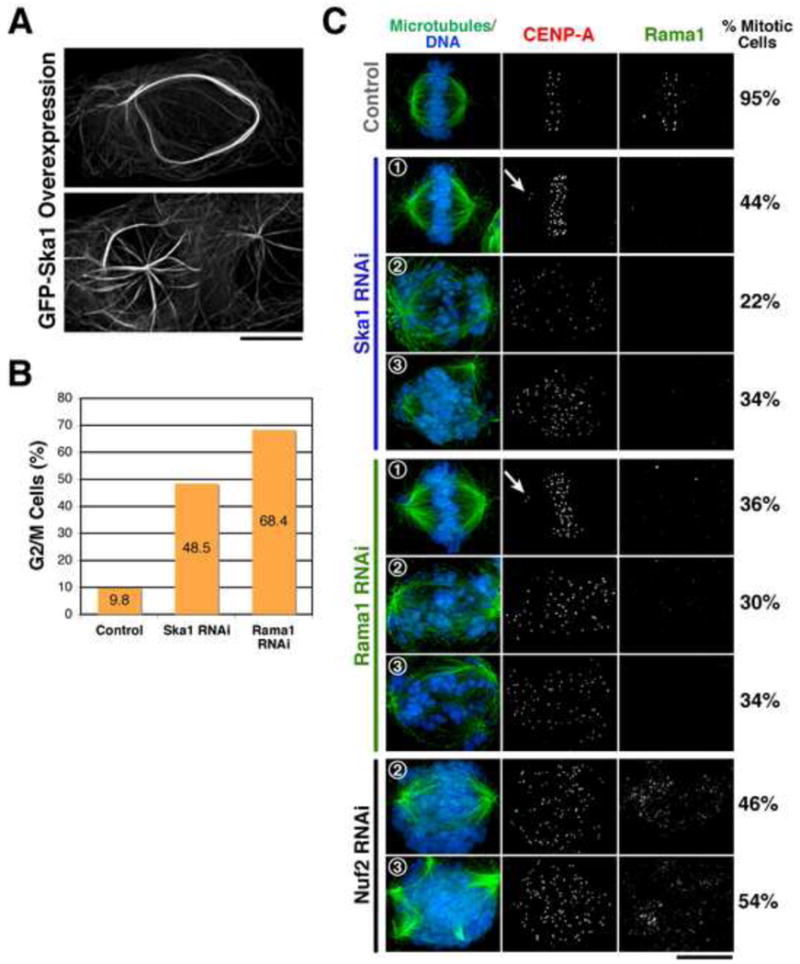
(A) Overexpression of GFP-Ska1 results in the localization to, and bundling of, interphase microtubules. HeLa cells were transiently transfected with a GFP-Ska1 plasmid and imaged 32 hrs after transfection. (B) Depletion of Ska1 or Rama1 results in a penetrant mitotic arrest. Graph shows percent of G2/M cells based on the fluorescent activated cell sorting and staining against DNA. Visual examination of these cells confirmed that they were arresting in mitosis (not shown). (C) Depletion of Ska1 and Rama1 results in severe chromosome segregation defects. Depleted cells were imaged for microtubules (using anti-tubulin antibodies), DNA, CENP-A (using anti-GFP antibodies in a cell line stably expressing GFP-CENP-A), and Rama1 (using anti-Rama1 antibodies). 1) Class 1 phenotype with the majority of chromosomes aligned at the metaphase plate. Arrows point to a single pair of off axis chromosomes in the depleted cells. 2) Class 2 phenotype with chromosomes severely mis-aligned. 3) Class 3 phenotype with mis-aligned chromosomes and multi-polar spindles. For each condition, 600 mitotic cells were counted to quantify the percent of cells with each of the indicated phenotypes. Scale bars, 10 μm.
To examine the function of the Ska1 complex during mitosis and characterize the Rama1 subunit, we depleted Ska1 and Rama1 by RNAi. In both cases, these depletions were effective based on the loss of Rama1 localization to kinetochores (see Fig. 2C). Consistent with previous reports (Hanisch et al., 2006), depletion of Ska1 resulted in a mitotic arrest based on fluorescent activated cell sorting (FACS) for DNA content (Fig. 2B) and visual examination of the cell population (not shown). Depletion of Rama1 also resulted in a penetrant mitotic arrest (Fig. 2B). Co-depletion of the mitotic checkpoint component Mad2 eliminated the arrest indicating that it was a consequence of checkpoint activation (not shown; Hanisch et al., 2006).
Immunofluorescence analysis of Ska1 or Rama1 depleted cells revealed several classes of mitotic defects. In 44% of Ska1 depleted mitotic cells and 36% of Rama1 depleted mitotic cells, the majority of chromosomes were aligned at the metaphase plate, with some off axis mis-aligned chromosomes observed (Fig. 2C, class 1 phenotype). This is the phenotype that was described previously for depletion of Ska1 and Ska2 (Hanisch et al., 2006). Although we are unable to detect kinetochore-localized Rama1 in these Ska1 or Rama1 depleted cells by immunofluorescence (Fig. 2C), this phenotype may reflect incomplete depletion of the Ska1 complex. Other depleted cells displayed much more severe phenotypes suggesting a critical role for the Ska1 complex in kinetochore-microtubule attachments. About a third of mitotic cells had extensively disorganized chromosomes, reminiscent of depletion of the Ndc80 complex (Fig. 2C, class 2 phenotype). The remaining cells displayed highly disorganized chromosomes, but additionally showed an increased number of spindle poles (Fig. 2C, class 3 phenotype). Multi-polar spindles occurred with a much higher frequency than in control depleted cells (34% vs. 4%, for Ska1 depletion and control cells, respectively). In addition, the extra spindle asters appeared to have widely varying sizes and were not evenly situated within the cell, unlike what is observed for the rare multi-polar control cells. A similar range of phenotypes were observed following both Ska1 and Rama1 depletion.
The class of phenotypes characterized by multiple spindle poles may reflect a defect in microtubule organization, or a defect in kinetochore function that indirectly causes a change in spindle structure. Given the similarity of the severely mis-aligned chromosomes to those observed following depletion of the Ndc80 complex, we also depleted the Ndc80 complex subunit Nuf2 using identical conditions (Fig. 2C). In this case, we found that 54% of cells showed multi-polar spindles with very similar morphologies to those observed following Ska1 or Rama1 depletion. The multi-polar spindles observed Nuf2 or Rama1 depletion does not appear to result from a failure in cytokinesis since the total number of chromosomes in these cells – as assessed by quantitating the number of kinetochores - does not change (not shown). In addition, only two poles of each multi-polar spindle show localization of the centriole marker centrin (Supplemental Fig. 1). In total, our functional analysis indicates critical roles for the Ska1 complex both in microtubule organization and mitotic chromosome segregation.
Reconstitution of the Ska1 complex
The dual localization of the Ska1 complex to kinetochores and microtubules, the critical role for the Ska1 complex at the outer kinetochore to mediate proper chromosome segregation, and the defects in microtubule organization observed following overexpression of Ska1, suggested the possibility that this complex associates directly with microtubule polymers. To test this, as well as analyze the organization of the Ska1 complex, we expressed its subunits either individually or in combination in bacteria. We purified GST-Ska1, GST-Ska2, and Rama1 individually from bacteria (Fig. 3A). In addition, we purified a dimer of Ska1-6xHis and untagged Ska2 by co-expressing these proteins (Fig. 3A). Resin-based binding assays demonstrated that Rama1 directly associates with GST-Ska1 and the Ska1-6xHis/Ska2 dimer, but not to GST-Ska2 alone (Fig. 3B). Finally, we were able to reconstitute the full Rama1/Ska1/Ska2 complex by combining purified Rama1 with purified Ska1/Ska2 and then conducting gel filtration chromatography (Fig. 3A). The reconstituted Ska1 complex contains approximately equimolar amounts of Ska1, Rama1, and Ska2 based on the relative intensities of these proteins in Coomassie stained gels (Fig. 3A; data not shown). We will refer to this Rama1/Ska1/Ska2 complex as the full Ska1 complex.
Figure 3. Reconstitution of the human Ska1 complex.
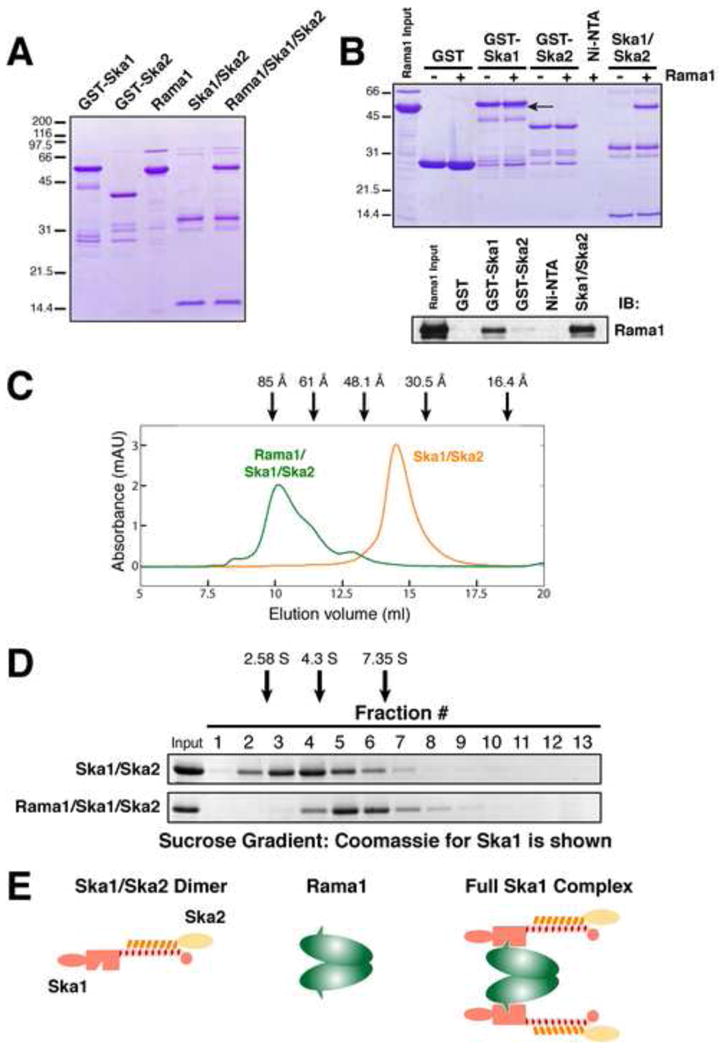
(A) Coomassie stained gel showing purified GST-Ska1, GST-Ska2, untagged Rama1 (isolated as a GST fusion and cleaved using PreScission protease), a Ska1/Ska2 dimer (isolated using a 6xHis tagged Ska1), and a 3 subunit Rama1/Ska1/Ska2 complex (isolated by mixing the Rama1 subunit with the Ska1/Ska2 dimer). (B) Rama1 binds to the Ska1 complex through the Ska1 subunit. Top, Coomassie stained gel showing a resin-based binding assay for Rama1 to either glutathione agarose beads containing GST (as a control), GST-Ska1, or GST-Ska2, or Ni-NTA agarose beads alone (as a control), or containing the Ska1/Ska2 dimer). Rama1 (arrow) runs at a similar size to GST-Ska1 on SDS-PAGE gels. Bottom, Western blot to visualize Rama1 in these binding assays. The Western blot confirms the binding to GST-Ska1, but not GST-Ska2. (C) Graph showing the elution profile of the full Rama1/Ska1/Ska2 complex or the Ska1/Ska2 dimer on a Superdex 200 size exclusion column (based on OD280 absorbance). Arrows indicate the migration of standards with known Stokes radii: Thyroglobulin (85 Å), Ferritin (61 Å), Aldolase (48 Å), and Ovalbumin (30.5 Å). (D) Coomassie stained gel showing the profile of the full Rama1/Ska1/Ska2 complex or the Ska1/Ska2 dimer in a 5-20% sucrose gradient. Arrows indicate the migration of standards with known S values: Chymotrypsinogen A (2.58 S), BSA (4.3 S) and Aldolase (6.5 S). (E) Diagram showing organization of the Ska1 complex based on the reconstitution experiments in this figure.
To determine the size and shape of the Ska1 complex, we analyzed the reconstituted proteins by gel filtration and sucrose gradients. Based on comparison to standards, analysis of the full Ska1 complex and the Ska1/Ska2 dimer by gel filtration indicated Stokes radii of 78 and 44 Å, respectively (Fig. 3C). Similarly, the sucrose gradient analysis indicated S values of 5.7 and 3.2 for the full Ska1 complex and the Ska1/Ska2 dimer, respectively (Fig. 3D). Calculations using the Stokes radii and the S values of these complexes (see Siegel and Monty, 1966), suggest native molecular weights of 184 kDa for the full Ska1 complex and 56 kDa for the Ska1/Ska2 dimer. The Ska1/Ska2 dimer has a predicted molecular weight of 43.7 kDa suggesting that this minimal complex contains a single molecule of each subunit. In contrast, the full Ska1 complex has a combined predicted molecular weight of 90 kDa suggesting that the native complex contains two molecules of each subunit. In addition to defining the molecular organization (diagrammed in Fig. 3E), the Stokes radii determined for these complexes indicate elongated shapes. For comparison, aldolase has a similar Stokes radius to the Ska1/Ska2 dimer (48 Å), but a molecular weight that is 3 times larger (161 kDa vs. 56 kDa), while thyroglobulin has a Stokes radius similar to the full Ska1 complex (85 Å), but a molecular weight that is greater than 3 times larger (650 kDa vs. 184 kDa). In total, these studies defined the organization of the Ska1 complex and provide reconstituted protein for the biochemical studies described below.
The Ska1 complex binds directly to microtubules
To test the binding of the reconstituted Ska1 complex to microtubules, we conducted microtubule co-sedimentation assays using limiting protein concentrations of the individually reconstituted Ska1 complex subunits (50 nM; Fig. 4A). None of the subunits sedimented in the absence of microtubules. However, GST-Ska1 co-sedimented with microtubules in a concentration dependent manner. GST-Ska2 also bound directly to microtubules, albeit at very low levels. In contrast, Rama1 did not bind directly to microtubules on its own.
Figure 4. The Ska1 complex binds cooperatively to microtubules in vitro.
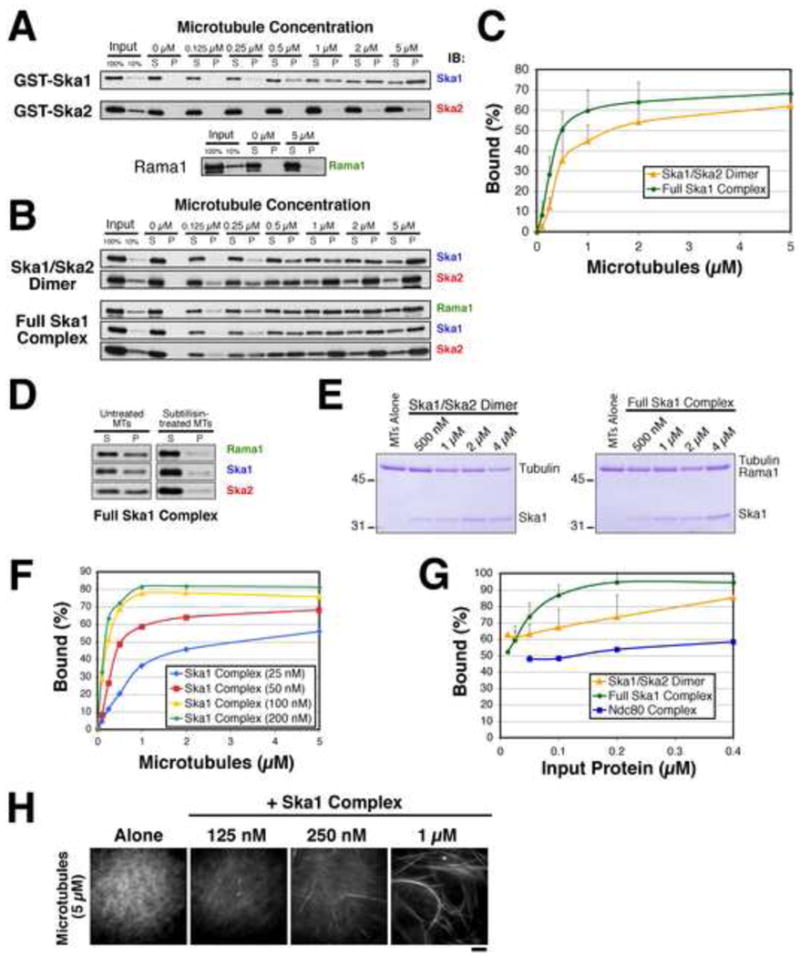
(A) Western blot using the indicated antibodies showing the co-sedimentation of 50 nM GST-Ska1, GST-Ska2, or Rama1 with microtubules at the indicated concentrations of polymerized tubulin. At least two samples were examined for each protein. (B) Co-sedimentation of the Ska1/Ska2 dimer, or the full Ska1 complex as in (A) using 50 nM input protein. (C) Graph plotting the quantification of the microtubule binding data shown in (B). The average of multiple samples were plotted for Ska1/Ska2 (n = 3) and Rama1/Ska1/Ska2 (n = 7). Error bars indicate standard deviation. (D) Binding of the Ska1 complex to microtubules requires the acidic C-terminus of tubulin. Treatment of GMPCPP microtubules with subtilisin greatly reduces the binding of the Ska1/Ska2/Rama1 complex based on Western blotting of a microtubule co-sedimentation assay relative to untreated microtubules. (E) Stoichiometry of microtubule binding. Coomassie stained gels showing the pellet fractions containing 0.5 μM microtubules and the indicated concentrations of the Ska1/Ska2 dimer or the full Ska1 complex. Rama1 runs at the same molecular weight as tubulin in SDS-PAGE gels (F) The Ska1 complex shows cooperative microtubule binding behavior. Graph showing microtubule binding as in (C), but with a range of protein input concentrations. (G) Graph showing the binding of the indicated concentrations of the Ska1/Ska2 dimer, the Ska1 complex, or the Ndc80 complex with 5 μM microtubules. Error bars indicate standard deviation. (G) The Ska1 complex bundles microtubules in vitro. Fluorescent images showing rhodamine-labeled microtubules in the presence of the indicated concentrations of the Ska1 complex. Scale bar, 10 μm.
To determine the microtubule binding affinity of the Ska1 complex, we varied the microtubule concentration and determined the concentration required for half maximal binding (Fig. 4B and 4C), which corresponds to the apparent microtubule binding affinity. The Ska1/Ska2 dimer bound to microtubules with an apparent affinity of ∼0.6 μM. Although Rama1 did not bind directly to microtubules on its own, it was able to co-sediment with microtubules when present in the full Ska1 complex. Addition of the Rama1 subunit resulted in a slight increase in the overall microtubule binding affinity to ∼0.3 μM. The microtubule binding affinity of the full Ska1 complex is comparable to that of the Dam1 complex and the KMN network, both of which bind to microtubules with an apparent affinity of ∼0.5 μM (Cheeseman et al., 2001a; Cheeseman et al., 2006).
We next examined the microtubule binding properties of the Ska1 complex in greater detail. Previous work has shown that a number of microtubule binding proteins, including the Dam1 complex (Westermann et al., 2005) and the Ndc80 complex (Wilson-Kubalek et al., 2008), associate with microtubules through the acidic C-terminus of tubulin. To determine whether the Ska1 complex shows a similar mode of microtubule binding, we treated polymerized microtubules with the protease subtilisin to remove the tubulin C-terminus. These experiments indicated a significant reduction in binding activity upon cleavage of the acidic C-terminus (Fig. 4D) indicating a similar mode of binding to known microtubule associated proteins.
Next, we determined the stoichiometry of binding of the Ska1 complex to microtubules. For these experiments, we used saturating amounts of Ska1 complex in the presence of 0.5 μM microtubules. Analysis of Coomassie stained gels of the pellet fractions of these reactions indicated a ratio of ∼1 Ska1/Ska2 dimer or full Ska1 complex bound per tubulin dimer in the microtubule (Fig. 4E). In total, these results demonstrate that the Ska1 complex binds directly to microtubules through the Ska1 subunit with properties consistent with established microtubule binding components of the kinetochore.
The Ska1 complex binds cooperatively to microtubules
The results described above indicate that the Ska1 complex directly associates with microtubule polymers. There are several ways in which such an interaction can occur. First, the Ska1 complex may function as an individual, independent unit to facilitate kinetochore-microtubule interactions. Second, the Ska1 complex may form an oligomeric assembly on microtubules. An oligomeric assembly requires interactions between neighboring Ska1 complexes on the microtubule and would likely be reflected as cooperative binding in the co-sedimentation assay. Thus, in addition to examining the microtubule binding behavior of the Ska1 complex at a fixed, limiting concentration, we also examined the concentration dependence of the microtubule binding activity. Small changes in the input concentration of the Ska1 complex resulted in dramatic changes in the apparent microtubule binding activity (Fig. 4F) reaching a maximal binding affinity of ∼0.2 μM at 100 nM protein. We also examined the microtubule binding activity of a range of concentrations of both the Ska1 complex and the Ska1/Ska2 dimer in the presence of excess (5 μM) microtubules. The Ska1/Ska2 dimer displayed a minor concentration dependent increase in microtubule binding (Fig. 4G). This change was similar to the Ndc80 complex, which does not show strong cooperativity at the concentrations shown in Fig. 4G (also see (Cheeseman et al., 2006). In contrast, the complete Ska1 complex displayed strongly cooperative microtubule binding activity (Fig. 4G). These results indicate that the microtubule binding activity of the Ska1/Ska2 dimer is not cooperative on its own, but in the presence of the Rama1 subunit, the full Ska1 complex shows a highly cooperative microtubule binding behavior.
Our previous work on the Ndc80 complex revealed a cooperative microtubule binding behavior at higher input concentrations (Cheeseman et al., 2006). In the case of the Ndc80 complex, one explanation for this cooperative behavior is the microtubule bundling that is observed at high concentrations. To determine whether this was the case for the Ska1 complex, we examined its microtubule bundling activity. We found that high concentrations of the Ska1 complex bundled microtubule polymers (Fig. 4H). However, we did not observe bundling at lower concentrations where cooperative microtubule binding was observed in the co-sedimentation assays. In addition, the Ska1/Ska2 dimer displayed microtubule bundling activity at a similar range of concentrations (not shown), but did not show cooperative microtubule binding (Fig. 4G). In total, these experiments demonstrate that the Ska1 complex binds directly to microtubules and displays a cooperative microtubule binding activity. This latter activity suggests self-association of Ska1 complex molecules on the same microtubule.
The Ska1 complex forms oligomeric assemblies on microtubules
The cooperative binding activity that we identified above suggested that the Ska1 complex may interact with itself in the presence of microtubules. To examine the presence of oligomeric Ska1 complex structures on microtubules, and to directly visualize the association of the Ska1 complex with microtubules, we next imaged the full Ska1 complex bound to microtubule polymers by negative stain electron microscopy (EM). The Ska1 complex showed a decoration of the microtubules that is consistent with the formation of oligomeric assemblies (Fig. 5). The cooperative behavior of the Ska1 complex was also observed by EM. While some microtubules were fully decorated, other microtubules in the same field remain unbound (Fig. 5, bottom image). Together with the co-sedimentation assays, these results demonstrate a microtubule-dependent oligomerization of the Ska1 complex to generate assemblies on microtubules.
Figure 5. The Ska1 complex forms ring-like assemblies on microtubules.
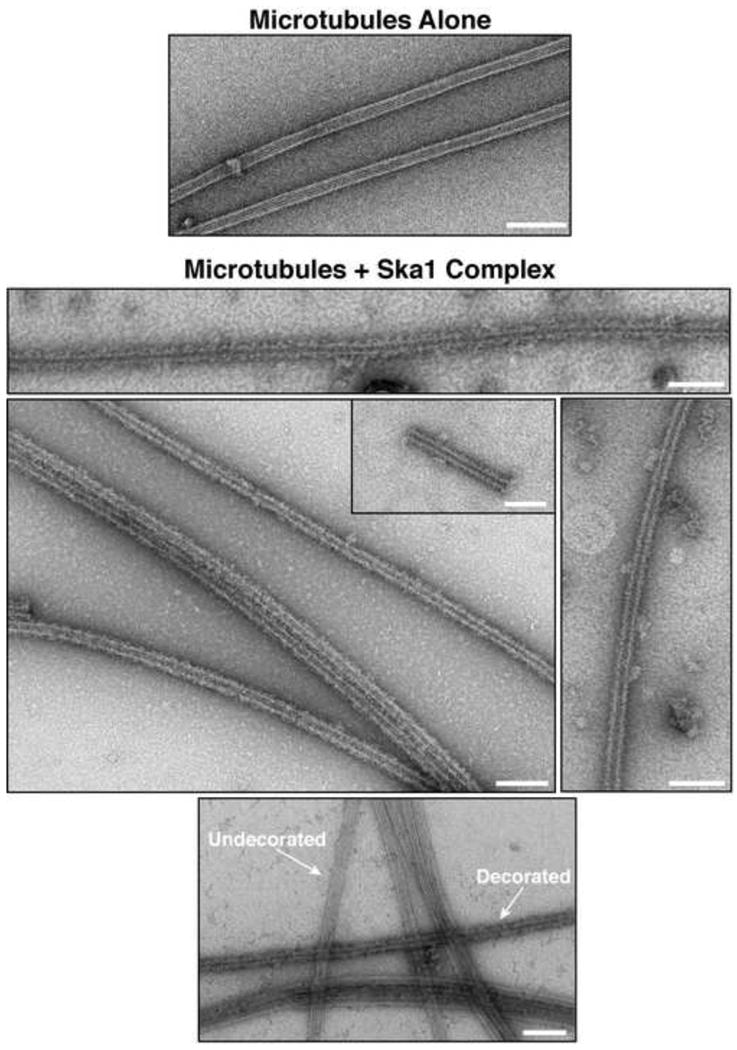
Electron microscopy of negatively stained taxol stabilized microtubules either alone or with Ska1 complex bound. In the bottom image, microtubules with Ska1 complex bound and undecorated microtubules are visible in the same field (indicated by arrows). Scale bars, 100 nm.
The Ska1 complex can couple a cargo to microtubule depolymerization
The above results strongly suggest that the Ska1 complex plays a direct role in mediating kinetochore-microtubule interactions. Given the oligomeric assemblies that this complex forms on microtubules, we next sought to determine the ability of the Ska1 complex to mediate the association of a cargo with a depolymerizing microtubule end. To test if the Ska1/Ska2 dimer or the full Ska1 complex can couple microtubule depolymerization to cargo motion, we used 0.5 μm microspheres coated with the corresponding complexes and additionally added 2.5-6 nM soluble protein (see Experimental Procedures). The beads were allowed to bind to microtubule polymers that were grown from coverslip-bound Chlamydomonas axonemes (Fig. 6A). Microtubules were induced to depolymerize (Grishchuk et al., 2005), and we recorded bead motions using DIC microscopy (Fig. 6B; Supplemental movies 1 and 2). A considerable fraction of these beads displayed depolymerization directed movement (Fig 6C). The average run length for the Ska1/Ska2 dimer and full Ska1 complex beads was 2 and 3.5 μm, respectively (Fig. 6E), indicating significant processivity. Thus, both the minimal Ska1/Ska2 dimer and the full Ska1 complex can robustly support motion of a bead cargo with a shortening microtubule end. Although the full Ska1 complex initiates fewer bead motions (Fig. 6C), the Rama1 subunit promoted an increased processivity to the movement (Fig. 6D).
Figure 6. The Ska1 complex coated microspheres display microtubule depolymerization driven motility.
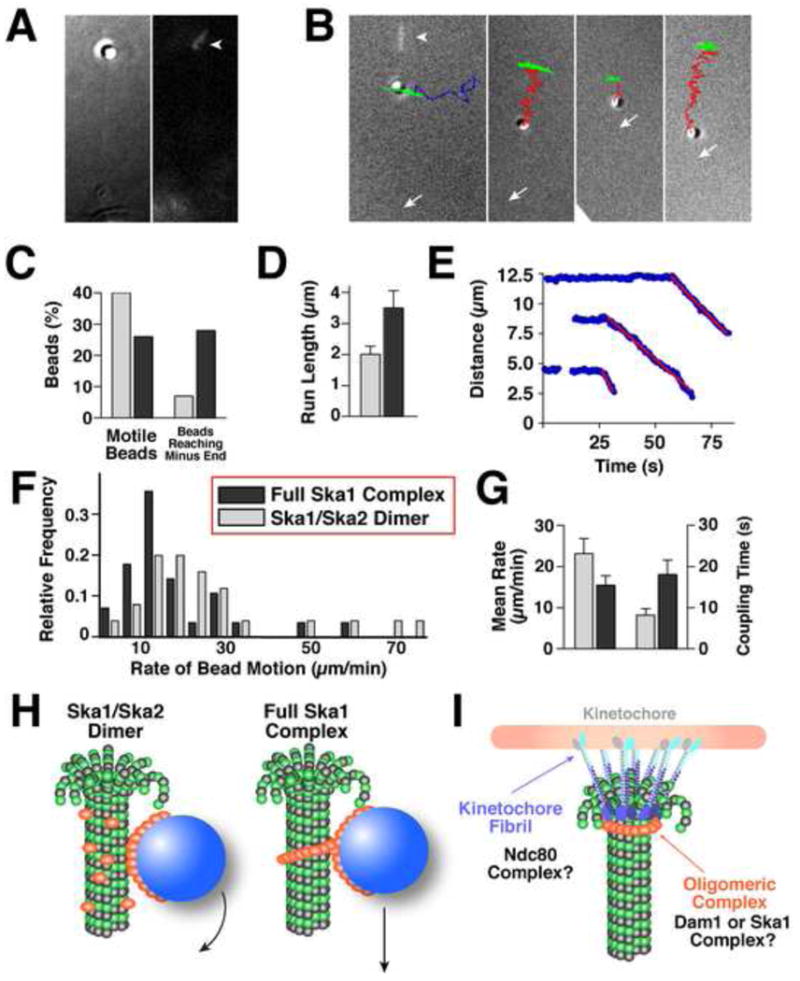
(A) DIC image (left panel) of a microtubule-bound bead with a corresponding fluorescent image (right panel) showing the stabilized microtubule cap (arrowhead). (B) Example trajectories of moving Ska1 complex-coated beads overlayed with the terminal corresponding DIC image. The trajectory of a bead that is stably attached to the wall of a static microtubule polymer (green) resembles a short arc, because the unattached microtubule plus end swings randomly as the overall microtubule length remains constant. Bead detachment from a microtubule is evidenced by a change of this swinging motion into a random “walk” (blue trajectory). Microtubule depolymerization-driven bead motions (red trajectories) are linearly directed and the amplitude of their swinging motions diminishes as the microtubule shortens. Arrows indicate positions of the coverslip-attached microtubule minus ends. First panel additionally shows an overlayed fluorescent image of the rhodamine-labeled microtubule cap before photo-ablation (arrowhead). (C) Graph showing the percent of beads that moved >0.5 μm and percent of moving beads that reached the microtubule end (full Ska1 complex, n = 112; Ska1/Ska2 dimer, n = 68). (D) Graph showing the mean distances traveled (+/- standard error of the mean - SEM) for Ska1 complex and Ska1/Ska2 dimer coated beads. (E) Graph showing the relative distance between the microsphere and microtubule minus end for the processively moving beads shown in (B). Red lines are the best linear fit for the descending segments of these curves. (F) Histogram distribution showing the motility rates of the Ska1/Ska2 dimer (n = 25) and full Ska1 complex (n = 28) coated microspheres. (G) Graph showing the average rates of bead motions and the time of bead attachment to the shortening microtubule end. Error bars are SEM. (H-I) Speculative model for Ska1 complex assembly and function. (H) The bead motions of the Ska1/Ska2 dimer are similar to Dam1 complex that does not form a ring-like structure, and may be due to the rolling of the beads. The full Ska1 complex moves at a similar rate to ring-like Dam1 complex. In this model, the Ska1 complex is shown as an oligomeric structure on microtubules. The oligomeric nature of Ska1 is based on the strong cooperativity in microtubule binding, the structures visualized on microtubules by EM, and the movement of the Ska1 along microtubules at a rate that suggests it creates an impediment to microtubule depolymerization. (I) Distinct microtubule coupling activities at the kinetochore include a fibril-like structure such as the Ndc80 complex and an oligomeric assembly such as the Dam1 or Ska1 complex.
Previous studies of Dam1 complex-mediated motility have revealed two distinct modes of motion of the Dam1 complex-coated beads (Grishchuk et al., 2008b). Dam1-coated beads can be transported by the shortening microtubule end even in the absence of soluble Dam1, which precludes formation of the microsphere-associated ring-like structures. Under these conditions, the beads move faster than the standard rate of microtubule disassembly, presumably because the bending of the shortening microtubule protofilaments is enhanced by the rolling bead motions. In contrast, under conditions where rings of the Dam1 complex are formed, the beads move significantly slower than the normal rate of microtubule depolymerization due to the bead-associated Dam1 ring impeding microtubule disassembly (Grishchuk et al., 2008a). To gain insight into the mechanism of the beads motions supported by the Ska1/Ska2 dimer and full Ska1 complex, we measured their rates by determining the slopes of the linear fits in distance vs. time plots (Fig. 6E). Based on this quantification, the rate of motion of Ska1/Ska2 dimer-coated beads varied significantly (Fig. 6F and 6G), with the mean rate similar to the average rate of free microtubule disassembly (∼25 μm/min). In contrast, the full Ska1 complex-coated beads displayed a tighter rate distribution (Fig. 6F and 6G); on average these beads moved 30% slower than the shortening rate of a free microtubule. This difference in the rate of the Ska1/Ska2 dimer in the presence and absence of the Rama1 subunit indicates that the full Ska1 complex can support bead attachment to the shortening microtubule end for a substantially longer period of time (Fig. 6G). Our data suggests that the complete Ska1 complex forms an oligomeric assembly based on the strong cooperativity in microtubule binding and the assemblies visualized on microtubules by EM. Combining this model of oligomeric Ska1 assemblies with the kinetic bead motility data suggests that the Ska1/Ska2 complex couples microspheres to depolymerizing microtubules similarly to the Dam1 complex not assembled into rings (Fig. 6H), while the full Ska1 complex functions kinetically similar to a ring-like Dam1 complex (Fig. 6H).
Discussion
Here, we have isolated a 3 subunit Ska1 complex that is an essential component of the human kinetochore and is critical for proper chromosome segregation. This complex directly associates with microtubules as oligomeric assemblies, and can couple a cargo to microtubule depolymerization. Our work has demonstrated that there are two distinct activities present within the Ska1 complex; a microtubule binding activity imparted by the Ska1 and, to a lesser extent, Ska2 subunits, and an oligomerization promoting activity provided by the Rama1 subunit and catalyzed by the microtubule lattice.
Is the Ska1 complex a metazoan functional counterpart to the fungal Dam1 complex?
Previous work has examined the Dam1 complex in budding yeast and fission yeast (Cheeseman et al., 2001b; Janke et al., 2002; Jones et al., 2001; Li et al., 2002; Liu et al., 2005; Sanchez-Perez et al., 2005; Tanaka et al., 2007). These studies have demonstrated a critical role for the Dam1 complex in meditating kinetochore-microtubule attachments and coupling chromosome movement to microtubule depolymerization. The Dam1 complex is particularly well suited to maintain such interactions in budding yeast where each kinetochore is stably attached to a single microtubule (Grishchuk et al., 2008a). However, despite the important function of the Dam1 complex in fungi, counterparts to the Dam1 complex have not been identified in multi-cellular organisms. Here, we have identified a 3 subunit Ska1 complex that is critical for chromosome segregation in human cells. Based on the analyses presented here, there are numerous functional similarities between the Ska1 and Dam1 complexes. However, it is important to note that the Ska1 complex is not a homologue of the Dam1 complex. These complexes show very different subunit composition (the Dam1 complex contains 10 subunits while the Ska1 complex contains 3 subunits) and there is no significant detectable sequence identity between subunits of the Ska1 complex and subunits of the Dam1 complex. Thus, these likely represent evolutionarily distinct complexes. Interestingly, while the Dam1 complex is present in fungi, but absent from metazoans, the Ska1 complex is conserved in diverse multi-cellular organisms including vertebrates, plants, and nematodes, but is absent from fungi.
While there is no obvious sequence similarly between the Ska1 and Dam1 complexes, the results presented here demonstrate that there are clear functional and biochemical similarities between these complexes. First, the Ska1 complex and Dam1 complex both show dual localization to kinetochores and spindle microtubules (Fig. 1; Cheeseman et al., 2001b). Second, the localization of both the Dam1 complex and Ska1 complex to kinetochores is dependent on the Ndc80 complex and on microtubules (Fig. 1; Hanisch et al., 2006; Janke et al., 2002; Li et al., 2002). Third, overexpression of a microtubule binding subunit (Ska1 or Dam1) causes defects in microtubule organization (Fig. 2; Hofmann et al., 1998). Fourth, loss of function of the complex results in a checkpoint-dependent mitotic arrest and causes severe chromosome mis-segregation (Fig. 2; Cheeseman et al., 2001b; Hanisch et al., 2006; Hofmann et al., 1998). Fifth, both complexes bind to microtubules through the acidic C-terminus of tubulin with a similar apparent microtubule binding affinity (Fig. 4; Westermann et al., 2005). Sixth, both complexes form cooperative assemblies on microtubule polymers (Fig. 4 and 5; Miranda et al., 2005; Westermann et al., 2005). And finally, both the Dam1 complex and Ska1 complex can processively move a cargo along a microtubule in a depolymerization-coupled manner (Fig. 6; Asbury et al., 2006; Grishchuk et al., 2008b; Westermann et al., 2006). The slow rate of the depolymerization-driven motion of the full Ska1 complex bound beads is highly reminiscent of the motions of beads coupled to microtubules by the Dam1 ring. Thus, despite the lack of sequence identity, the Ska1 complex has the properties that make it well suited to function as a metazoan counterpart to the Dam1 complex with a critical role at the outer kinetochore to couple chromosome movement with microtubule depolymerization.
Coupling chromosome movement to microtubule dynamics
A key player in forming kinetochore-microtubule attachments is the KNL-1/Mis12 complex/Ndc80 (KMN) complex network (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006; Wei et al., 2007). However, it was unclear whether the KMN network is sufficient to maintain interactions with dynamic microtubule polymers. Several recent studies identified additional human proteins that were suggested to function at the kinetochore-microtubule interface including Bod1 (Porter et al., 2007), Cep57 (Emanuele and Stukenberg, 2007), and Ska1 (Hanisch et al., 2006). Based on data presented here, the Ska1 complex is a key player at the outer kinetochore to mediate chromosome segregation. In contrast, our preliminary studies are not consistent with Cep57 or Bod1 playing a direct role in binding to microtubules at kinetochores (our unpublished work).
There are several possibilities for the specific functions of the Ska1 complex at kinetochores. Despite a lack of sequence similarity to the Dam1 complex, the Ska1 complex may be a true functional counterpart to the Dam1 complex. In this case, like the Dam1 complex, the Ska1 complex would form a microtubule-encircling ring in vivo that couples chromosomes to microtubule depolymerization (Fig 6I). Future work will determine the biophysical properties of the Ska1 complex including the diffusion properties of Ska1 complex assemblies and its ability to track along the ends of shortening microtubules. While the Ska1 complex displays many similar properties to the Dam1 complex, it may be optimized for the distinct features of metazoan kinetochores including different binding partners or a distinct molecular ultrastructure such as the presence of multiple kinetochore-bound microtubules. Recent work suggests that vertebrate cells can facilitate connections with depolymerizing microtubules using fibril-like connections to microtubule protofilaments (McIntosh et al., 2008). Indeed, the fibril-like Ndc80 complex can couple a microsphere to microtubule depolymerization, but with low efficiency (McIntosh et al., 2008). Ska1 complex oligomers may be adapted to play an integral part in this fibrous mechanism. While numerous fibrils may maintain connections to depolymerizing microtubules and drive chromosome movement, unlike a microtubule-associated oligomer, there is always a possibility that some protofilaments may fail to establish lasting attachments to kinetochore fibrils. Such protofilaments would continue to depolymerize and the microtubule would lose its integrity and associations with its kinetochore cargo. However, a combination of the fibril-like attachment and a oligomeric complex might mechanically stabilize this structure thereby helping to ensure the fidelity of motion (diagrammed in Fig. 6I). The ability of the Ska1 complex to cooperatively form oligomeric assemblies on the microtubule lattice is pivotal for this proposed function. Future work will examine the connections and relationships between the Ska1 and Ndc80 complexes.
Experimental Procedures
Cell culture and siRNA transfection
cDNAs for the human Ska1 complex proteins were obtained as IMAGE clones. Stable clonal cells lines expressing GFPLAP-Ska1, Ska2, and Rama1 were generated as described previously (Cheeseman et al., 2004). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin, and L-glutamine (Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. RNAi experiments were conducted as described previously (Kline et al., 2006). siRNAs against Nuf2 (DeLuca et al., 2002), and pools of 4 pre-designed siRNAs against Ska1 (GGACUUACUCGUUAUGUUA, UCAAUGGUGUUCCUUCGUA, UAUAGUGGAAGCUGACAUA, and CCGCUUAACCUAUAAUCAA), and Rama1 (GGAAGAGCCCGUAAUUGUA, GAUCGUACUUCGUUGGUUU, AAUCCAGGCUCAAUGUAA, CAUCGUAUCCCAAGUUCUA) were obtained from Dharmacon.
Immunofluorescence and Microscopy
Immunofluorescence in human cells was conducted as described previously (Kline et al., 2006). For immunofluorescence against microtubules, DM1α (Sigma) was used at 1:500. For visualization of kinetochore proteins, mouse anti-HEC1 (9G3; Abcam, Cambridge, MA) was used at 1:1000, and human anti-centromere antibodies (ACA; Antibodies, Inc., Davis, CA) were used at 1:100. HeLa cells expressing YFP-CENP-A (Kops et al., 2005) were counterstained with goat anti-GFP (a generous gift of Andres Ladurner) at 1:1000. Affinity purified rabbit polyclonal antibodies were generated against full length Ska1, Ska2, and Rama1 as described previously (Desai et al., 2003). Polyclonal antibodies against Ska1, Ska2, and Rama1 were used at 1 μg/ml. Cy2, Cy3, and Cy5-conjugated secondary antibodies (Jackson Laboratories) were used at 1:100. DNA was visualized using 10 μg/ml Hoechst.
Images were acquired on a DeltaVision Core deconvolution microscope (Applied Precision) equipped with a CoolSnap HQ2 CCD camera. 30 to 40 Z-sections were acquired at 0.2-μm steps using a 100×, 1.3 NA Olympus U-PlanApo objective with 1×1 binning. Images were deconvolved using the DeltaVision software. Equivalent exposure conditions and scaling was used between controls and RNAi-depleted cells.
Protein purification and biochemical assays
GFPLAP tagged Ska1, Ska2, and Rama1 were isolated from HeLa cells as described previously (Cheeseman and Desai, 2005). Purified proteins were identified by mass spectrometry using an LTQ XL Ion trap mass spectrometer (Thermo) using MudPIT and SEQUEST software as described previously (see Cantin et al., 2007; Washburn et al., 2001).
The Ska1 complex proteins were induced and isolated essentially as described previously (Cheeseman et al., 2006). For the expression and purification of the recombinant proteins, GST fusions with full length Ska1, Ska2, and Rama1 were generated in pGEX-6P-1. GST-Ska1 and GST-Ska2 were eluted from the glutathione agarose by the addition of 10 mM glutathione, while Rama1 was eluted from this resin using cleavage with GST-PreScission protease. For co-expression of Ska2 and Ska1-6xHis, these were sub-cloned into the polycistronic expression system (pET3aTr/pST39; Tan, 2001). The 6xHis tagged fusion was eluted from Ni-NTA agarose with 250 mM imidazole. In each case, following elution from the glutathione agarose or Ni-NTA agarose, the protein was concentrated in an Amicon Ultra column with a 30 kDa cutoff and separated on a Superdex 200 size exclusion column into 25 mM Hepes pH 7.0, 1 mM EDTA, 150 mM KCl, 1 mM DTT. For the purification of the full Ska1 complex, Ska2/Ska1-6xHis was eluted from the Ni-NTA agarose and bound to GST-Rama1 isolated on Gluthione agarose resin. The resin was washed, and the full complex was eluted with GST-PreScission. The complex was then concentrated and fractionated on the Superdex 200 column. Microtubule binding and bundling assays using the purified proteins were conducted as described previously (Cheeseman et al., 2006). To determine the binding of the Ska1 complex in the absence of the acidic C-terminus of tubulin, GMPCPP stabilized microtubules were treated with 250 μg/ml type XXIV subtilisin (Sigma) for 60 min at 30°C.
Electron microscopy
Microtubules decorated with the full Ska1 complex and the Ska1/Ska2 dimer were stained and visualized using negative-stain electron microscopy (EM) on continuous carbon that had been glow discharged. 4 μl of 1 μM microtubules were applied to the grid for 1 min, and briefly washed across using BRB80 buffer. 4 μl of the Ska1 complex at ∼0.2 mg/ml was then applied to the microtubule bound grid for 1 min. The sample was washed with BRB80 to remove unbound protein and then washed and stained twice with 10 μl 2% uranyl acetate. The stained sample was left to sit on the grid for 1 min, then blotted to dry. Images were acquired on a Technai Spirit electron microscope at 120 kV using a CCD camera.
In vitro motility of protein-coated beads
The assays were carried out as described in (Grishchuk et al., 2005) and the beads were coated with proteins as in (Grishchuk et al., 2008b). Briefly, streptavidin-coated polystyrene beads (Bangs Laboratories, Inc.) were functionalized with saturating amounts of biotinylated anti-6xHis antibodies (Abcam) and then Ska1/Ska2 dimer or full Ska1 complex proteins containing a 6xHis-tagged Ska1 subunit. Efficient conjugation was confirmed with anti-Ska1 antibodies followed by fluorescent secondary antibodies. For motility assays, protein-coated beads were washed to remove the unbound protein and then added to experimental chamber in BRB80 supplemented with 1 mM DTT, 0.5 mg/ml casein, 1 mg/ml BSA and 2.5-6 nM of the corresponding soluble Ska1 complex proteins. Images were acquired every 0.3-0.4 sec with low light DIC and Texas Red filter, as described (Grishchuk et al., 2008b).
Supplementary Material
Acknowledgments
We are grateful to Arshad Desai for his generous support during the early stages of this project, Mary Porter for a kind gift of Chlamydomonas axonemes, Paula Grissom for technical assistance, and Angelika Amon, Arshad Desai, Andreas Hochwagen, and Terry Orr-Weaver for critical reading of the manuscript. This work was supported by Smith Family Foundation Award for Excellence in Biomedical Research to I.M.C. I.M.C. is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin GT, Shock TR, Park SK, Madhani HD, Yates JR., 3rd Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal Chem. 2007;79:4666–4673. doi: 10.1021/ac0618730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001a;155:1137–1146. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular Architecture of the Kinetochore-Microtubule Interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Müller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. Journal of Cell Biology. 2001b;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore Microtubule Dynamics and Attachment Stability Are Regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Stukenberg PT. Xenopus Cep57 Is a Novel Kinetochore Component Involved in Microtubule Attachment. Cell. 2007;130:893–905. doi: 10.1016/j.cell.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci U S A. 2008a doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A. 2008b;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. Embo J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. Journal of Cell Biology. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, He X, Giddings TH, Winey M. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc Natl Acad Sci U S A. 2001;98:13675–13680. doi: 10.1073/pnas.241417098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR, 3rd, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR, 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. Embo J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils Connect Microtubule Tips with Kinetochores: A Mechanism to Couple Tubulin Dynamics to Chromosome Motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Porter IM, McClelland SE, Khoudoli GA, Hunter CJ, Andersen JS, McAinsh AD, Blow JJ, Swedlow JR. Bod1, a novel kinetochore protein required for chromosome biorientation. J Cell Biol. 2007;179:187–197. doi: 10.1083/jcb.200704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochimica et Biophysica Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006 doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


