Abstract
Mutations in each of the transcriptional co-activator genes - CBP, p300, Cited2, Cart1 and Carm1 - result in neural tube defects in mice. The present study thus furnishes a complete and comparative temporal and spatial expression map of CBP/p300 and associated transcriptional co-activators, Cited2, Cart1 and Carm1 during the period of murine neural tube development (embryonic days 8.5 to 10.5). Each co-activator except Cart1 was expressed in the dorsal neural folds on E8.5. Although CBP and p300 are functionally interchangeable in vitro, their respective expression patterns diverge during embryogenesis before neural fold fusion is complete. CBP gene expression was lost from the neural folds by E8.75 and was thereafter weakly expressed in the maxillary region and limb buds, while p300 exhibited strong expression in the first branchial arch, limb bud and telencephalic regions on E9.5. Cart1 exhibited strong expression in the forebrain mesenchyme from E9.0 through E10.5. Although CBP, p300, Carm1 and Cited2 share temporal expression on E8.5, these co-activators have different spatial expression in mesenchyme and/or the neuroepithelium. Nevertheless, co-localization to the dorsal neural folds on E8.5 suggests a functional role in elevation and/or fusion of the neural folds. Target genes, and pathways that promote cranial neural tube fusion that are activated by CBP/p300/Carm1/Cited2/Cart1-containing transcriptional complexes await elucidation.
Keywords: craniofacial, neural tube, mouse, embryogenesis, CBP, p300, Cited2, p300, Cart1, Carm1
Models of neural tube defects (NTDs) include mice with mutations in genes encoding the transcriptional activators or co-activators CREB binding protein (CBP), p300, Cited2 (alternative designation Mrg1), Carm1, and Cart1 (Bamforth et al., 2001, Barbera et al., 2002, Petrij et al., 1995, Tanaka et al., 2000, Weninger et al., 2005, Yadav et al., 2003, Yao et al., 1998, Zhao et al., 1996). The 250 kDa CBP and the closely related 264 kDa p300 are functionally redundant histone deacetylases (McManus and Hendzel, 2001, Yao et al., 1998) that are essential for proliferation and embryonic development (Kitabayashi et al., 2001, Li et al., 2002). Homozygous deletion in transgenic mice of either the CBP or p300 genes results in lethality on gestational day 9–11, with defects in neurulation, cell proliferation and cardiac development (Tanaka et al., 2000; Yao et al., 1998). Mutations in CBP in humans result in Rubenstein-Taybi syndrome (Petrij et al., 1995) characterized, in part, by a defined craniofacial phenotype. CBP has a high affinity binding site for another nuclear co-activator, Cited2 (cAMP-responsive element-binding protein [CBP]/p300-interacting transactivator with glutamic acid [E] and aspartic acid [D]-rich tail). The interaction of CBP and p300 with Cited2 is necessary for activation of transcription factors such as AP-2α (Bamforth et al., 2001, Braganca et al., 2003). Owing to this close functional linkage, deletion of the Cited2 gene also results in heart and neural tube defects (Bamforth et al., 2001, Barbera et al., 2002, Volcik et al., 2004, Weninger et al., 2005). Cart1 (cartilage homeoprotein 1) is a 37 kDa paired class homeobox-containing transcription factor that is selectively expressed in chondrocytes (Zhao et al., 1994, Zhao et al., 1993). The importance of expression in the developing craniofacial complex is highlighted by postnatal lethality of Cart1-deficient fetuses that suffer from acrania and meroanencephaly and die soon after birth (Zhao et al., 1996). Methylation of CBP/p300 by co-activator-associated arginine methyltransferase 1 (Carm1) inhibits the recruitment of CREB (cAMP response element binding protein). Methylation of CBP/p300 blocks its interaction with CREB resulting in repression of cAMP signaling, therefore Carm1 is probably a co-repressor of cAMP signaling (Xu et al., 2001). Despite the functional association of Carm1 with CBP, Carm1-deficient mice have not been reported to suffer from neural tube defects. However, the homozygous mutation of Carm1 causes perinatal lethality and a much smaller body size than heterozygous or wild type siblings (Yadav et al., 2003). Proteins examined in this study are nuclear transcriptional co-activators and the neural tube defects caused by deficiency of their cognate genes would predict expression in neural folds associated with neural tube fusion. Limited spatial expression data has been published (Dunwoodie et al., 1998, Partanen et al., 1999, Zhao et al., 1996), but no parallel comparison of these genes during the critical period of neural tube development has been conducted. The present study thus furnishes a complete and comparative temporal and spatial expression map of CBP/p300 and associated transcriptional co-activators, Cited2, Cart1 and Carm1 during embryonic days (E) 8.5–10.5.
Digoxigenin (DIG)-labeled RNA probes were transcribed from CBP, Cart1, Cited2, and Carm1 templates for in situ hybridization experiments. Despite numerous attempts using differents sequences from the p300 cDNA sequence (NM_177821.5) as templates for DIG-riboprobes, specific in situ hybridization signals corresponding to p300 expression could not be obtained. After attempts at amplifying the p300 3′-untranslated sequence for use as a template were also unsuccessful, an anti-p300 antibody (Partanen et al., 1999) was utilized for whole embryo immunohistochemistry in order to, at least, demonstrate protein distribution. In order to ensure reproducibility and specificity, in situ hybridizations and immunostaining was replicated no less than three times with each probe on each gestational day. Stage-matched negative controls were performed for each gene/protein examined using sense transcript riboprobes or pre-blocked antibody, respectively.
Distribution of CBP and p300 during murine embryogenesis (E8.5–10.5)
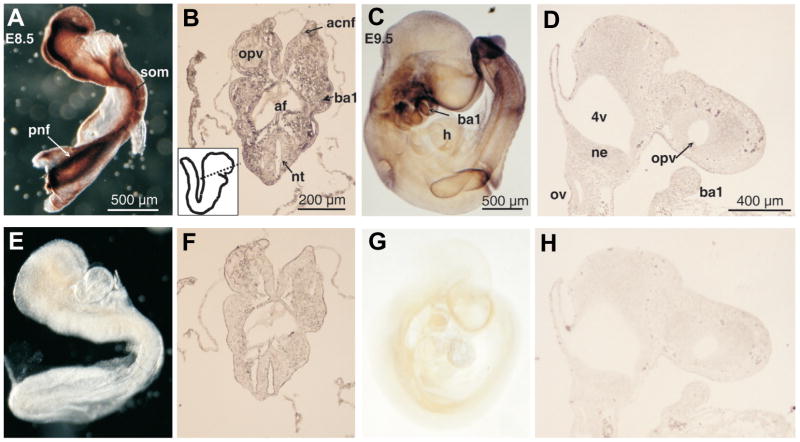
CBP transcripts were distributed in the dorsal neural folds along the entire rostral-caudal axis of the embryo on E8.5 (Fig. 1A). Examination of transverse sections (Fig. 1B) revealed that the expression was strongest in the neuroepithelium itself at both the anterior and posterior neural folds, and extended into the dorsal mesenchyme at both the presumptive first branchial arch and in the posterior region of the embryo. On E8.75, CBP transcripts were expressed in the frontonasal region around the telencephalon, and the first and second branchial arches (Fig. 1C). The presumptive cardiac region was noticeably devoid of any CBP expression (Fig. 1C). Sagittal sections of the E9.0 cranial region found that low levels of CBP expression was limited to the neuroepithelium of the midbrain and forebrain (Fig. 1D). By E9.5, low levels of CBP expression were restricted to the frontonasal region, the first and second branchial arches and the forming forelimb bud (Fig. 2A). Sagittal sections exhibited CBP expression limited to the neuroepithelium and the branchial arch mesenchyme (Fig. 2B) consistent with that observed on E8.5. On E10.5, CBP expression persisted in the first and second branchial arches, the frontonasal region around the optic and telencephalic vesicles, and in the limb buds (Fig. 2C). CBP expression continued to be notably absent in the heart on E10.5 (Fig. 2C). Analysis of tissue sections of E10.5 embryos revealed that CBP was strongly expressed in neuroepithelium in the cranial region and the branchial arches (Fig. 2D). Significant expression was not detected in sense probe-hybridized embryos (Figs. 1E–H and 2E–H). The p300 protein was expressed at high levels in the neural folds (Fig. 3A) along the entire rostral to caudal axis on E8.5 as well as in somites (Fig. 3A). Transverse sections, however, showed that in contrast to CBP, the p300 expression was pronounced in the mesenchyme as well as in the neuroepithelium on E8.5, especially in the cranial neural tube (Fig. 3B). Subsequent to E8.5, p300 expression markedly decreased and was not detected in sections from E8.75–9.0 embryos (not shown). By E9.5, p300 expression was restricted to the first and second branchial arches, the limb bud and caudal somites (Fig. 3C). Unlike sections from E8.5, sections taken fro E9.5 embryos revealed weak expression in the neuroepithelium, surrounding mesenchyme and the first branchial arch (Fig. 3D). Specific expression was not detected on E10.5 in either whole embryos or in sagittal sections. Non-specific staining was not detected in negative controls (Fig. 3E–H). Partanen et al. (1999) have shown that on E8.5, both CBP and p300 transcripts were broadly distributed throughout the developing neural tube. Specifically, in the caudal region of the embryos, cells at all mediolateral positions of the newly formed neural plate expressed CBP and p300 transcripts. Interestingly, expression was restricted to the dorsal part of the neural fold/tube in the head and trunk regions. However, while expression in developing cardiac tissue was observed (Partanen et al., 1999), we did not detect any cardiac expression at any stage studied.
Fig. 1. Distribution of CREB binding protein (CBP) transcripts in E8.5 and E9.0 mouse embryos and tissue sections as determined by in situ hybridization.
Mouse embryos (A,C,E,G) or tissue sections (B,D,F,H) were hybridized with CBP RNA antisense (A–D) or sense (E–H) probes. Panels showing E8.5 whole embryos are adjacent to stage-matched transverse sections (A–B, E–F), while E9.0 embryos are adjacent to stage-matched sagittal tissue sections of the of the anterior cranial region (C–D, G–H). The boxed inset in (B) shows a schematic representing the plane of section indicated by the dotted line. The boxed region of the E9.0 whole embryo (C) is shown in a sagittal tissue section (D). anf, anterior cranial neural folds: ba1/ba2, first/second branchial arch; cnf, cranial neural folds; fb, forebrain; h, heart; mv, mesencephalic vesicle; ne, neuroepithelium; nt, neural tube; opv, optic vesicle; plv, presumptive left ventricle; pnf, posterior neural folds.
Fig. 2. Distribution of CREB binding protein (CBP) transcripts in E9.5 and E10.5 mouse embryos and tissue sections as determined by in situ hybridization.
Mouse embryos (A,C,E,G) or tissue sections (B,D,F,H) were hybridized with CBP RNA antisense (A–D) or sense (E–H) probes. Panels showing E9.5 (A–B, E–F) and E10.5 (C–D, G–H) whole embryos are adjacent to panels showing the anterior regions of stage-matched sagittal sections. 4v, fourth ventricle; ba1/ba2, first/second branchial arch; flb, forelimb bud; h, heart; mv, mesencephalic vesicle; ne, neuroepithelium; opv, optic vesicle; tv, telencephalic vesicle.
Fig. 3. Distribution of p300 protein expression in E8.5 and E9.5 mouse embryos as determined by whole-mount immunohistochemistry.
E8.5 and E9.5 whole embryos (A,C) or sections (B,D) were incubated with anti-p300 polyclonal antibodies. Specificity of the antibody was demonstrated by preincubating the antibody with synthetic peptide before exposure to the embryos for one hour at 4°C at a 10 times molar excess (E–H). Note the exceptionally strong p300 signal emanating from the neural folds on E8.5. E8.5 embryos were not cleared (A,E), while E9.5 embryos were cleared (C,G) in order to optimally demonstrate p300 staining. Note that clearing causes the appearance of double images, such as the branchial arches on E9.5 (C) due to transparency of embryonic tissue. The boxed inset in (B) shows a schematic representing the plane of section as indicated by the dotted line. 4v, fourth ventricle; acnf, anterior cranial neural folds; af, anterior foregut; ba1, first branchial arch; h, heart; ne, neuroepithelium; nt, neural tube; opv, optic vesicle; ov, otic vesicle; pnf, posterior neural folds; som, somite.
Distribution of Cited2 during murine embryogenesis (E8.5–10.5)
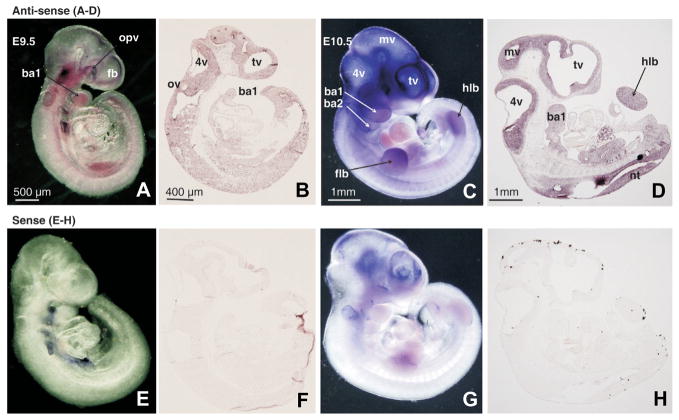
Cited2 lacks a DNA-binding domain, thus its role as a transcriptional regulator is mediated through its interactions with other cofactors such as CBP/p300. We investigated the expression of Cited2 within the neural tube during early murine embryogenesis. Unlike CBP or p300 (Figs. 1–3), expression of Cited2 on E8.5 was restricted to the cranial dorsal neural folds, the presumptive septum transversum, weak expression in the neuroepithelium and the mesenchyme (data not shown). By E9.0, Cited2 expression was detected in the mesenchyme around the telencephalon and in the first branchial arch (data not shown). Comparisons of hybridizations of numerous comparable sections revealed that Cited2 expression, detected in the somites, was reproducible although not evident before or after E9.5. On E9.5 Cited2 expression, was localized to the telencephalic, otic and optic vesicles, the first branchial arch, and limb buds (Fig. 4A). Sectional analysis revealed that the neuroepithelium was the primary site of Cited2 expression in the cranial region (Fig. 4B). By E10.5, Cited2 expression was pronounced in the cranial neural tube, the telencephalon, first and second branchial arches and the limb buds (Fig. 4C). In situ hybridization of Cited2 in sagittal sections revealed strongest expression in the neuroepithelium around the fore-, mid- and hindbrain, the first branchial arch, as well as the dorsal trunk neural tube (Fig. 4D). Sense probe-hybridized embryos were generally signal free (Fig. 5E–H). While a relatively high background was consistent in the sense-hybridized E10.5 whole embryo (Fig. 4G), staining generated by the anti-sense-hybridized probe was consistently and significantly higher, and such background staining was absent in the hybridized sections (Fig. 4H). Distribution of Cited2 expression was consistent with previous reports showing limited expression in a 7-somite embryo (Dunwoodie et al., 1998), and also consistent with earlier reports (Barbera et al., 2002, Weninger et al., 2005) that Cited2 expression is elevated in regions where the neural folds fuse (Dunwoodie et al., 1998). Cited2-deficient mice present with exencephaly and increased apoptosis in the forebrain-midbrain junction on E9.0 and E9.5 (Bamforth et al., 2001, Barbera et al., 2002), further supporting the vital nature of the Cited2 protein for proper neural tube closure.
Fig. 4. Distribution of Cited2 transcripts in E9.5 and E10.5 mouse embryos and tissue sections as determined by in situ hybridization.
E9.5 and E10.5 mouse embryos and sagittal sections were hybridized with the Cited2 RNA antisense probe (A–D) or with the corresponding sense probe as a negative control (E–H). 4v, fourth ventricle; ba1/2, first/second branchial arch; fb, forebrain; flb, forelimb bud; hlb, hindlimb bud; mv, mesencephalic vesicle; nt, neural tube; opv, optic vesicle; ov, otic vesicle; tv, telencephalic vesicle.
Fig. 5. Distribution of Cart1 transcripts in E9.5 mouse embryos and tissue sections as determined by in situ hybridization.
Mouse embryos or sections from E9.5 were hybridized with the Cart1 RNA antisense probe (A,B) or with the sense probe as a negative control (C,D). The expression detected in the orofacial region of the E9.5 embryo boxed in A was confirmed in the equivalent region of a sagittal section (B). ba1, first branchial arch; fb, forebrain; ne, neuroepithelium; opv, optic vesicle; tv, telencephalic vesicle.
Distribution of Cart1 during murine embryogenesis (E8.5–10.5)
Cart1 did not exhibit reproducible expression during E8.5 that could be detected in transverse sections (data not shown). On E9.0 and E9.5, however, Cart1 expression was tightly localized to the frontonasal mesenchyme around the optic and telencephalic vesicles (Fig. 5A). Sectional in situ hybridization demonstrated Cart1 expression in the cranial neuroepithelium and the frontonasal mesenchyme (Fig. 5B). On E10.5, Cart1 expression was tightly restricted to the frontonasal region and the anterior region of the first branchial arch (Fig. 6A). Expression was also visible in the limb buds (Fig. 6A). In situ hybridization of serial sagittal sections revealed that Cart1 was restricted to the lateral region of the medial nasal prominence and the medial region of the first branchial arch (Fig. 6B–D). Negative controls that were hybridized with sense Cart1 probes exhibited no specific staining (Fig. 5C,D and \6E–H). Previous spatial studies have shown that at the Cart1 gene transcript was restricted to mesenchymal cells of the E8.5 forebrain (Zhao et al., 1996). However, on E9.5, this expression pattern was expanded to mesenchymal cells in the frontonasal region as well as those in the mesenchyme surrounding the optic vesicles. No expression was visible in the midbrain, hindbrain or neuroepithelium of the neural tube. The results from this study are precisely consistent with our observations.
Fig. 6. Distribution of Cart1 transcripts in E10.5 mouse embryos and tissue sections as determined by in situ hybridization.
E10.5 mouse embryos (A,E) or sections (B–D, F–H) were hybridized with the Cart1 RNA antisense probe (A–D) or with the sense probe as a negative control (EH). Panels (B–D, F–H) show adjacent sagittal sections of the cranial region from medial to lateral positions in the same embryo. 4v, fourth ventricle; ba1, first branchial arch; mnp, medial nasal process; mv, mesencephalic vesicle; opv, optic vesicle; tv, telencephalic vesicle.
Distribution of Carm1 during murine embryogenesis (E8.5–10.5)
Carm1 exhibited expression on both E8.5 (Fig. 7A) and E8.75 (not shown) that was most evident in the anterior and posterior regions of the embryo. Transverse sections of E8.5 embryos revealed strong expression, ubiquitously distributed throughout the neuroepithelium, surrounding mesenchyme, presumptive cardiac region, and first branchial arch (Fig. 7B). Expression did not extend to the ventral mesenchyme of the posterior region of the E8.5 embryo (Fig. 7B). In situ hybridization of whole E9.5 embryos, revealed Carm1 expression in the maxillary region, first and second branchial arches, the frontonasal region around the optic vesicle and telencephalon, the otic vesicle and developing limb buds (Fig. 7C). In situ analysis of sections from E9.5 embryos confirmed cranial expression in the neuroepithelium around the otic vesicle, hindbrain and forebrain, and in the mesenchyme of the frontonasal region and branchial arch (Fig. 7D). By E10.5, strong Carm1 expression was detected in the first and second branchial arches, frontonasal region and fore and hindlimb buds (Fig. 8A). Expression in the mesencephalic neural tube, especially evident in the ventral midbrain, was also detected (Fig. 8A). In situ analysis of sections from E10.5 embryos confirmed strong expression in both the neuroepithelium and adjacent cranial mesenchyme, as well as strong expression in the first branchial arch. Negative controls that were hybridized with sense Carm1 probes exhibited little or no staining (Figs. 7E–H and 8C,D). Previously published results (Chen et al., 2002) have shown that on E8.25, Carm1 expression was observed in the fore- and hindbrain, neural folds, somites, and posterior lateral plate of the developing mouse embryo. This report also observed that on E8.75, Carm1 expression became prominent in the neural tube and somites (Chen et al., 2002).
Fig. 7. Distribution of Carm1 transcripts in early mouse embryos at E8.5 and 9.5 as determined by in situ hybridization.
Mouse embryos (A,C) or tissue sections (B,D) at E8.5- and E9.5 were hybridized with the Carm1 RNA antisense probe (A–D) or with the sense probe (E–H). The boxed inset in (B) shows a schematic representing the plane of section as indicated by the dotted line. (D) A cranial sagittal section of the E9.5 embryo. 4v, fourth ventricle; acnf, anterior cranial neural folds; ba1, first branchial arch; flb, forelimb bud; fn, frontonasal mesenchyme; nt, neural tube; ov, otic vesicle; opv, optic vesicle; plv, presumptive left ventricle; pnt, posterior neural tube.
Fig. 8. Distribution of Carm1 transcripts in E10.5 mouse embryos and tissue sections as determined by in situ hybridization.
Mouse embryos (A,C) or sagittal sections (B,D) at E10.5 were hybridized with the Carm1 RNA antisense probe (A,B) or with the sense probe (C,D). 4v, fourth ventricle; ba1, first branchial arch; fb, forebrain; flb, forelimb bud; hlb, hindlimb bud; tv, telencephalic vesicle.
For genes that are regulated by CBP- or p300-dependent transcription factors, rates of transcription will depend, in part, on cellular levels and distribution of CBP/p300. In the present study, we show that the expression patterns of CBP and p300 are highly divergent, both spatially and temporally. While CBP maintains expression in the neuroepithelium from E8.5–E10.5, p300 exhibits greater expression in the adjacent mesenchyme at E8.5 and, thereafter, expression decreases significantly and is undetectable by E10.5. While CBP and p300 are functionally redundant, lethality of the CBP/p300 double heterozygous mutation has lead to the conclusion that CBP/p300 gene dosage is important for normal embryogenesis (Yao et al., 1998). This conclusion is supported by differences in the spatial expression of CBP and p300 in numerous developing tissues. Differential spatial expression of CBP and p300, combined with relatively low levels of p300 expression during later stages of embryonic development, is likely to result in these two co-activators functioning in a rate limiting capacity during transcriptional activation (Chakravarti et al., 1996, Hanstein et al., 1996, Montminy, 1997). The actual target genes, or pathways that promote cranial neural tube fusion that are activated by CBP/p300/Carm1/Cited2-containing transcriptional complexes await elucidation. Transcriptional activation of signaling pathways that contribute to ontogeny of the neural tube, or development of other embryonic structures, may require combinations of CBP, p300, Carm1 and Cited2 in transcriptional complexes during organogenesis.
Materials & Methods
Cloning of cDNA sequences for riboprobes
Human full-length CITED2 was subcloned from pcDNA3-CITED2 into pBluescript KS2+ (Stratagene, Cedar Creek, TX). First-strand cDNA was synthesized from E11.0 mouse embryo total RNA using Superscript II reverse transcriptase (both RNA and enzyme from Invitrogen, Carlsbad, CA) using gene-specific reverse primers for Cart1 and Carm1. Subsequent PCR amplifications were carried out using Platinum Pfx enzyme (Invitrogen, Carlsbad, CA) with specific forward and reverse primers for Cart1 and Carm1. The sequences of the forward and reverse primers are as follows: Cart1, 5′ CGC GAA TTC GTA TGG AGT TTC TGA GCG AGA AG 3′ (forward) and 5′ GCG TCT AGA GGT ACC CAT GGC CCA TGA AAT ATT GGC 3′ (reverse); Carm1 5′ CGC TCT AGA GGC TCC ATA ATG ACC GTG TG 3′ (forward) and 5′ CGC GAA TTC CCC ATA GTG CAT GGT GTT GG 3′ (reverse). Both primer pairs included EcoRI and XbaI sites to allow directional cloning of Cart1 and Carm1 DNA fragments after the expected PCR products were excised from agarose gels, purified and ligated into vectors. The mouse Cart1 probe, corresponding to nucleotides 173–1150 of GenBank Accession number NM172553, was subcloned into pSPT18 (Roche Diagnostics, Indianapolis, IN). Carm1, corresponding to nucleotides 1216–1845 of GenBank Accession number NM021531, was also cloned into pSPT18. For CBP, a 1000 bp Pst1 fragment corresponding to the 3′ end of the CBP cDNA sequence from pRcRSV-CBP was subcloned into the Pst1 site of pSPT19 (Roche Diagnostics, Indianapolis, IN). After cloning, the identities of cloned template sequences were confirmed by sequencing. Antisense and sense anti-digoxigenin-labeled probes were generated using a DIG RNA labeling kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions.
Whole embryo in situ hybridization
ICR mice (Harlan, Indianapolis, IN) were maintained at a temperature of 22°C with an alternating 12-hour light/dark cycle and were provided access to food and water ad libitum. Mature male and female mice were mated overnight and the presence of a vaginal plug the following morning was taken as evidence of mating (E0.5). Embryos were collected on E8.5, E8.75, E9.0, E9.5, and E10.5. Five embryos (selected from a pool generated from no less than three dams) were processed for each time point, and for each coactivator. For each of these five embryos, in situ hybridization or immunohistochemistry was performed in triplicate. Whole embryo in situ hybridizations were performed as previously described (Nagy et al., 2003, Wilkinson and Nieto, 1993) with the following modifications: embryos from E8.5, E8.75 and E9.0 were processed in 15 mm diameter netwells (74 μm netwells, Electron Microscope Services, Hatfield, PA) to minimize damage. Bound probes were detected by an alkaline phosphatase conjugated anti-digoxigenin antibody at a dilution of 1:2000 (Anti-Digoxigenin-AP Fab Fragments, Roche Diagnostics) in 1% sheep serum (overnight, 4°C). Bound signals were visualized with 2% (vol/vol) NBT/BCIP in NTMT substrate buffer (Roche Diagnostics) for 2–3 hours. Cleared embryos were photographed using a Nikon SMZ1500 stereomicroscope (Fryer Company, Cincinnati, OH).
Sectional in situ hybridization
Embryos were washed in phosphate buffered saline (PBS) after fixation, then cryoprotected in 30% sucrose in PBS for at least 30 min, then stored in 15% sucrose in OCT (optimal cutting temperature compound, Tissue-Tek 4583, Sakura Finetek USA, Inc., Torrance, CA) overnight before embedding in OCT. Seven to ten μm transverse sections of the embryos were cut and mounted on uncoated, uncharged, RNase-free slides using a Leica CM1900 cryostat (Leica Inc., Bannockburn, IL). Sections were dried overnight before use or stored at minus 80°C. Sections were fixed in 4% (wt/vol) paraformaldehyde in PBS plus 0.1% Tween 20 (PBT) (15 min) and washed twice in PBT (5 min each) before permeabilization in 1 μg/ml proteinase K in PBT (15 min). Proteinase treatment was stopped by washing twice in 2 mg/ml glycine in PBT (5 min each) followed by washing twice in PBT (5 min each). Sections were fixed again in 0.2% glutaraldehyde, 4% paraformaldehyde (PFA) in PBT (20 min) followed by washing three times in PBT (5 min each). Sections were then hybridized to sense and anti-sense riboprobes and probe detection was performed under the same conditions as described for whole embryos. While all other hybridizations were performed at a probe concentration of 1 μg/ml and a hybridization temperature of 65°C, Cited2 hybridizations were performed at a probe concentration of 600 ng/ml and a hybridization temperature of 70°C. Bound probes were detected after a three day exposure to alkaline phosphatase conjugated anti-digoxigenin antibody. Sections were then photographed under differential interference contrast optics using a Nikon Eclipse E600 microscope equipped with a Nikon DXM1200 digital camera and Nikon Act-1 imaging software.
Whole embryo and sectional immunohistochemistry
Expression of p300 was detected using a rabbit polyclonal anti-p300 antibody (anti-p300-N15, sc-584, Santa Cruz Biotechnology, Santa Cruz, CA) directed to the N-terminal 15 amino acids of the p300 protein, using a previously published protocol (Davis et al., 1991). This antibody has been shown to be specific for the p300 protein by immunoblotting (Partanen et al., 1999). Embryos were fixed for 3 days in 4% PFA in PBS at 4°C. For whole mount immunohistochemistry, embryos were washed in PBS, dehydrated in methanol (Nagy et al., 2003) and bleached with 6% hydrogen peroxide in methanol for 5 hours at room temperature before storage in methanol at −20°C until further processing. Embryos were rehydrated through 5 min washes in 75%, 50% and 25% (vol/vol) methanol in PBS at room temperature, washed twice in PBS for 5 min, and then blocked in two 30 min washes with PBSMT (0.5% Triton X-100, 2% non-fat milk in PBS) at room temperature. For control samples, the anti-p300 antibody was pre-bound for 90 min to a 10X excess of synthetic peptide (the antigenic peptide originally used to raise the anti-p300-N15 antibody, sc-584 P, Santa Cruz) corresponding to the N-terminal amino acids of p300 in PBSMT. Embryos were incubated with blocked or unblocked antibody at 1 μg/ml in PBSMT overnight at 4°C followed by two five-minute washes in PBSMT and then five washes in PBSMT of one hour each at 4°C. Embryos were then incubated with a secondary antibody (goat anti-rabbit HRP conjugate, sc-2004, Santa Cruz) at 0.4 μg/ml overnight in PBSMT followed by two five-minute washes and five one-hour washes in PBSMT at 4°C. The embryos were finally washed in PBSBT (0.1% BSA Sigma cat. No. 7030, 0.5% Triton X-100 in PBS) for 10 min at room temperature. Expression of p300 was detected using a DAB peroxidase kit (Vector Laboratories, Burlingame, CA). The manufacturer’s protocol was followed except that only a single drop of hydrogen peroxide was added to the substrate mixture. The embryos were incubated in the substrate for 2–3 min (E8.5, and E8.75) or 10 min (E9.5 and E10.5). The staining reaction was stopped by two 10 min washes in PBSBT at room temperature. Stained embryos were rinsed in PBS and fixed in 4% PFA in PBS for 2–3 days at 4°C. When necessary, embryos were cleared in 1:2 benzyl alcohol: benzyl benzoate.
Sections were prepared as for sectional in situ hybridization. Sections were washed in PBS (3 times, 5 min), fixed in 3.7% formaldehyde in PBS (20 min) and washed in PBS again. Tissue sections were permeabilized in 0.1% Triton X-100 in PBS (2 times, 5 min). Sections were blocked in 5% inactivated sheep serum/0.1% BSA (Sigma cat. no. 7030)/0.1% Triton X-100 in PBS for 30 min at 37°C. Either pre-blocked or unblocked anti-p300 antibody was added in 10% serum/PBS at a 1 μg/ml dilution and incubated overnight at 4°C. The antibody was washed off in PBS (3 times, 5 min) and secondary antibody was added at 0.4 μg/ml in PBS for 60 min at 37°C. After washing in PBS (2 times, 5 min), sections were washed in PBSBT for 10 min and DAB detection was performed as for whole embryos.
Acknowledgments
Plasmids pcDNA3-CITED2 and pRcRSV-CBP were kindly provided by Dr. S. Bhattacharya (Department of Cardiovascular Medicine, University of Oxford, Wellcome Trust Center for Human Genetics, Oxford, UK) and Dr. S.R. Grossman (Dana-Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts 02115, USA) respectively. This work was supported in part by PHS grants HD053509 and DE18215 to RMG, a grant from the Cleft Palate Foundation to VB, the COBRE Program of the National Center for Research Resources (P20RR017702), and the Commonwealth of Kentucky Research Challenge Trust Fund.
Abbreviations used in this paper
- CBP
CREB binding protein
- CREB
cAMP-responsive element binding protein
- NTD
neural tube defect
References
- BAMFORTH SD, BRAGANCA J, ELORANTA JJ, MURDOCH JN, MARQUES FI, KRANC KR, FARZA H, HENDERSON DJ, HURST HC, BHATTACHARYA S. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- BARBERA JP, RODRIGUEZ TA, GREENE ND, WENINGER WJ, SIMEONE A, COPP AJ, BEDDINGTON RS, DUNWOODIE S. Folic acid prevents exencephaly in Cited2 deficient mice. Hum Mol Genet. 2002;11:283–293. doi: 10.1093/hmg/11.3.283. [DOI] [PubMed] [Google Scholar]
- BRAGANCA J, ELORANTA JJ, BAMFORTH SD, IBBITT JC, HURST HC, BHATTACHARYA S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278:16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- CHAKRAVARTI D, LAMORTE VJ, NELSON MC, NAKAJIMA T, SCHULMAN IG, JUGUILON H, MONTMINY M, EVANS RM. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- CHEN SL, LOFFLER KA, CHEN D, STALLCUP MR, MUSCAT GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- DAVIS CA, HOLMYARD DP, MILLEN KJ, JOYNER AL. Examining pattern formation in mouse, chicken and frog embryos with an En-specific antiserum. Development. 1991;111:287–298. doi: 10.1242/dev.111.2.287. [DOI] [PubMed] [Google Scholar]
- DUNWOODIE SL, RODRIGUEZ TA, BEDDINGTON RS. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- HANSTEIN B, ECKNER R, DIRENZO J, HALACHMI S, LIU H, SEARCY B, KUROKAWA R, BROWN M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KITABAYASHI I, AIKAWA Y, NGUYEN LA, YOKOYAMA A, OHKI M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001;20:7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Q, XIAO H, ISOBE K. Histone acetyltransferase activities of cAMP-regulated enhancer-binding protein and p300 in tissues of fetal, young, and old mice. J Gerontol A Biol Sci Med Sci. 2002;57:B93–B98. doi: 10.1093/gerona/57.3.b93. [DOI] [PubMed] [Google Scholar]
- MCMANUS KJ, HENDZEL MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol. 2001;79:253–66. [PubMed] [Google Scholar]
- MONTMINY M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- NAGY A, GERTENSTEIN M, VINTERSTEN K, BEHRINGER R. Manipulating the Mouse Embryo - A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2003. [Google Scholar]
- PARTANEN A, MOTOYAMA J, HUI CC. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int J Dev Biol. 1999;43:487–494. [PubMed] [Google Scholar]
- PETRIJ F, GILES RH, DAUWERSE HG, SARIS JJ, HENNEKAM RC, MASUNO M, TOMMERUP N, VAN OMMEN GJ, GOODMAN RH, PETERS DJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- TANAKA Y, NARUSE I, HONGO T, XU M, NAKAHATA T, MAEKAWA T, ISHII S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- VOLCIK KA, ZHU H, FINNELL RH, SHAW GM, CANFIELD M, LAMMER EJ. Evaluation of the Cited2 gene and risk for spina bifida and congenital heart defects. Am J Med Genet A. 2004;126:324–325. doi: 10.1002/ajmg.a.20578. [DOI] [PubMed] [Google Scholar]
- WENINGER WJ, FLORO KL, BENNETT MB, WITHINGTON SL, PREIS JI, BARBERA JP, MOHUN TJ, DUNWOODIE SL. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- WILKINSON DG, NIETO MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- XU W, CHEN H, DU K, ASAHARA H, TINI M, EMERSON BM, MONTMINY M, EVANS RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- YADAV N, LEE J, KIM J, SHEN J, HU MC, ALDAZ CM, BEDFORD MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO TP, OH SP, FUCHS M, ZHOU ND, CH’NG LE, NEWSOME D, BRONSON RT, LI E, LIVINGSTON DM, ECKNER R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- ZHAO GQ, EBERSPAECHER H, SELDIN MF, DE CROMBRUGGHE B. The gene for the homeodomain-containing protein Cart-1 is expressed in cells that have a chondrogenic potential during embryonic development. Mech Dev. 1994;48:245–254. doi: 10.1016/0925-4773(94)90063-9. [DOI] [PubMed] [Google Scholar]
- ZHAO GQ, ZHOU X, EBERSPAECHER H, SOLURSH M, DE CROMBRUGGHE B. Cartilage homeoprotein 1, a homeoprotein selectively expressed in chondrocytes. Proc Natl Acad Sci USA. 1993;90:8633–8637. doi: 10.1073/pnas.90.18.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Q, BEHRINGER RR, DE CROMBRUGGHE B. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat Genet. 1996;13:275–283. doi: 10.1038/ng0796-275. [DOI] [PubMed] [Google Scholar]