Abstract
Diagnosis of a case of septicemic plague acquired in rural California was delayed because of a series of confounding events, resulting in concern about reliance on community hospitals as sentinels for detecting potential bioterrorism-related events. An epizootic study confirmed the peri-domestic source of Yersinia pestis infection.
INTRODUCTION
When 22 letters of anthrax were maliciously transmitted through the postal service after the September 11, 2001 terrorist attacks on the World Trade Center, the US federal government dedicated substantial resources toward the prevention and amelioration of future bioterrorism attacks. Critical to bioterrorism preparedness is the prompt detection of index cases of such diseases and reporting them to public health authorities, wherever they might occur.1 This process depends on the maintenance of a minimum standard of microbiological expertise at the community hospital level.2 This report describes a case of community-acquired septicemic plague in a known endemic region in which diagnosis was delayed because of a series of confounding events, reflecting a possible systemic problem in microbiological capacity at the community hospital level.
CASE REPORT
A 79-year-old woman, who lived alone in a rural, mountainous region of eastern California, was admitted to a local community hospital with altered mental status after being found by a neighbor. Plague was well known to be endemic in this region. She had been suffering from severe low back pain in recent months because of degenerative disc disease, which kept her confined to her home and unable to care for her rural, mountainous property located in a known sylvatic plague-endemic area. The patient’s house was noted to be neglected, with evidence of deer mouse (Peromyscus maniculatus) and wood rat (Neotoma spp.) droppings throughout.
On arrival at the local emergency department, her temperature was 38.2°C, her pulse was 110 beats/min, her blood pressure was 121/60 mm of Hg, and her oxygen saturation was 80% on room air. She was arousable but oriented only to person. Auscultation of the lungs showed crackles at the left base. The abdomen was soft with normal bowel sounds. There was no lymphadenopathy. Neurologic exam showed no focal deficits. She had leukocytosis (46,800 cells/μL, 19% bands), renal dysfunction (serum creatinine, 1.9 mg/dL), and elevated transaminases (AST, 336 U/L; ALT, 59 U/L). A chest radio-graph was interpreted as showing left lower lung atelectasis.
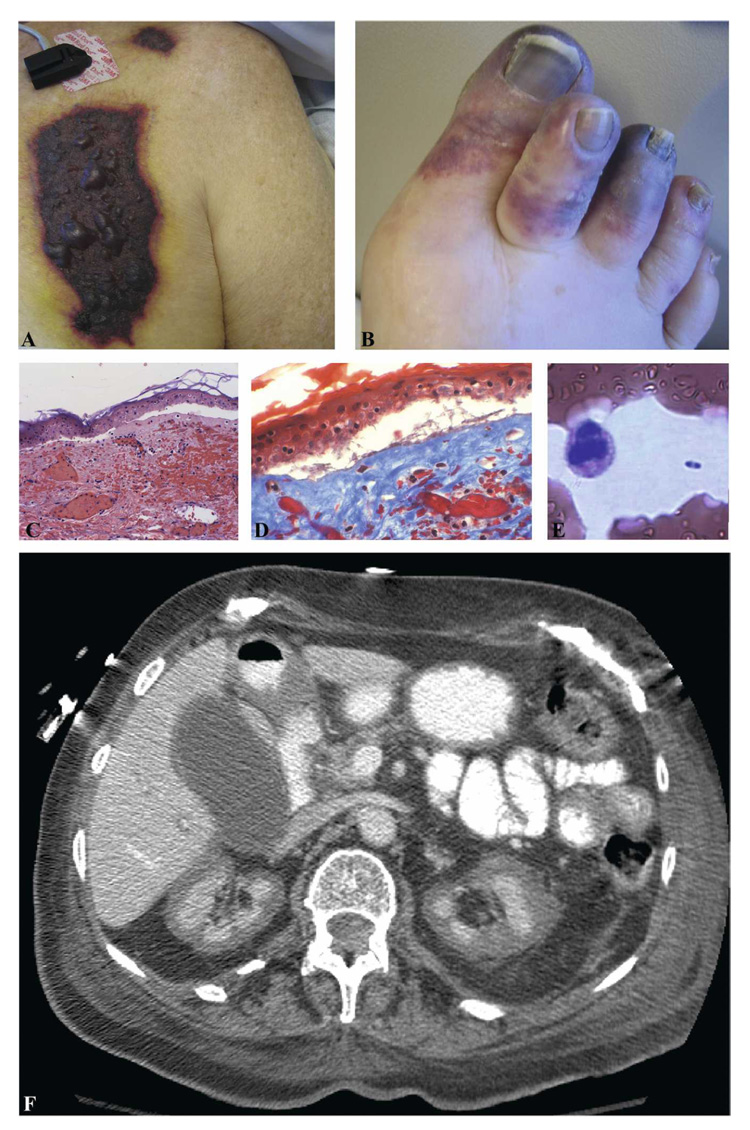
The patient received a diagnosis of community-acquired pneumonia (CAP); a dose of ceftriaxone followed by scheduled ampicillin/sulbactam and levofloxacin were given. She initially defervesced and had improved mentation. By 48 hours after admission, renal failure had worsened (serum creatinine, 4.2 mg/dL). Acidosis and thrombocytopenia (platelets, 54,000 cells/µL) developed. She was transferred to our academic center on hospital Day 3 for further management, where she soon needed intubation and hemodynamic support. Results from the other hospital were reported to include one blood culture bottle growing a gram-positive bacillus interpreted as a contaminant. Lactate dehydrogenase level was elevated (3,296 IU/mL). A moderate number of schistocytes were seen on peripheral smear, the prothrombin time was 1.2 times control, and the partial thromboplastin time was 1.3 times control. Blood cultures on admission to our hospital were negative. With the combination of clinical and laboratory findings, a tentative diagnosis of thrombotic thrombocytopenic purpura was entertained, for which plasmapheresis and plasma exchange were initiated. Several plaques of necrotic skin with surrounding erythema were noted on her chest and upper extremities (Figure 1A). Several toes were gangrenous (Figure 1B). Histopathologic examination showed dermal necrosis, acute inflammation, and small vessel thrombosis but no visible organisms (Figure 1C, D). Computed tomography of the abdomen showed bilateral renal cortical necrosis (Figure 1F), consistent with a severe, generalized Schwartzman reaction.3 The antibiotic regimen was changed to meropenem and gentamicin when the admission urine culture from the other hospital was reported to be growing > 105 CFU/mL Pseudomonas aeruginosa.
FIGURE 1.
A, Several plaques of necrotic skin with surrounding erythema were noted on the patient’s chest and upper extremities (only chest lesions). B, A punch biopsy of the chest lesion was performed. A subepidermal cleft was present over the dermis with marked erythrocyte extravasation and dermal purpura; the dermis was necrotic (×200). C, A trichrome stain confirmed the presence of fibrin thrombi in the superficial vessels (×400). Special stains for bacteria organisms were negative. D, Several toes on the patient’s right foot were purplish and gangrenous. E, Computed tomography of the abdomen showed bilateral renal cortical necrosis. F, The patient’s initial peripheral blood smear contained approximately one bipolar-staining bacterium every three to five ×40 fields.
The microbiological diagnosis in this case was not quickly defined. The Gram stain of the outside blood culture was initially reported as showing “gram-positive rods” on hospital Day 4. After inquiry by physicians at the hospital to which the patient was referred, this interpretation was changed to “gram-negative rod” on hospital Day 6 by a laboratory technologist who had returned from leave. Automated biochemical analysis (Vitek) of this isolate was consistent with Yersinia pestis, but another biochemical analysis (API) on the same day was interpreted as more likely poorly viable E. coli or Proteus spp. A subculture of the original isolate was received and subsequently analyzed by our clinical microbiology laboratory starting on hospital Day 7. Slow-growing, small colonies were noted to be cocco-bacillary, bipolar staining, nonmotile, non-lactose fermenting gram-negative rods. Review of the patient’s initial peripheral blood smear showed one bipolar-staining bacterium every three to five 40× fields (Figure 1E). On hospital Day 9, a presumptive identification of Y. pestis was suggested by standard and API biochemical testing and verified by additional biochemical testing. The San Diego County Public Health Laboratory (a Laboratory Response Network Reference Laboratory [LRN]) was immediately notified and identified the isolate as Y. pestis using direct fluorescence antibody testing and polymerase chain reaction testing of the isolate according to federally classified methods of the LRN. Isolates of Y. pestis and cases of plague must be reported within 1 hour to the California Department of Public Health.
The evening that the identification was made, a nurse thought that she saw live fleas in the patient’s bed, resulting in moving the patient to another room and sealing off and fumigating the patient’s room. Ultimately these “fleas” were identified as fruit flies.
The patient received 10 days of effective Y. pestis coverage, had an extended hospital stay, remained anuric, elected not to undergo renal replacement therapy, and died from uremia.
EPIZOOTIC STUDY
Several rodent genera involved either in the enzootic maintenance or amplification of plague epizootics, most notably Spermophilus, Neotoma, Ammospermophilus tamias, and Peromyscus, had been known to be present in the predominantly pinyon-juniper and sage-scrub habitat. California Department of Health Service signs had been previously posted in recreation sites throughout the county warning people to avoid rodents because of “plague.”
Eight days after the confirmation of septicemic plague, the California Department of Public Health Vector-Borne Disease Section carried out a 2-day field study at the case patient’s residence and at two adjacent residences. Live traps for rodents set inside and under the home as well as on the land surrounding all three residences yielded 35 rodents made up of nine species from six genera. Abandoned/inactive ground squirrel burrows along the driveway approaching the patient’s residence provided visible evidence of a plague epizootic. Additionally, the hind leg from a mostly decomposed cottontail rabbit (Sylvilagus spp.) found adjacent to the house and a freshly dead California ground squirrel (S. beecheyi) carcass found on the road 0.6 km north of the home were collected for testing. Blood from the patient’s two dogs, a neighbor’s dog, and from rodents was collected on Nobuto blood-sampling strips and submitted (along with flea pools from the rodents) for serologic testing.
Although serologic results showed no rodents testing positive from the adjacent properties, anecdotal evidence suggested a plague epizootic was heterogeneously distributed throughout the community. From the patient’s property, two of three S. beecheyi tested positive for antibodies to Y. pestis with high titers (1:2,048) suggestive of recent infection. One of the case patient’s two dogs tested positive for antibodies to Y. pestis with a titer of 1:64. None of the other species tested positive for antibodies to plague. However, both the S. beecheyi carcass and rabbit hind leg tested positive by polymerase chain reaction for Y. pestis. In addition, five flea pools from both serologically positive and negative rodents were positive by polymerase chain reaction for Y. pestis (Table 1). Interestingly (although not unusual), the flea pool from a serologically positive California ground squirrel was negative by polymerase chain reaction, whereas the serologically negative California ground squirrel had a plague-positive flea pool.
TABLE 1.
Summary of epizootic study
| Species | No. tested for anti-Y. pestis antibodies | No. positive for anti-Y. pestis antibodies | IFA titer | Comment |
|---|---|---|---|---|
| Spermophilus beecheyi | 3 | 2 | 1: 2048 | One flea pool from a serologically positive animal was negative. One flea pool from a serologically negative animal was positive. |
| Canis familiaris | 3 | 1 | 1: 64 | Patient’s dog |
| Ammospermophilus leucurus | 5 | 0 | NA | Two of two flea pools collected from four animals were positive. |
NA = not applicable.
DISCUSSION
This case is noteworthy for the classic clinical presentation of septicemic plague manifesting as the Black Death.4 In a community hospital located in a region where plague was known to be endemic, the capability of this hospital to make a simple microbiological diagnosis was inadequate.5 The patient presented with necrotic lesions and sepsis. There was little clinical suspicion for the diagnosis of plague, and the patient was diagnosed with CAP, despite the lack of typical findings. The clinical laboratory, a small multidisciplinary operation, did not have adequate staffing to interpret a routine Gram stain on a blood culture. On transfer to a tertiary care hospital, appropriate treatment was administered based on initially inadequate information, and the confirmation of septicemic plague was delayed because of difficulties transporting bacterial isolates between institutions.
Yersinia pestis, the etiologic agent of plague, belongs to the Enterobacteriaceae family, with the potential for microbiological confusion leading to diagnostic delay as seen in this case. Growth on media is slow, often requiring 48 hours on enriched media, although experienced microbiologists can distinguish suggestive colony morphology earlier.2 Although Y. pestis is readily suspected by standard automated biochemical strip testing, suspected isolates must be reported to and confirmed by a regional health authority.2
This patient’s exposure is consistent with the epidemiology and ecology of plague. In vivo Y. pestis proliferates rapidly, with the potential of fulminating into the septicemic form of plague associated with high-grade bacteremia, which allows for visualization of bacteria on routine Wright-Giemsa–stained peripheral blood smear, as seen in this case. The high-grade bacteremia was responsible for the bilateral renal cortical necrosis caused by a generalized Schwartzmann reaction.3 Septicemic plague can occur without any obvious painful lymph-adenopathy (bubo). Hematogenous dissemination to the lungs leads to pneumonic plague, which has the potential to be transmitted person-to-person through aerosolization.7
Plague has been classified as a bioterrorism disease because of its potential for aerosolization to disperse to large populations with the potential for subsequent person-to-person spread.6 Although the incidence of plague is highest in countries of the developing world, such as in Tanzania and Madagascar, the western United States is well known to be endemic for Y. pestis. Large-scale panic in response to mere possibility of plague, as illustrated in the globally reported but unconfirmed pseudo-epidemic of plague in India in the 1990s,8–10 illustrates the potential of such diseases for causing serious social disruption. Despite the potential for human-to-human spread of plague from patients with pneumonic plague, the last documented case of human-to-human plague transmission in the United States was in 1924,7,11 although this mode of transmission still occurs in endemic regions in less-developed countries.12
Among the general population, there is substantial fear of plague and diseases caused by other class A bioterrorism agents (anthrax, tularemia, botulinum toxin, smallpox, viral hemorrhagic fevers).13 Any suggestion of a potential bioterrorism event is likely to lead to substantial social disruption. For this reason, the federal government has devoted substantial resources for bioterrorism preparedness. A central feature of comprehensive preparedness for a bioterrorist event is early detection and diagnosis, including the clinical recognition of specific diseases and syndromes of such diseases at community-based sentinels, in particular hospitals. Although a physician’s ability to recognize these diseases has been shown to be limited,5 having an adequately functioning clinical microbiology laboratory with appropriately trained microbiology technologists is critical to any sentinel system.14,15 As a number of recent authors have pointed out, the current era of health care expenditure cost cutting and clinical microbiology laboratory outsourcing places the maintenance of basic and rapidly responsive clinical microbiology diagnostic expertise at risk.16,17
Here we present a case of primary septicemic plague, complicated by pulmonary involvement, in a 79-year-old woman who acquired the disease in a known plague-endemic region in California. A number of issues confused the clinical picture, including the initial misidentification of a positive blood culture by a laboratory technologist working in a primarily automated all-in-one community hospital laboratory. This case highlights the potential for poor general community capacity for detecting infectious diseases of bioterrorism potential, even in a known endemic region.
Acknowledgments
The authors thank Peter Fedullo and members of the UCSD Medical Center pulmonary and critical care team, Ravindra Mehta and Mita Shah, of the Division of Nephrology, William Read, Thomas Kipps, Dan Tong, and the UCSD Medical Center Department of Medicine housestaff, clinical fellows, and hospital staff for excellent contributions to the care of this patient; Chris Peter and the San Diego County Public Health Services for prompt diagnostic testing; Charles Smith, Curtis Fritz, Jim Tucker, and Tina Feiszli for field and technical support; the California Department of Public Health Microbial Diseases Laboratory for diagnostic animal and arthropod testing, and Bruno Chomel of the UC Davis School of Veterinary Medicine for Nobuto testing.
Contributor Information
David A. Margolis, Divisions of Infectious Diseases and Dermatology, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California
Joseph Burns, California Department of Public Health, Vector-Borne Disease Section, Ontario, California.
Sharon L. Reed, Divisions of Infectious Diseases and Dermatology, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California
Michele M. Ginsberg, San Diego Health Department, San Diego, California
Terrence C. O’Grady, Divisions of Infectious Diseases and Dermatology, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California
Joseph M. Vinetz, Divisions of Infectious Diseases and Dermatology, Department of Medicine, University of California, San Diego School of Medicine, La Jolla, California
REFERENCES
- 1.Khan AS, Ashford DA. Ready or not—preparedness for bioterrorism. N Engl J Med. 2001;345:287–289. doi: 10.1056/NEJM200107263450411. [DOI] [PubMed] [Google Scholar]
- 2.Robinson-Dunn B. The microbiology laboratory’s role in response to bioterrorism. Arch Pathol Lab Med. 2002;126:291–294. doi: 10.5858/2002-126-0291-TMLSRI. [DOI] [PubMed] [Google Scholar]
- 3.Finegold MJ. Pathogenesis of plague. A review of plague deaths in the United States during the last decade. Am J Med. 1968;45:549–554. doi: 10.1016/0002-9343(68)90171-x. [DOI] [PubMed] [Google Scholar]
- 4.Koplan J. CDC’s strategic plan for bioterrorism preparedness and response. Public Health Rep. 2001;116 Suppl 2:9–16. doi: 10.1016/S0033-3549(04)50132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosgrove SE, Perl TM, Song X, Sisson SD. Ability of physicians to diagnose and manage illness due to category A bio-terrorism agents. Arch Intern Med. 2005;165:2002–2006. doi: 10.1001/archinte.165.17.2002. [DOI] [PubMed] [Google Scholar]
- 6.Relman DA. Bioterrorism—preparing to fight the next war. N Engl J Med. 2006;354:113–115. doi: 10.1056/NEJMp058078. [DOI] [PubMed] [Google Scholar]
- 7.Kool JL. Risk of person-to-person transmission of pneumonic plague. Clin Infect Dis. 2005;40:1166–1172. doi: 10.1086/428617. [DOI] [PubMed] [Google Scholar]
- 8.John TJ. India: is it plague? Lancet. 1994;344:1359–1360. [PubMed] [Google Scholar]
- 9.Bharadwaj R, Kagal A, Deshpandey SK, Joshi SA, Khare PM, Junnarkar AR, Phadke MA. Outbreak of plague-like illness caused by Pseudomonas pseudomallei in Maharashtra, India. Lancet. 1994;344:1574. doi: 10.1016/s0140-6736(94)90382-4. [DOI] [PubMed] [Google Scholar]
- 10.Twigg G. Plague in India. Lancet. 1995;345:258. [PubMed] [Google Scholar]
- 11.Viseltear AJ. The pneumonic plague epidemic of 1924 in Los Angeles. Yale J Biol Med. 1974;47:40–54. [PMC free article] [PubMed] [Google Scholar]
- 12.Mwengee W, Butler T, Mgema S, Mhina G, Almasi Y, Bradley C, Formanik JB, Rochester CG. Treatment of plague with gentamicin or doxycycline in a randomized clinical trial in Tanzania. Clin Infect Dis. 2006;42:614–621. doi: 10.1086/500137. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson NE, Steele L, Crawford CY, Huebner NL, Fonseka JC, Bonander JC, Kuehnert MJ. Bioterrorism web site resources for infectious disease clinicians and epidemiologists. Clin Infect Dis. 2003;36:1458–1473. doi: 10.1086/374560. [DOI] [PubMed] [Google Scholar]
- 14.Peterson LR, Hamilton JD, Baron EJ, Tompkins LS, Miller JM, Wilfert CM, Tenover FC, Thomson RB., Jr Role of clinical microbiology laboratories in the management and control of infectious diseases and the delivery of health care. Clin Infect Dis. 2001;32:605–611. doi: 10.1086/318725. Jr. [DOI] [PubMed] [Google Scholar]
- 15.Pien BC, Saah JR, Miller SE, Woods CW. Use of sentinel laboratories by clinicians to evaluate potential bioterrorism and emerging infections. Clin Infect Dis. 2006;42:1311–1324. doi: 10.1086/503260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch R, Hryniewicz W, Van Eldere J. Report of working group 2: healthcare needs in the organisation and management of infection. Clin Microbiol Infect. 2005;11 Suppl 1:41–45. doi: 10.1111/j.1469-0691.2005.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Eldere J. Changing needs, opportunities and constraints for the 21st century microbiology laboratory. Clin Microbiol Infect. 2005;11 Suppl 1:15–18. doi: 10.1111/j.1469-0691.2005.01084.x. [DOI] [PubMed] [Google Scholar]