Abstract
Several closely related Mn(II)-oxidizing alpha-Proteobacteria were isolated from very different marine environments: strain SI85-9A1 from the oxic/anoxic interface of a stratified Canadian fjord, strain HTCC 2156 from the surface waters off the Oregon coast, and strain AE01 from the dorsal surface of a hydrothermal vent tubeworm. 16S rRNA analysis reveals that these isolates are part of a tight phylogenetic cluster with previously characterized members of the genus Aurantimonas. Other organisms within this clade have been isolated from disparate environments such as surface waters of the Arctic and Mediterranean seas, a deep-sea hydrothermal plume, and a Caribbean coral. Further analysis of all these strains revealed that many of them are capable of oxidizing dissolved Mn(II) and producing particulate Mn(III/IV) oxides. Strains SI85-9A1 and HTCC 2156 were characterized further. Despite sharing nearly identical 16S rRNA gene sequences with the previously described Aurantimonas coralicida, whole genome DNA-DNA hybridization indicated that their overall genomic similarity is low. Polyphasic phenotype characterization further supported distinguishing characteristics among these bacteria. Thus SI85-9A1 and HTCC 2156 are described as two new species within the family ‘Aurantimionadaceae’: Aurantimonas manganoxydans sp. nov. and Aurantimonas litoralis sp. nov. This clade of bacteria is widely distributed around the globe and may be important contributors to Mn cycling in many environments. Our results highlight the difficulty in utilizing 16S rRNA-based approaches to investigate the microbial ecology of Mn(II) oxidation.
Keywords: Manganese, Aurantimonas, Rhizobiales, alpha-Proteobacteria
Introduction
Manganese plays key roles in several biogeochemical cycles of global significance. As the catalytic center of photosystem II and a prominent player in cellular mechanisms of oxidative stress, Mn is also an essential micronutrient. Manganese has three environmentally relevant oxidation states, II, III and IV. Mn(III) is thermodynamically unstable unless complexed to ligands such as desferrioxamine, pyoverdine and pyrophosphate (Faulkner and others 1994; Kostka and others 1995; Parker and others 2004; Trouwborst and others 2006) or as a component of insoluble Mn(III/IV) oxides (Tebo and others 2004). At pH 7, Mn-oxides can catalyze the formation of humic substances and organic N complexes. They can also oxidatively degrade humic and fulvic acids forming biologically usable low molecular weight organic compounds (Sunda and Kieber 1994). Mn oxides can promote the oxidation of metals such as Fe and have strong affinity and sorptive capacities for many trace elements (Tebo and others 2004; Tebo and others 1997).
Unlike Fe(II), chemical oxidation of Mn(II) is generally slow under oxic conditions in the pH range of natural waters (pH 6–8). Microorganisms catalyze Mn(II) oxidation and are thought to be responsible for Mn(II) oxidation rates in the environment that are up to 4–5 orders of magnitude greater than expected abiotically (Nealson and others 1988; Tebo 1991; Wehrli and others 1995). The Mn cycle is therefore largely driven by the activity of microorganisms, yet despite the well-recognized environmental importance of this element, major questions concerning the identity, physiology, and ecology of the key microorganisms that drive the oxidative segment of the Mn cycle remain unanswered.
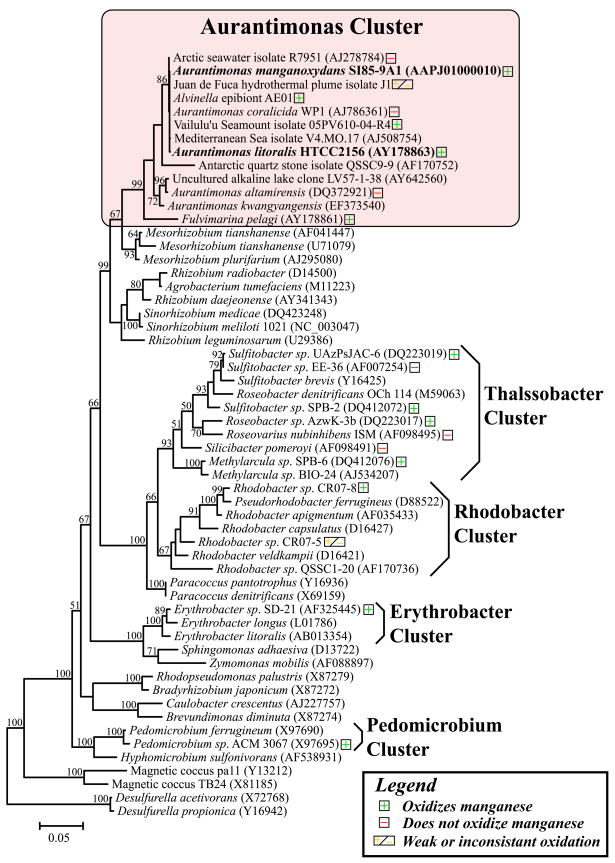
Bacteria that oxidize Mn(II) are phylogenetically diverse and include alpha, beta and gamma-Proteobacteria such as Leptothrix discophora and Pseudomonas putida the low GC Gram-positive bacteria (Firmicutes) such as Bacillus sp. strain SG–1, and the Actinobacteria (Tebo and others 2005). The fact that alpha-Proteobacteria oxidize manganese is of particular importance as these organisms are globally distributed in a diverse array of marine and terrestrial environments. Confirmed manganese oxidizers within the alpha-Proteobacteria include Roseobacter spp., Pedomicrobium sp. ACM 3067, Erythrobacter sp. strain SD-21, Aurantimonas sp. strain SI85-9A1, Sulfitobacter spp., Methylarcula spp., and Rhodobacter spp. (Fig. 1) (Caspi and others 1996; Francis and others 2001; Hansel and Francis 2006; Larsen and others 1999; Tebo and others 2005; Templeton and others 2005).
Figure 1.
Phylogeny of Mn(II) oxidizing alpha-Proteobacteria based on the16S rRNA gene. Mn(II) oxidizers are indicated with “+”, isolates that have been tested and do not oxidize Mn(II) are indicated with “−”, and “+/−” indicates weak or inconsistent Mn(II) oxidation.
The genus Aurantimonas is part of the relatively new family ‘Aurantimonadaceae’ that lies within the order Rhizobiales. Four Aurantimonas species from this family have been described before, Aurantimonas coralicida (Denner and others 2003), Aurantimonas altamirensis (Jurado and others 2006), Aurantimonas ureilytica (Weon and others 2007) and Aurantimonas frigidaquae (Kim and others 2008). These organisms were isolated from very distinct environments, these being a diseased coral, a terrestrial cave wall, an air sample and a water-cooling system respectively.. Fulvimarina and Martelella are two other genera described within the ‘Aurantimonadaceae’ represented by the described species Fulvimarina pelagi which was also isolated from a marine environment (Cho and Giovannoni 2003) and Martelella mediterranea which was isolated from a subterranean saline lake (Rivas and others 2005).
Multiple possible physiological functions of bacterial Mn(II) oxidation have been proposed, yet the actual physiological function remains enigmatic. Possible functions include energy production (chemolithotrophy), access to refractory natural organic matter, protection from radiation and oxidative stress, and storage of an electron acceptor for anaerobic respiration (Tebo and others 2005). Aurantimonas sp. strain SI85-9A1, isolated from the oxic/anoxic interface in Saanich Inlet, is intriguing as a potential Mn(II)-oxidizing chemolithoautotroph because it contains the CO2-fixing enzyme ribulose-1,6-bisphosphate carboxylase/oxygenase (Caspi and others 1996) and its growth is stimulated by Mn(II) (Dick and others 2008). The genome sequence of SI85-9A1 revealed versatile metabolic potential and confirmed the presence of all genes required for carbon fixation via the Calvin-Benson cycle (Dick and others 2008), but conclusive evidence for a link between Mn(II) oxidation and carbon fixation has yet to be found.
In this communication we report on the isolation of a collection of ‘Aurantimonas-like’ species, discuss their significance in global manganese cycling, and characterize two of these isolates as new members of the ‘Aurantimonadaceae’ family. We also report that many, but not all, members of the ‘Aurantimonadaceae’ are capable of Mn(II) oxidation, including three previously isolated organisms and two new isolates. Two strains, SI85-9A1 and HTCC 2156, were chosen for further characterization. Physiological and genomic differences distinguish these strains from the previously described Aurantimonas species: therefore, we propose their classification as two new species of this genus, Aurantimonas manganoxidans, sp. nov. and Aurantimonas litoralis, sp. nov. respectively.
Materials and methods
Isolation and cultivation of characterised strains
Strain SI85-9A1 was isolated in 1985 from Sannich Inlet, Vancouver Island, British Columbia, Canada from a water sample taken at 125 m depth. SI85-9A1 was originally isolated on M medium (Tebo and others 2006). For all the experiments described here, SI85-9A1 was grown in liquid culture batches in either M medium or J medium (Tebo and others 2006) or on solid media of the same composition. These marine media types were supplemented with a mixture of sterile-filtered 10 mM glycerol, 10 mM formate, 100 μl of a 3 mg mL−1 ferric ammonium citrate solution (FAC) and 100 μM of MnCl2. The FAC and MnCl2 were always made fresh prior to inoculation. Inocula consisted of 1/1000 volumes of 3 mL starter cultures that were grown for 48 hours in J medium without MnCl2.
Strain HTCC 2156 was isolated from surface waters of the Pacific Ocean off the Oregon coast and was grown in liquid culture using high throughput culturing (HTC) approaches described by Connon and Giovannoni (2002) and Rappé et al. (2002). This liquid culture was spread on plates of marine medium 2216 (Difco) plates and single colonies were purified after incubation for 10 days at 30 °C. For all other characterization experiments presented in this publication and maintenance of the isolate, HTCC 2156 was grown using marine medium 2216 (Difco).
Isolation, cultivation and source of other bacterial strains used in this study
Strain AE01-7 (and other epibionts) was isolated from the surface of Alvinella pompejana tubeworms during the Extreme 2001 cruise (October/November, 2001) to the East Pacific Rise (EPR) aboard the R/V Atlantis. A. pompejana worms were collected with DSV Alvin (dive 3713) from a depth of 2494 m at P-vent, located on the EPR at 9°N. Shipboard, dorsal hairs were removed from the worm using a sterile scalpel, then washed and homogenized in filter-sterilized natural seawater. This homogenate was spread on J medium agar plates that contained no organic carbon source (Tebo and others 2006) and incubated at 37 °C. Brown, Mn oxide-encrusted colonies became apparent after 11 days and were transferred to J-plates at room temperature. These isolates are maintained on M medium (Tebo and others 2006) at room temperature.
Other strains with highly similar 16S rRNA gene sequences to SI85-9A1 were obtained from the following sources: Aurantimonas altamirensis (Jurado and others 2006), BCCM/LMG bacteria collection (LMG 23375); Aurantimonas coralicida (Denner and others 2003), L. Richardson; F. pelagi HTCC 2506, 2615, and 2619 (Cho and Giovannoni 2003), S.J. Giovannoni; isolate ARK 126 (Mergaert and others 2001), BCCM/LMG Bacteria collection (LMG 23055); isolate Eplume4.J1 (Kaye and Baross 2000), J. Baross; isolate Aurantimonas VS05-110, A. Templeton. Mn oxidation by all isolates was tested on J, M, and K plates and confirmed with the LBB spot test (Tebo and others 2006).
Phylogenetic analysis, G + C content analysis and DNA-DNA hybridization
DNA extraction, PCR, and sequencing of 16S rRNA genes were as described previously (Francis and Tebo 2002). Whole-genome DNA-DNA hybridization was performed in 2 X SSC and 10 % formamide at 69°C by the Deutsche Saamlung von Mikrooorganismen und Zellkulturen GmbH (DSMZ). Sequence alignment and phylogenetic analysis using 1400 bp was performed using the ARB software environment (Ludwig and others 2004). Alignments were manually inspected and edited to remove gaps and ambiguously aligned regions. A maximum likelihood phylogenetic tree was constructed to show the clustering of Aurantimonas within the alpha-Proteobacteria. The tree was constructed with 57 sequences aligned to the arb-silva ref 91 database and was calculated with fastDNAml repeating the calculation until the tree with the greatest likelihood was found 3 times. Bootstrap values for each node were based on 100 bootstrap re-samplings. G + C content of strain HTCC 2156 was determined by HPLC according to Mesbah et al (1989) using a platinum EPS reverse-phase C18 column (150 mm, 4.6 mm, 5 μm pore size, Alltech). G + C content from strain SI85-9A1 was determined through genome sequencing (Dick and others 2008).
Phenotypic characterization
Unless otherwise indicated, the standard methods for phenotypic characterization as described by Smibert and Krieg (1994) were used in this study. Strain SI85-9A1 was normally grown in M medium at room temperature for up to 2 weeks or until there was obvious Mn(II) oxidation. Strain HTCC 2156 was always grown for a period of 5 days in Difco marine broth 2216 at 30 °C. The range of temperature for growth was measured between 4 and 45 °C. Tolerance to NaCl was tested in the range of 0 to 20 % (w/v) NaCl while the concentration of other salts in the M-media remained constant. Growth response to pH for strain SI85-9A1 was measured at room temperature over the range of 5.5 to 10 with a 50 mM final concentration of buffers MES, MOPS, HEPES, TAPS and CHES. Where the buffering ranges overlapped, duplicates of each buffer were prepared at the same pH to investigate any differences in growth due to the different buffers. For HTCC 2156 the pH range was measured between 4 and 12 at 30 °C. These tests were also conducted on Aurantimonas altamirensis. Anaerobic growth was tested using an Oxoid anaerobic system. Growth on thiosulfate and elemental sulfur was also tested for strain SI85-9A1 (1 or 10 mM concentrations in J medium at pH 7.5 incubated at room temperature). Motility was examined using wet mounts of exponential phase cells under dark field microscopy.
Basic biochemical tests for both strains were performed using API 20NE strips and API ZYM strips (bioMérieux). For strain SI85-9A1, biochemical tests were performed in duplicate for both the manufacturers supplied medium or the basal salts for J medium (Tebo and others 2006) These tests were also investigated in the presence and absence of manganese oxides to control for any artifacts that could be produced by the Mn oxides. Carbon source utilization was carried out using BIOLOG GN2 MicroPlates using the procedures recommended by the manufacturer (Biolog, CA) and outlined in Ruger and Krambeck (1994). Inocula of strain SI85-9A1 were prepared from cells swabbed off K media plates with and without manganese. These tests were also performed in duplicate both with the manufacturers supplied medium and basal J medium. Carbon source utilization was also carried out in the presence or absence of manganese oxides. For strain HTCC 2156, custom made 48 well microplates containing 47 different carbon sources supplemented the use of BIOLOG GN2 plates (Cho and Giovannoni 2003). These tests were also conducted on Aurantimonas altamirensis.
Susceptibility to antibiotics was determined using the disc diffusion method. The following antibiotics were tested (the highest concentration in μg tested per disc in parentheses): Kanamycin (50), ampicillin (10), chloramphenicol (25), erythromycin (15), neomycin (25), streptomycin (50), tetracycline (30), lincomycin (15), penicillin G (50), bacitracin (10), geneticin (50), capreomycin sulfate (50), polymyxin B sulfate (50), cephapirin (50), rifampicin (50), carbenicillin (50), spectinomycin (50), puromycin (25), vancomycin (30), gentamycin (10) and cyclohexamide (50).
Cellular fatty acid analysis and pigment extraction
For strain SI85-9A1, cells were grown in liquid M medium for 2 weeks until they started to oxidize manganese. Cells were harvested by centrifugation and three 40 mg samples were prepared - one from the ‘no manganese’ control flask, and two from the flask with manganese. Of these two samples one was washed in 10 mM buffered ascorbate (100 mM HEPES, pH 7.5) to remove Mn-oxides. For strain HTCC 2156 the cells were grown on marine agar 2216 at 30 °C for 5 days. All samples were analysed by GC according to the requirements of the Microbial Identification System (MIDI). Fatty acid profiles were analysed by Microbial ID, Newark, DE, USA.
Cellular pigments were extracted from 100 mg of cells using both 100 % methanol and a methanol/acetone mixture (1:1, vol/vol). For strain SI85-9A1, the same three sample types used for the analysis of fatty acids were tested. The absorbance spectra were determined using a scanning UV-visible spectrophotometer (Cary 100 Scan, Varian or Biospec-1601, Shimadzu).
Results and Discussion
Isolation, phylogeny, and Mn(II) oxidation by Aurantimonas-like species
This study was prompted by the isolation of new Aurantimonas-like species from several very different environments under very distinct cultivation conditions. A number of isolates (“AE01” isolates), selected based on their ability to catalyze the formation of Mn oxides, were obtained from the surface of an Alvinella pompejana tube worm on a deep-sea hydrothermal vent chimney at 9°N on the East Pacific Rise (EPR). A. pompejana is a polychaete tube worm that inhabits the walls of black smoker chimneys at deep-sea hydrothermal vent sites (depth ~ 2500 m) along the EPR. This habitat lies at the interface of hot, reducing, metal-laden hydrothermal fluids and cold, oxygenated deep-sea waters. In contrast, strain HTCC 2156 was isolated from surface waters of the Pacific Ocean off the coast of Oregon by high throughput culturing (HTC) approaches without any screening for Mn(II) oxidation.
Portions of the 16S rRNA genes of five AE01 isolates were sequenced and found to be identical. Nearly full-length 16S rRNA gene sequences were retrieved from one representative of the AE01 isolates (AE01-7) as well as HTCC 2156. Phylogenetic analysis of the 16S rRNA gene sequences revealed that AE01-7 and HTCC2156 both fell within a tight cluster of alpha-Proteobacteria that includes previously described members of the genus Aurantimonas, the Mn(II)-oxidizing bacterium strain SI85-9A1 (Caspi and others 1996), and several undescribed isolates from widespread geographic locations (Fig. 1). The genus Aurantimonas currently includes four species that have been described previously, Aurantimonas coralicida (Denner and others 2003) and Aurantimonas altamirensis (Jurado and others 2006), Aurantimonas ureilytica (Weon and others 2007) and Aurantimonas frigidaquae (Kim and others 2008). These species were isolated from diseased coral, a terrestrial cave wall, an air sample and a water-cooling system respectively.
Aurantimonas is one of three recently described genera in the family ‘Aurantimionadaceae’ and order Rhizobiales, the second and third being Fulvimarina and Martelella. Fulvimarina is represented by several F. pelagi strains that were isolated from surface marine waters (Sargasso Sea) by HTC (Cho and Giovannoni 2003). Martelella is represented by the strain Martelella mediterranea that was isolated from a subterranean saline lake (Rivas and others 2005). Numerous isolates (published but not formally described) that cluster phylogenetically within the ‘Aurantimionadaceae’ have been reported from environments around the globe, including a deep-sea hydrothermal plume in the north eastern Pacific (Kaye and Baross 2000) and surface waters of the Greenland Sea (strain R7591/ARK 126, (Mergaert and others 2001)) and an isolate from a microbial mat from the Nafauna Seamount in Samoa (strain VS05-110, submitted).
Other unpublished 16S rRNA gene sequences that fall within this clade have recently been deposited into the databases including a number of sequences from Artic and Antarctic regions (including sublithic communities), 2 sequences from the western Mediterranean, sequences associated with mangrove soil communities, endophytic bacteria associated with alpine mosses in China and alpine dolomite rocks in Switzerland, a sequence from an alkaline lake in Turkey, a sequence from bacterial communities associated with discolouration of ancient wall paintings and finally sequences from bacterial communities associated with hydrocarbon contaminated regions such as the NW Spanish shoreline (prestige oil spill), an Estonian oil shale chemical isolation plant, Indonesian coastal waters and human impacted zones in Antarctica.
To determine whether Mn(II) oxidation is a common feature of the ‘Aurantimonadaceae’, we screened all published members (apart from A. ureilytica and A. frigidaquae) for the ability to form Mn oxide encrusted colonies on agar plates. Three of the seven Aurantimonas strains tested consistently oxidized Mn(II), three strains did not show any signs of Mn(II) oxidation, and one strain weakly or inconsistently oxidized Mn(II) (Fig. 1). Three Fulvimarina strains were also tested and all three robustly oxidized Mn(II). Two Aurantimonas strains, SI85-9A1 and HTCC 2156, were chosen for further characterization.
Characterization of strains SI85-9A1 and HTCC 2156
DNA base composition and DNA-DNA hybridization
Levels of overall genomic similarity were determined by DNA-DNA hybridization. Despite sharing nearly identical 16S rRNA sequences, whole genome DNA-DNA hybridization of SI85-9A1 and HT2156 to each other and their closest relatives revealed very low similarity (Table 2), suggesting that SI85-9A1 and HTCC 2156 each represent new species within the genus Aurantimonas. The GC content of strain SI85-9A1 was 67 % and for HTCC 2156 it was 68.5 % (Table 3).
Table 2.
Whole genome DNA-DNA similarity between the published strains Aurantimonas altamirensis and Aurantimonas coralicida with Aurantimonas spp. SI85-9A1 and HTCC 2156.
| DNA-DNA Hybridization | A. altamirensis | A. coralicida | SI85-9A1 | HTCC 2156 |
|---|---|---|---|---|
| A. altamirensis | x | 56* | n/d | n/d |
| A. coralicida | 56* | x | 21.8 | 9.45 |
| SI85-9A1 | n/d | 21.8 | x | 56.4 |
| HTCC 2156 | n/d | 9.45 | 56.4 | x |
from Jurado et al. 2006
Table 3.
Strain comparison between the published strains Aurantimonas altamirensis and Aurantimonas coralicida with Aurantimonas spp. SI85-9A1 and HTCC 2156. + = positive, (+) = weakly positive, − = negative.
| Characteristic | A. altamirensis | A. coralcida | SI85-9A1 | HTCC 2156 |
|---|---|---|---|---|
| pH optimum | 6.8* | 7.6 | 6.8 to 7.5 | 7.5 to 8.0 |
| pH Range | 5.5 to 9.5* | n/d | 5.5 to 10 | 5.5 to 10 |
| Temp Range(°C) | 10 to 40 | 4 to 37 | 10 to 40 | 4 to 40 |
| Temp Optimum (°C) | 28 | 28 | 27 to 30 | 30 |
| Salt optimum (% NaCl) | 0 to 2 | 3.2 | 2.5 | 2.0 to 2.5 |
| Growth at 10% salt | − | − | + | + |
| Gram Stain | negative | negative | negative | negative |
| Pigment absorbance (nm) | 447, 447–471, 424–427 inflection | 447, 470–471, 424–427 inflection | 450, 471–477, 424–429 inflection | 452, 339 |
| DNA G+C mol% | 71.8 | 66.3 | 67 | 68.5 |
| Manganese oxidation | −* | − | + | + |
| Carbon Source Utilization | ||||
| Acetic Acid | +* | + | (+) | − |
| Adenine | + | − | n/d | n/d |
| α-Keto Glutaric Acid | +* | − | + | + |
| α-D-Glucose | + | − | + | + |
| D-Galactose | + | + | + | + |
| D-Mannitol | + | − | + | + |
| D-Melibiose | (+) | + | − | + |
| Formic Acid | +* | − | + | + |
| Gentiobiose | − | + | − | + |
| Gluconic Acid | + | + | + | + |
| Glycerol | +* | + | + | + |
| i-Erythritol | + | − | + | − |
| L-Arabinose | + | + | + | + |
| L-Asparagine | +* | (+) | + | − |
| L-Glutamic Acid | +* | + | + | + |
| L-Pyroglutamic Acid | −* | − | + | − |
| L-Rhamnose | + | + | + | + |
| L-Serine | +* | + | − | + |
| Malonic Acid | + | − | − | − |
| Maltose | + | (+) | − | − |
| m-Inositol | +* | − | + | − |
| N-Acetyl-D-Galactosamine | + | (+) | − | − |
| Fatty Acids (%) | ||||
| C16:0 | 11.3 | 6.7 | 7.16 | 2.3 |
| C16:1ω7c | − | 1.3 | 4.36 | − |
| C18:0 | 0.7 | 1.5 | − | 1.3 |
| C18:1 2-OH | 3.5 | 2 | 3 | 0.8 |
| C18:1ω7c | 74.4 | 76.9 | − | 63 |
| C18:1ω9t | − | − | 64.31 | − |
| C19:0ω7c cyclo | − | − | 21.16 | − |
| C19:0ω8c cyclo | − | 10.5 | − | 28 |
Extra experiments performed in this study to complement the work of Jurado et al. 2006.
n/d = not determined
Phenotypic characteristics
Strain SI85-9A1 is a Gram-negative motile (0.9 – 1.2 μm long and 0.5 – 0.8 μm wide) rod that grows optimally at 30 °C in marine media with NaCl concentrations of 2.5 % (w/v). SI85-9A1 can grow in the range of 4 – 37 °C but the most significant growth occurs between 20 and 35 °C. SI85-9A1 can grow without the addition of NaCl to the media and can tolerate up to 15 % (w/v). The pH optimum is between 6.8 and 7.5 with growth in the range of pH 5.5 and 10. SI85-9A1 cells do not grow anaerobically and no evidence for anaerobic respiration genes was detected in the genome. The cells are non-spore forming. SI85-9A1 forms circular, opaque, convex colonies with smooth edges and develop a central brown colour in the presence of manganese. Manganese oxidation occurs after 192 to 240 hours and the organism will only oxidize manganese when the incubation temperature is below 30 °C. Colonies are generally between 0.5 and 1.5 mm in diameter.
HTCC 2156 is a short Gram-negative non-motile rod 0.9 – 1.9 μm long and 0.6 – 1.2 μm wide. HTCC 2156 grows optimally at 30 °C and within the range of 4 –40 °C. HTCC 2156 is grown routinely in Difco Marine Broth 2216 with an optimum salt (NaCl) concentration of 2 % (w/v). HTCC 2156 can grow in up to 10 % NaCl (w/v) and in the absence of NaCl. The pH optimum is between 7.5 and 8.0 with growth in the range of pH 5.5 and 10. No flagella were observed on the negatively stained cells. No endospores or poly-β-hydroxybutyrate granules were produced. Colonies are 1.4 – 2.5 mm in diameter. Colonies are brownish yellow in colour, circular, convex and dry. HTCC 2156 does not grow anaerobically.
Both strains were catalase, oxidase and urease positive. Denitrification activity was not detected nor was indole production, arginine deaminase, gelatine and aesculin hydrolysis and acid production. Acid and alkaline phosphatase, esterase and Naphthol-AS-BI-phosphohydrolase were detected in both strains as was leucine and valine arylamidase activity and arginine dihydrolase activity.
Susceptibility to antibiotics also differs with HTCC 2156 being resistant to tetracycline and rifampicin, whereas SI85-9A1 is resistant to tetracycline and erythromycin. SI85-9A1 exhibited differences in antibiotic susceptibility when grown in the presence of manganese versus the absence of manganese. When Manganese is present it becomes susceptible to erythromycin and tetracycline and more susceptible to rifampicin (5 μg as opposed to 20 μg per disc).
The strains can be distinguished from one another through pigments, fatty acid profiles and carbon source utilization. SI85-9A1 shares pigments in common with the other Aurantimonas species described but HTCC 2156 has an additional peak at 339 nm (Table 3). For both strains the major fatty acid is C18:1 but the side-chain for HTCC 2156 appears in the ω7c position whereas for SI85-9A1 it appears at ω9t. The strains have the fatty acids C16:0 and C18:1 2-OH in common but the proportions differ (see Table 3). HTCC 2156 shares more fatty acids in common with the other Aurantimonas species compared to SI85-9A1 (Table 3). Both strains can utilize a wide variety of carbon sources that include pentoses, hexoses, oligosaccharides, sugar alcohols, organic acids and amino acids. The following are carbon sources that both strains utilize which are not mentioned in Table 3: β-Hydroxybutyric Acid, D, L-Lactic Acid, D-Arabitol, D-Fructose, D-Galacturonic Acid, D-Mannitol, D-Mannose, Glucuronamide, L-Alaninamide, L-Fucose, L-Proline, L-Rhamnose, Pyruvic Acid Methyl Ester, Succinic Acid, Succinic Acid Mono-methyl Ester and Uridine. Carbon sources that HTCC 2156 cannot utilize compared to SI85-9A1 (beyond those mentioned in Table 3) include: Bromosuccinic Acid, D-Galactonic Acid Lactone, D-Saccharic Acid, L-Alanine, L-Aspartic Acid and Quinic Acid. Carbon sources that SI85-9A1 cannot utilize compared to HTCC 2156 (beyond those mentioned in Table 3) include: α-Hydroxybutyric Acid, α-Ketobutyric Acid, α-Ketovaleric Acid, Itaconic Acid, Lactulose, Succinamic Acid, Thymidine, Tween 40 and Tween 80.
Analysis of the genomic sequence of strain SI85-9A1 suggests that in addition to heterotrophy, SI85-9A1 is also capable of carbon fixation via the Calvin Benson cycle (Dick and others 2008). Possible electron donors during autotrophic growth include Mn(II), carbon monoxide, and reduced sulfur species, but in no case has growth on any of these substrates as the sole energy source been demonstrated. Since Mn(II) stimulates heterotrophic growth and genes for utilization of CO and sulfur are present in the genome sequence (Dick and others 2008) this hints that SI85-9A1 may be a mixotroph. The laboratory conditions under which this putative mixotrophy and/or autotrophy might be operative still remain elusive. Aside from carbon fixation and lithotrophy genes, strain SI85-9A1 also has a number of genes associated with methylotrophy such as methanol, formate and formaldehyde oxidation (Dick and others 2008).
Distribution of manganese oxidizing Aurantimonas species and implications for Mn cycling
Apart from A. altamirensis, A. ureilytica and A. frigidaquae, the cultivated representatives of the Aurantimonadaceae family come from marine localities. These localities are widespread and include the Pacific Ocean, the Sargasso Sea, the Greenland Sea and the Mediterranean. While most Aurantimonas strains have been cultured from surficial waters there are several cases where these bacteria have been found in areas with high metal flux such as deep-sea hydrothermal vents and redox transitional zones such as oxic/anoxic boundaries found in stratified fjords.
Our finding that many Aurantimonas-like bacteria oxidize Mn(II) raises the possibility that these bacteria are key contributors to metal cycling and availability in these ecosystems. It is unknown whether Mn(II) oxidation is a direct response to pervading environmental conditions or an indirect process. In surface waters the role that Mn(II)-oxidising bacteria play in metal cycling is quite important as manganese oxides control not only the distribution of trace metals via adsorption but also affect redox chemistry associated with limiting nutrients like iron. Mn oxides also affect the stability of metal-organic complexes that then effect productivity in surficial waters. The fact that they can oxidize CO also indicates that they have a strong involvement in carbon cycling as well.
The dorsal hairs of A. pompejana from where the AE01 isolates were retrieved are home to a dense community of microorganisms whose symbiotic relationship to the worm is poorly understood (Campbell and Cary 2001; Haddad and others 1995). It has been hypothesized that one potential function of this epibiotic community is detoxification of metals, which are abundant in hydrothermal vent fluids (Alayse-Danet and others 1987). Previous studies noted the prevalence of Mn oxides (B.T. Glazer, personal communication) and Mn(II)-oxidizing bacteria (Prieur and others 1990) in the dense microbial communities that inhabit the dorsal surface of A. pompejana. Because Mn oxides are strong scavengers of many metals (Tebo and others 2005), these Mn oxides could sequester and thus control the availability of metals found in hydrothermal fluids. The isolation of the Mn(II)-oxidizing AE01 strains from this environment represents the first identification of the organisms that may be playing this crucial ecological role of metal sequestration in the A. pompejana epibiotic community.
Though members of the Aurantimonadaceae are clearly distributed widely in diverse environments, further work is required to determine their abundance in the environment and thus potential contribution to Mn cycling. Early indications from current work suggests that the 16S rRNA gene from Aurantimonas can be directly amplified from environmental genomic DNA extracts at a ratio of 1:2238 (Aurantimonas 16S rDNA copies to total Bacterial 16S rDNA copies) and that the Mn oxidase protein from Aurantimonas sp. SI85-9A1 and orthologs are easily identified and numerous off the coast of Oregon and in the Columbia River Estuary. If this is the case then Aurantimonas has a significant role to play in both Mn-redox cycling and in delivering trace metals to the coast of the Western United States.
Our results also highlight the difficulty in using 16S rRNA gene-based approaches to investigate the ecology of Mn(II)-oxidizing bacteria. The polyphyletic nature of Mn(II)-oxidizers has been recognized for some time, but the results we present here are a clear example. Despite the tight phylogenetic clustering of members of the genus Aurantimonas, some are able to oxidize Mn(II) and others apparently are not, and genome hybridization indicates extensive intra-ribotype diversity in this clade (Table 2). Comparative analysis of these closely related bacteria might elucidate the genetic, biochemical, and physiological factors that underpin this phenotypic difference among Aurantimonas species. The intra-ribotype variation could well be linked to environment and the need for the planktonic species such as HTCC 2156 and SI85-9A1 to exhibit a wider range of metabolic diversity.
Considering the phenotypic and genotypic characteristics described above and the differences between strain SI85-9A1 and HTCC 2156 with respect to the two other previously described Aurantimonas species, strain SI85-9A1 and HTCC 2156 represent novel species. The names Aurantimonas manganoxydans sp. nov. is proposed for SI85-9A1 and Aurantimonas litoralis for HTCC 2156.
Description of Aurantimonas manganoxidans sp. nov
Aurantimonas manganoxydans [man.ga.no’xydans. N.L. manganum manganese; N.L. part. adj. oxydans oxidizing; manganoxydans oxidizing manganese compounds].
Cells are Gram negative, motile short rods that occur singly or in groups often arranged in stacks. The cell dimensions are 0.9 – 1.2 μm long and 0.5 – 0.8 μm wide. The cells form circular, opaque, convex colonies with smooth edges and develop a central brown colour in the presence of manganese. Manganese oxidation occurs after 192 to 240 hours and the organism will only oxidize manganese when the incubation temperature is below 30 °C. The organism grows optimally at 30 °C in a marine media with NaCl concentrations of 2.5 % (w/v). SI85-9A1 can grow in the range of 4 −37 °C but the most significant growth occurs between 20 and 35 °C. SI85-9A1 can grow without the addition of NaCl to the media and can tolerate up to 15 % (w/v). The pH optimum is between 6.8 and 7.5 with growth in the range of pH 5.5 and 10. SI85-9A1 cells do not grow anaerobically, are non-spore forming and motile. Colonies are generally between 0.5 and 1.5 mm in diameter.
The strain is catalase, oxidase and urease positive. Denitrification activity is not detected nor is indole production, arginine deaminase, gelatine and aesculin hydrolysis and acid production. Acid and alkaline phosphatase, esterase and naphthol-phosphohydrolase are detected as is leucine and valine arylamidase activity and arginine dihydrolase activity.
Predominant fatty acids are C18:1ω9t (64.31 %) and C19:0ω7c cyclo (21.16 %) with minor fatty acids being C16:0 (7.16 %), C16:1ω7c (4.36 %) and C18:1 2-OH (3.0 %). The DNA G + C content is 67 mol % (by HPLC). Absorption spectral peaks are observed at 450, 471 – 477 nm with a slight inflection at 424 – 429 nm.
Metabolism is obligately aerobic and chemoheterotrophic. According to Biolog tests the following carbon sources are utilized: Acetic Acid, Bromosuccinic Acid, D-Galactonic Acid Lactone, α-D-Glucose, D-Saccharic Acid, i-Erythritol, L-Alanine, L-Asparagine, L-Aspartic Acid, L-Pyroglutamic Acid, m-Inositol, Quinic Acid, α-Keto Glutaric Acid, β-Hydroxybutyric Acid, D-Gluconic Acid, D, L-Lactic Acid, D-Arabitol, D-Fructose, D-Galactose, D-Galacturonic Acid, D-Mannitol, D-Mannose, Formic Acid, Glucuronamide, Glycerol, L-Alaninamide, L-Arabinose, L-Fucose, L-Glutamic Acid, L-Proline, L-Rhamnose, Pyruvic Acid Methyl Ester, Succinic Acid, Succinic Acid Mono-Methyl Ester and Uridine.
The type strain is SI85-9A1 (ATCC BAA-1229, DSM 21871). Isolated from Saanich Inlet, Vancouver Island, British Columbia, Canada.
Description of Aurantimonas litoralis sp. nov
Aurantimonas litoralis [li.to.ra’lis. L. adj. litoralis from the sea-shore].
HTCC 2156 is a short Gram-negative non-motile rod 0.9 – 1.9 μm long and 0.6 – 1.2 μm wide. HTCC 2156 grows optimally at 30 °C and within the range of 4 – 40 °C. HTCC 2156 is grown routinely in Difco Marine Broth with an optimum salt (NaCl) concentration of 2 % (w/v). HTCC 2156 can grow in up to 10 % NaCl (w/v) and in the absence of NaCl. The pH optimum is between 7.5 and 8.0 with growth in the range of pH 5.5 and 10. No flagella were observed on the negatively stained cells. No endospores or poly-β-hydroxybutyrate granules were produced. Colonies are 1.4 – 2.5 mm in diameter. Colonies are brownish yellow in colour, circular, convex and dry and can oxidize manganese. HTCC 2156 does not grow anaerobically.
The strain is catalase, oxidase and urease positive. Denitrification activity was not detected nor was indole production, arginine deaminase, gelatine and aesculin hydrolysis and acid production. Acid and alkaline phosphatase, esterase and naphthol-phosphohydrolase are detected as is leucine and valine arylamidase activity and arginine dihydrolase activity.
Predominant fatty acids are C18:1ω7c (63.0 %) and C19:0ω8c cyclo (28.0 %) with minor fatty acids being C16:0 (2.3 %), C18:0 (1.3 %) and C18:1 2-OH (0.8 %). The DNA G + C content is 68.5 mol% (by HPLC). Absorption spectral peaks are observed at 452 and 339 nm.
Metabolism is obligately aerobic and chemoheterotrophic. According to Biolog tests the following carbon sources are utilized: α-Hydroxybutyric Acid, α-Ketobutyric Acid, α-Ketovaleric Acid, L-Serine, D-Melibiose, Gentiobiose, Itaconic Acid, Lactulose, Succinamic Acid, Thymidine, Tween 40, Tween 80, α-D-Glucose, α-Keto Glutaric Acid, β-Hydroxybutyric Acid, D-Gluconic Acid, D, L-Lactic Acid, D-Arabitol, D-Fructose, D-Galactose, D-Galacturonic Acid, D-Mannitol, D-Mannose, Formic Acid, Glucuronamide, Glycerol, L-Alaninamide, L-Arabinose, L-Fucose, L-Glutamic Acid, L-Proline, L-Rhamnose, Pyruvic Acid Methyl Ester, Succinic Acid, Succinic Acid Mono-Methyl Ester and Uridine.
The type strain is HTCC 2156 (ATCC BAA-667, KCTC 12094). Isolated from coastal waters off Oregon, United States of America.
Table 1.
Strains used in this study.
| Strain | 16S rRNA Accession | Isolated from: | Reference |
|---|---|---|---|
| SI85-9A1 | U53824 | Oxic/anoxic interface of the Saanich Inlet fjord, Vancouver Island, BC, Canada | Caspi, 1996 |
| HTCC 2156 | AY178863 | Surface waters off the Oregon Coast, United States | This study |
| AE01 | FJ527311 | Dorsal surface of Alvinella pompejana tube worms on a hydrothermal vent chimney at 9°N, East Pacific Rise | This study |
| Aurantimonas coralicida | AJ786361 | Caribbean coral pathogen | Denner, 2003 |
| Aurantimonas altamirensis | DQ372921 | Terrestrial Spanish cave | Jurado, 2006 |
| Eplume4.J1 | AF251774 | Hydrothermal plume, Juan de Fuca Ridge | Kaye, 2000 |
| VS05-110 | FJ497698 | Microbial mat from the summit region of Nafauna Seamount Samoa | This study |
| R7591/ARK 126 (LMG23055) | AJ278784 | Surface waters of the Greenland Sea | Mergaert, 2001 |
| Fulvarmarina pelagi strains: HTCC 2506, 2615, 2619 | AY178861 | Surface waters of the Sargasso Sea | Cho, 2003 |
Acknowledgments
The project described was partially supported by the National Science Foundation grant OCE-0635493 and by grant number ES010337 from the National Institute of Environmental Health Sciences (NIEHS), NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
References
- Alayse-Danet AM, Desbruyeres D, Gaill F. The possible nutritional or detoxification role of the epibiotic bacteria of Alvinellid polychaetes: Review of current data. Symbiosis. 1987;4:51–62. [Google Scholar]
- Campbell BJ, Cary SC. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl Environ Microbiol. 2001;67:110–7. doi: 10.1128/AEM.67.1.110-117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Haygood MG, Tebo BM. Unusual ribulose-1,5-bisphosphate carboxylase/oxygenase genes from a marine manganese-oxidizing bacterium. Microbiology. 1996;142:2549–2559. doi: 10.1099/00221287-142-9-2549. [DOI] [PubMed] [Google Scholar]
- Cho JC, Giovannoni SJ. Fulvimarina pelagi gen. nov., sp. nov., a marine bacterium that forms a deep evolutionary lineage of descent in the order “Rhizobiales”. Int J Syst Evol Microbiol. 2003;53:1853–9. doi: 10.1099/ijs.0.02644-0. [DOI] [PubMed] [Google Scholar]
- Connon SA, Giovannoni SJ. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–85. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner EB, Smith GW, Busse HJ, Schumann P, Narzt T, Polson SW, Lubitz W, Richardson LL. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean Scleractinian corals. Int J Syst Evol Microbiol. 2003;53:1115–22. doi: 10.1099/ijs.0.02359-0. [DOI] [PubMed] [Google Scholar]
- Dick GJ, Podell S, Johnson HA, Rivera-Espinoza Y, Bernier-Latmani R, McCarthy JK, Torpey JW, Clement BG, Gaasterland T, Tebo BM. Genomic insights into Mn(II) oxidation by the marine alpha-proteobacterium Aurantimonas sp. strain SI85-9A1. Appl Environ Microbiol. 2008;74:2646–58. doi: 10.1128/AEM.01656-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner KM, Stevens RD, Fridovich I. Characterization of Mn(III) complexes of linear and cyclic desferrioxamines as mimics of superoxide dismutase activity. Arch Biochem Biophys. 1994;310:341–6. doi: 10.1006/abbi.1994.1176. [DOI] [PubMed] [Google Scholar]
- Francis CA, Co E-M, Tebo BM. Enzymatic manganese(II) oxidation by a marine α-proteobacterium. Applied and Environmental Microbiology. 2001;67:4024–4029. doi: 10.1128/AEM.67.9.4024-4029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis CA, Tebo BM. Enzymatic manganese(II) oxidation by metabolically-dormant spores of diverse Bacillus species. Applied and Environmental Microbiology. 2002;68:874–880. doi: 10.1128/AEM.68.2.874-880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad A, Camacho F, Durand P, Cary SC. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl Environ Microbiol. 1995;61:1679–87. doi: 10.1128/aem.61.5.1679-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weon HY, Kim BY, Yoo SH, Joa JH, Lee KH, Zhang YS, Kwon SW, Koo BS. Aurantimonas ureilytica sp. nov., isolated from an air sample. Int J Syst Evol Microbiol. 2007;57:1717–1720. doi: 10.1099/ijs.0.65035-0. [DOI] [PubMed] [Google Scholar]
- Hansel CM, Francis CA. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl Environ Microbiol. 2006;72:3543–9. doi: 10.1128/AEM.72.5.3543-3549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado V, Gonzalez JM, Laiz L, Saiz-Jimenez C. Aurantimonas altamirensis sp. nov., a member of the order Rhizobiales isolated from Altamira Cave. Int J Syst Evol Microbiol. 2006;56:2583–5. doi: 10.1099/ijs.0.64397-0. [DOI] [PubMed] [Google Scholar]
- Kaye JZ, Baross JA. High incidence of halotolerant bacteria in Pacific hydrothermal vent and pelagic environments. FEMS Microbiology Ecology. 2000;32:249–260. doi: 10.1111/j.1574-6941.2000.tb00718.x. [DOI] [PubMed] [Google Scholar]
- Kim MS, Hoa KT, Baik KS, Park SC, Seong CN. Aurantimonas fridgidaquae sp. nov., isolated from a water cooling system. Int J Syst Evol Microbiol. 2008;58:1142–1146. doi: 10.1099/ijs.0.65421-0. [DOI] [PubMed] [Google Scholar]
- Kostka JE, Luther GW, III, Nealson KH. Chemical and biological reduction of Mn(III)-pyrophosphate complexes - Potential importance of dissolved Mn(III) as an environmental oxidant. Geochim Cosmochim Acta. 1995;59:885–894. [Google Scholar]
- Larsen EI, Sly LI, McEwan AG. Manganese(II) adsorption and oxidation by whole cells and a membrane fraction of Pedomicrobium sp. ACM 3067. Arch of Microbiol. 1999;171:257–264. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert J, Verhelst A, Cnockaert MC, Tan T, Swings J. Characterization of facultative oligotrophic bacteria from polar seas by analysis of their fatty acids and 16S rDNA sequences. Syst and Appl Microbiol. 2001;24:98–107. doi: 10.1078/0723-2020-00012. [DOI] [PubMed] [Google Scholar]
- Mesbah M, Premachandran U, Whitman WB. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- Nealson KH, Tebo BM, Rosson RA. Occurrence and mechanisms of microbial oxidation of manganese. Adv Appl Microbiol. 1988;33:279–318. [Google Scholar]
- Parker DL, Sposito G, Tebo BM. Manganese(III) binding to a siderophore produced by a manganese(II)-oxidizing bacterium. Geochim Cosmochim Acta. 2004;68:4809–4820. [Google Scholar]
- Prieur D, Chamroux S, Durand P, Erauso G, Fera P, Jeanthon C, Le Borgne L, Mevel G, Vincent P. Metabolic diversity in epibiotic microflora associated with the Pompeii worms Alvinella pompejana and A. caudata (Polychaetae: Annelida) from deep-sea hydrothermal vents. Marine Biology. 1990;106:361–367. [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–3. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rivas R, Sánchez-Márquez S, Mateos PF, Martínez-Molina E, Velázquez E. Martelella mediterranea gen. nov., sp. nov., a novel alpha-proteobacterium isolated from a subterranean saline lake. Int J Syst Evol Microbiol. 2005;55:955–959. doi: 10.1099/ijs.0.63438-0. [DOI] [PubMed] [Google Scholar]
- Rüger HJ, Krambeck HJ. Evaluation of the BIOLOG substrate metabolism system for classification of marine bacteria. Syst Appl Microbiol. 1994;17:281–288. [Google Scholar]
- Smibert RM, Krieg NR. Phenotypic characterization. In: Gerhardt RGE, Murray WA, Wood WA, Kreig NR, editors. Methods for General and Molecular Microbiology. ASM Press; Washington, DC: 1994. pp. 611–654. [Google Scholar]
- Sunda WG, Kieber DJ. Oxidation of humic substances by manganese oxides yields low-molecular-weight organic substrates. Nature. 1994;367:62–64. [Google Scholar]
- Tebo BM. Manganese(II) oxidation in the suboxic zone of the Black Sea. Deep-Sea Res. 1991;38:S883–S905. [Google Scholar]
- Tebo BM, Bargar JR, Clement BG, Dick GJ, Murray KJ, Parker D, Verity R, Webb SM. Biogenic manganese oxides: properties and mechanisms of formation. Ann Rev Earth Planet Sci. 2004;32:287–328. [Google Scholar]
- Tebo BM, Clement B, Dick GJ. Biotransformations of Manganese. In: Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach lD, editors. Manual of Environmental Microbiology. ASM Press; Washington, DC: 2006. pp. 1223–1238. [Google Scholar]
- Tebo BM, Ghiorse WC, van Waasbergen LG, Siering PL, Caspi R. Bacterially-mediated mineral formation: Insights into manganese(II) oxidation from molecular genetic and biochemical studies. In: Banfield JF, Nealson KH, editors. Geomicrobiology: interactions between microbes and minerals. Mineralogical Society of America; Washington, D.C: 1997. pp. 225–266. [Google Scholar]
- Tebo BM, Johnson HA, McCarthy JK, Templeton AS. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 2005;13:421–428. doi: 10.1016/j.tim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Templeton AS, Staudigel H, Tebo BM. Diverse Mn(II)-oxidising bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol J. 2005;22:127–139. [Google Scholar]
- Trouwborst RE, Clement BG, Tebo BM, Glazer BT, Luther GW. Soluble Mn(III) in suboxic zones. Science. 2006;313:1955–1957. doi: 10.1126/science.1132876. [DOI] [PubMed] [Google Scholar]
- Wehrli B, Friedl G, Manceau A. Reaction rates and products of manganese oxdiation at the sediment-water interface. In: Huang CP, O’Melia CR, Morgan JJ, editors. Aquatic Chemistry: Interfacial and Interspecies Processes. American Chemical Society; Washington, D.C: 1995. [Google Scholar]