Abstract
The BCL6 transcriptional repressor protein has been shown to promote B cell lymphoma in transgenic mouse models. The mechanism by which BCL6 transforms primary B cells is unclear, although repression of the p53 tumor suppressor is thought to play a role. Here we show that BCL6 has critical oncogene functions that are independent of p53 repression. We find BCL6 cooperates with constitutive CD40 signaling to rapidly transform p53-deficient primary mouse B cells in vitro. Constitutive CD40 signaling alone does not transform p53-deficient B cells, indicating that BCL6 acts specifically as an immortalizing oncogene in this system. The BCL6 transformed B cells are polyclonal and form polyclonal tumors. At the initiation of the cultures, BCL6 does not significantly alter cell cycle progression, but does promote increased cell survival. Early cultures of BCL6 expressing B cells show marked repression of ATR and p27kip1 but not other BCL6 target genes, suggesting that the ATR and p27kip1 genes have key early roles in mediating BCL6 transformation function. BCL6 transformed cell lines show further decreases of ATR and p27kip1 expression plus strong decreases in Blimp1 and PDCD2 expression. This study provides important clues about the critical target genes used by BCL6 to transform primary B cells and indicates that the CD40 signaling pathway can collaborate with BCL6 in the transformation of primary B cells. Lastly, our study demonstrates a rapid in vitro system to analyze the transformation function of BCL6.
Keywords: B cell lymphoma, BCL6, oncogene, transformation
The BCL6 gene encodes a transcriptional repressor protein that regulates B cell differentiation and B cell lymphomagenesis. BCL6 protein is expressed at low levels in resting B cells, induced to very high levels in germinal center B cells and is then turned off as B cells differentiate into plasma cells 1-3. B cells deficient in BCL6 expression are unable to form germinal centers 4, 5. Chromosomal translocations involving the BCL6 gene occur in almost one-third of cases of non-Hodgkin’s lymphoma, and BCL6 translocations are most common in Diffuse Large Cell Lymphoma 6. Translocations of the BCL6 gene involve promoter substitution and have the effect of conferring constitutive expression of BCL6 to the B cell 6-8. Constitutive BCL6 expression blocks normal B cell differentiation into plasma cells, and maintains the B cells in a rapidly proliferating germinal center-like state that is susceptible to oncogenic transformation. Transgenic mice that over-express BCL6 develop B cell lymphomas late in life 9, 10. In one study, the development of lymphomas was greatly increased and accelerated by exposure of the mice to a DNA damaging agent 10. In a second study, the lymphomas that develop are monoclonal in origin despite the expression of the BCL6 transgene in all B cells 9. These data indicate that transformation of primary B cells by BCL6 is not a direct process and requires secondary oncogenic mutations. Earlier, we found that BCL6 over-expression could immortalize p53-deficient B cells but not wild-type B cells 11, consistent with the idea that BCL6 requires other oncogenic mutations for transformation. Although BCL6 can repress p53 gene expression in human B cells 12, the functional significance of p53 repression by BCL6 has not been tested with regard to primary B cell transformation. We have found that BCL6 does not repress p53 transcription in mouse B cells, casting doubt on the general importance of p53 repression by BCL6. However, altered expression of BCL6 in p53-deficient mouse B cells may mimic the function of BCL6 in human B cells. Here we present data that constitutively expressed BCL6 can transform primary B cells if p53 function is ablated and if the CD40 signaling pathway is activated.
Materials and Methods
Retrovirus constructs and retrovirus infections
The retroviral vectors pMIEG3 and pMIEG3-BCL6 have been described previously 5, 11. The CD40-ligand (CD40L) expressing retrovirus was made by PCR amplifying cDNA made from anti-CD3 activated mouse T cells with the following primers for CD40-ligand: sense 5′-CTTTCAGTCAGCATGATAGAAACA-3′ and anti-sense 5′-TCAGAGTTTGAGTAAGCCAAAAGA-3′, which amplify the complete coding region of mouse CD40-ligand. CD40-ligand gene was then re-amplified with primers containing XhoI and NotI sites, and then cloned via XhoI and NotI sites into a retroviral vector expressing human CD4 13. Anti-human CD4 Ab was obtained from BD Biosciences. B cells were stimulated and infected with retroviruses as previously described 11.
Quantitative PCR
Quantitative PCR (QPCR) was performed on an ABI Prism 7000 machine (Applied Biosystems) using SYBR-green PCR mix (Roche), and data was analyzed with ABI software. The delta-delta Ct method was used to analyze relative differences in gene expression, using beta-tubulin levels as the standard. Q-PCR primer sequences were: ATR, (Forward) CTGAGAACATTCCCTGATCCTACA and (Reverse) TGCAGTAGAGCGGCAATATGC; beta-tubulin, CTGGGAGGTGATAAGCGATGA (Forward) and CGCTGTCACCGTGGTAGGT (Reverse); Blimp-1, CCCCGACATGCCCTTCTA (Forward) and GGACGCTTTCAAAAAGTCTTCTG (Reverse); PDCD2, GGTCATGCCTCAGCTGTTGA (Forward) and AGACACCCCAGTCGATGCTT (Reverse); BCL-XL, CTGGGACACTTTTGTGGATCTCT (Forward) and AAGCGCTCCTGGCCTTTC (Reverse); Cyclin D2, CAGACGTGCGGGATGTTG (Forward) and TGACGAACACGCCTCTCTCTT (Reverse); p21cip1, CAAAGTGTGCCGTTGTCTCTTC (Forward) and TGAGCGCATCGCAATCA (Reverse); p27kip1, GGCCCGGTCAATCATGAA (Forward) and TTGCGCTGACTCGCTTCTT (Reverse); p53, TCCTCCCCTCAATAAGCTATTCTG (Forward) and TGGCGCTGACCCACAAC (Reverse).
Results
Because BCL6 is known to repress p53 in human B cells 12, and because our previous data showed a role for BCL6 in p53-deficient B cells 11, we tested if BCL6 could repress p53 in mouse B cells. To test this, we stimulated wild-type spleen B cells with LPS for 24 hours, then infected the cells with either Green Fluorescent Protein (GFP)-expressing control or with BCL6-GFP-expressing retroviruses, isolated the retrovirus-expressing cells by fluorescence-activated cell sorting on the basis of GFP expression, and prepared RNA from the cells. Following the preparation of cDNA, we measured p53 mRNA levels by quantitative PCR. Averaging results from five different preparations of cells, we found that setting relative p53 mRNA levels in the control cells to 1, p53 mRNA levels with BCL6 were 0.9 +/- 0.4 (S.E.). Thus, the expression of p53 was not significantly decreased by BCL6 and repression of p53 by BCL6 is not conserved in mouse B cells. This indicates that regulation of p53 transcription by BCL6 is not conserved in mouse B cells. Margalit et al found that BCL6 transcription is regulated by p53 in human but not mouse cells 14, and our data coupled with the Margalit et al data indicate the reciprocal regulation of p53 and BCL6 seen in humans is not evolutionarily conserved. Nonetheless, an advantage of analyzing BCL6 function in p53-deficient mouse B cells is that it allows us to dissect p53-independent functions of BCL6 involved in B cell transformation.
CD40 signaling is critical for the growth of BCL6-expressing germinal center B cells, and earlier we showed that BCL6-immortalized B cells require stimulation through the CD40 pathway for growth 11. The importance of CD40 signaling is further demonstrated by findings that some B cell lymphomas can express self-stimulatory CD40 ligand (CD40L)15-17. We therefore examined whether primary B cells could be transformed to autonomous growth by co-expression of BCL6 and CD40L. To test this, we transduced lipopolysaccharide (LPS)-activated p53-deficient B cells with GFP-expressing plus CD40L-expressing retroviruses or with BCL6-GFP-expressing plus CD40L-expressing retroviruses. We then purified cells expressing both CD40L and GFP by cell sorting, and assessed the growth of the sorted B cells over time (Figure 1A). Following cell sorting and placement back into culture, GFP plus CD40L (“ML”) B cells die off over the course of two weeks. While GFP-BCL6 plus CD40L (“BL”) B cells initially died at a similar rate as “ML” B cells when first placed in culture, they soon stabilized and started steady growth from then onwards. After an initial three-week culture period, the BL B cells could thereafter be maintained as an exponentially growing cell line. In over ten different experiments, BL B cell lines were established consistently, while cell lines never grew out of ML cultures. Tests with wild-type B cells in this system showed dampened proliferation and survival, consistent with our previous experiments 11. Importantly, transformed BL B cell lines were never generated with wild-type B cells, showing the importance of p53 mutation in this system, as in our “immortalization” system 11. BL B cell lines were also generated consistently from unsorted “bulk” B cell cultures. Figure 1B shows the strong outgrowth of CD40L plus GFP-BCL6 positive cells from a bulk BL B cell culture over a few weeks, where the CD40L plus GFP-BCL6 positive cells started as only 2% of the original culture. BL B cell lines are polyclonal, as shown by genomic DNA analysis of retrovirus insertion (Figure 2A), and thus do not represent an outgrowth of one rare mutant cell type.
Figure 1. BCL6 and CD40-ligand cooperate with loss of p53 to produce autonomously growing B cell lines.
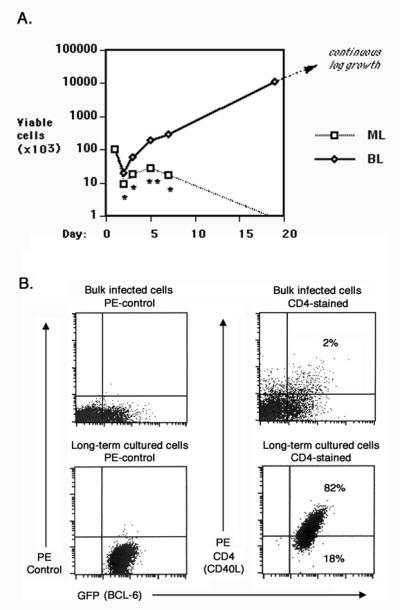
A. Spleen cells from p53-deficient mice were co-transduced with two combinations of retroviruses: MIEG3 plus CD40-ligand (“ML”) or MIEG3-BCL6 plus CD40-ligand (“BL”). Retroviruses expressing CD40-ligand (CD40L) also co-express human CD4, and MIEG3 and MIEG3-BCL6 retroviruses both express Green Fluorescent Protein (GFP). ML and BL retrovirus infected cells positive for both GFP and human CD4 were isolated by fluorescence activated cell sorting and then placed in culture at 2 ×105 cells per ml (day 1). At the indicated times, three wells of each type were harvested and viable cells were counted. ML cells were completely dead by three weeks of culture, whereas BL cells initiated log phase growth by this time. Asterisks indicate probability (P) values for differences in cell yields, where * is P < 0.05 and ** is P < 0.01. This experiment is representative of multiple similar experiments. B. Flow cytometry analysis of p53-deficient B cells infected with MIEG3-BCL6 plus CD40-ligand retroviruses before culture and after long term culture, showing that only BCL6 plus CD40L double positive cells exhibit long-term proliferation. The dot plots on the left show GFP expression along with staining with a PE-conjugated isotype control antibody, while the dot plots on the right show GFP expression plus staining with anti-human CD4 antibody.
Figure 2. Polyclonality of BCL6 transformed cell lines.
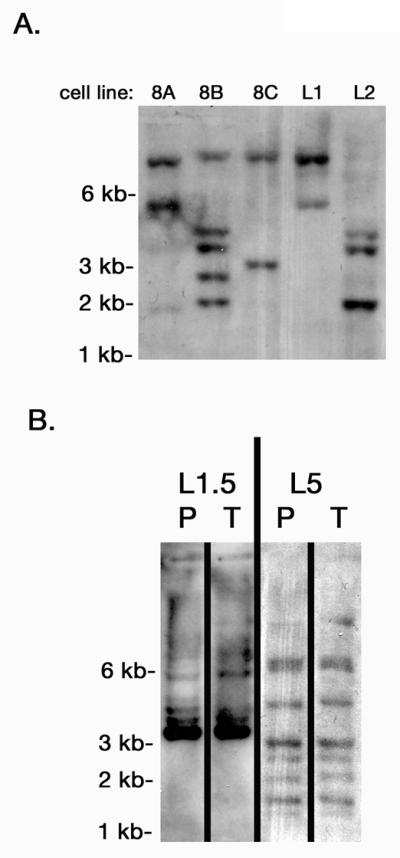
Southern blots with EcoRI-digested genomic DNA from the indicated BL B cell lines were probed with a 32P-labeled DNA fragment derived from the GFP gene of the pMIEG3-BCL6 vector. The different sized bands show different retroviral insertions representing unique B cell clones. A. Five different BL B cell lines. B. Two different BL B cell lines showing similar clones in the parental cell line (P) as in the cells isolated from a tumor (T) derived from the parental cell line.
BL B cells grow as tumors in mice and thus represent true transformed B cells. Three different BL B cell lines were injected intraperitoneally into Rag1-/- (lymphocyte cell deficient) mice, 3 mice per cell line, 5 × 106 cells per mouse. Two to four weeks later, tumors were visible on all injected mice (9/9), and large numbers of tumor cells were recovered from either the ascites or the spleen. Figure 2B shows that tumors cells recovered from mice show very similar polyclonality as the input cells, indicating that multiple clones within each BL B cell line are transformed and can grow as tumor cells.
In order to understand how BCL6 cooperated with CD40L to transform primary p53-deficient B cells, we assessed survival and cell cycle progression in purified ML and BL cells following 24 hours in cell culture (Figure 3). We observed that expression of BCL6 led to a significant increase in the survival of the cultured cells, while in contrast BCL6 did not dramatically alter cell cycle progression of the B cells. We did observe a slight increase in cells in the G2/M phase with BCL6 plus CD40L.
Figure 3. BCL6 enhances B cell survival but not cell cycle progression early in BL cultures.
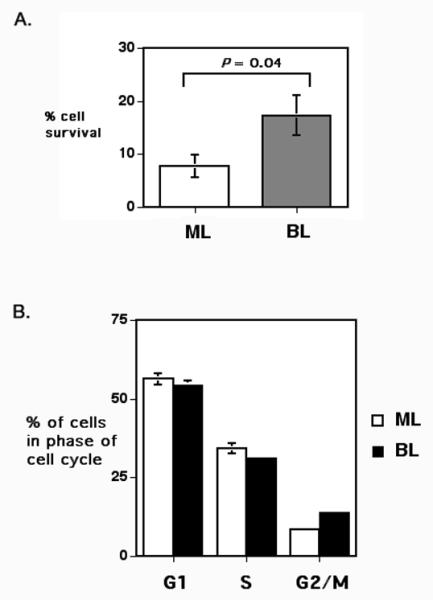
Double positive cells from ML and BL retrovirus-infected cultures were isolated by fluorescence activated cell sorting, as described in Figure 1. Following sorting, the cells were placed in culture for 24 h and then analyzed for viability (A) and DNA content (B). The data shown are the average of three independent experiments. Viable cells were assessed via Trypan blue exclusion and counted under a light microscope. Cellular DNA content was measured by flow cytometry following Propidium Iodide staining of permeabilized Rnase-treated cells.
To understand the function of BCL6 in these cells at the molecular level, and how BCL6 influenced cell survival, we used Q-PCR to assess the expression of several well-characterized BCL6 target genes in purified ML and BL cells after 24 hours of culture (Figure 4A) 18-26. We analyzed BCL6-mediated repression of seven target genes in the presence of CD40L (Figure 4A). Q-PCR analysis revealed that many BCL6 targets were not repressed in the presence of CD40L. Of the seven BCL6 target genes tested, only ATR and p27kip1 were repressed by BCL6 in presence of CD40L signaling. Previous data have shown that CD40 signaling can interfere with BCL6-mediated repression 27, and our data indicate that this interference varies for different target genes. More importantly, our data suggest that only p27kip1 and ATR are critical BCL6 target genes for the early transforming effects of BCL6 in primary B cells. To further define the target genes involved in B cell transformation by BCL6, we analyzed the expression of the same set of target genes in a panel of BL cell lines (Figure 4B). We found that in the outgrown transformed cell lines, there was decreased expression of other BCL6 target genes, such as Blimp1 and PDCD2, as well as larger decreases in the expression of p27kip1 and ATR. These data indicate that cells expressing decreased levels of ATR, Blimp1, p27kip1 and PDCD2 have a competitive growth advantage and/or a more stable growth phenotype.
Figure 4. BCL6 target gene expression in early BL cultures and transformed BL B cell lines.
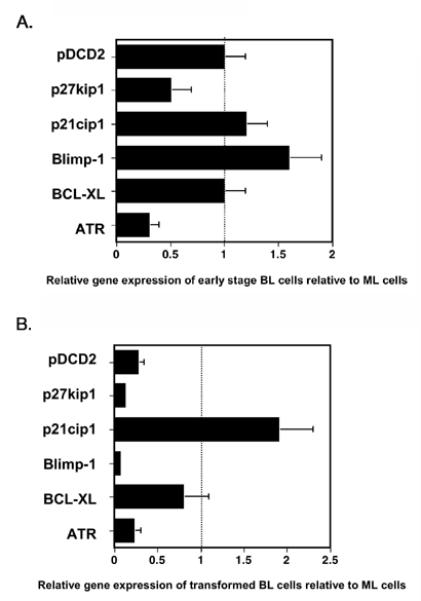
Relative mRNA expression was measured by real-time quantitative PCR (QPCR). For each cell type, mRNA expression was initially normalized to beta-tubulin expression levels. Graph show relative mRNA levels in the BCL6-expressing cells with the mRNA levels in ML cells set as “1” for each individual gene. A. ML cells prepared as above were cultured for 24 hr following sorting for GFP and hCD4 expression, and then used in the QPCR analysis as a control for 24 hr cultured BL cells. Data shown is representative of two independent experiments. Error bars represent standard error from triplicate QPCR reactions. B. The mRNA levels of transformed BL B cell lines are shown relative to mRNA levels of ML B cells. Gene expression data was averaged from at least six different transformed BL cell lines, and error bars represent the variability in expression between cell lines expressed as standard error.
Discussion
The mechanism by which BCL6 acts as an oncogene is critical for understanding the pathogenesis of Diffuse Large Cell Lymphoma, a common and aggressive form of non-Hodgkin’s lymphoma. Although target genes for BCL6 have been characterized in the past few years, still very little is known about how BCL6 functions as an oncogene in primary B cells. Previously we found that BCL6 promoted B cell immortalization 11, and this finding is both reinforced and expanded upon in this study. We find that the immortalization function of BCL6 synergizes with the strong growth signals provided by CD40L, apparently by inhibiting the senescence induced by strong CD40L signaling alone. Previous studies have found relationship between BCL6 and the CD40 pathway. Saito et al found that CD40 signaling in B cells normally down-regulates BCL6 expression, and that this pathway is disrupted in lymphomas with certain mutations in the BCL6 promoter 28. Polo et al found a post-translational effect of CD40 signaling on BCL6, where a signaling cascade induced by CD40 blocked the transcriptional repression activity of BCL6 27. In our study, we are using retrovirus-expressed BCL6, which is resistant to the effects of CD40 signaling on normal BCL6 transcription. However, the post-translational effect of CD40 signaling on BCL6 function may affect our B cell transformation system by decreasing the overall transcriptional repression activity of BCL6. Alternatively, the B cells that become transformed in our system may develop a way to circumvent the CD40-mediated disruption of BCL6 activity and thus allow the critical transformation functions of BCL6 to operate. Further studies will be required to address this issue.
In this study, we have presented the first evidence for the role of specific target genes in primary B cell transformation by BCL6. We have found that of several target genes involved in B cell differentiation, cell cycle progression, and cell survival, only ATR and p27kip1 were strongly regulated by BCL6 early in the transformation process. ATR is a protein that induces cell cycle check-points in response to DNA damage and also regulates apoptosis. Repression of ATR in our system indicates that repression of ATR-mediated function on cell cycle and apoptosis is a key function for BCL6 in its role as oncogene 25, 26. BCL6-mediated repression of the p27kip1 cell cycle inhibitor also shows the importance of repressing cell cycle inhibitors for BCL6 oncogene function. Our data also indicate that repression of Blimp1 and PDCD2 are important for the transformation process. Blimp1 is a transcription factor that promotes terminal differentiation of B cells into plasma cells as well as inhibits cell cycle progression 20, 21. PDCD2 is a pro-apoptotic protein, and thus repression of PDCD2 by BCL6 should enhance survival of the transformed cell lines 23.
Although we have found that BCL6 represses genes that inhibit cell cycle progression, we did not observe a large increase in cells in the S or G2/M phases of the cell cycle with BL cells. We did observe a small increase in the G2/M phase with BL cells compared to ML cells, which may be explained by repression of p27kip1 and ATR. However, we observed a more dramatic increase in overall cell survival with BCL6. These data may indicate that repression of p27kip1 and ATR by BCL6 allows B cells to survive that would otherwise die due to cell cycle inhibition, up-regulation of p53 and induction of apoptosis, leading to cell transformation. Curiously, however, we did not observe significant repression of the pro-apototic gene PDCD2 in these early BCL6-expressing B cells.
Besides these insights into the mechanism of transformation by BCL6, we have developed a novel in vitro system for study of the transforming function of BCL6 in primary B cells. Unlike transgenic systems for studying the transforming potential of BCL6, our in vitro system transforms B cells in a relatively short amount of time and relies on cells with known genetic alterations. The in vitro nature of our system allows for easy manipulation, and the rapid growth in our system has benefits over transgenic systems in which BCL6-driven B cell lymphomas develop relatively late in life and have one or more uncharacterized genetic mutations. Thus, our system will be useful to further characterize the oncogenic function of BCL6 in transforming primary B cells.
Acknowledgements
We would like to thank Dr. Mark Kaplan for the gift of the human CD4-expressing retrovirus construct.
This work was supported by National Institutes of Health Grant AI46410 to A.L.D. A.L.D. also receives support from the Indiana Genomics Initiative (INGEN) of Indiana University. INGEN is supported in part by Lilly Endowment Inc.
References
- 1.Onizuka T, Moriyama M, Yamochi T, Kuroda T, Kazama A, Kanazawa N, Sato K, Kato T, Ota H, Mori S. BCL-6 gene product, a 92- to 98-kD nuclear phosphoprotein, is highly expressed in germinal center B cells and their neoplastic counterparts. Blood. 1995;86:28–37. [PubMed] [Google Scholar]
- 2.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 3.Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–68. [PubMed] [Google Scholar]
- 4.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–48. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–20. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 6.Staudt LM, Dent AL, Shaffer AL, Yu X. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int Rev Immunol. 1999;18:381–403. doi: 10.3109/08830189909088490. [DOI] [PubMed] [Google Scholar]
- 7.Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large- cell lymphoma. Science. 1993;262:747–50. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 8.Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. Embo J. 1995;14:6209–17. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–55. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Baron BW, Anastasi J, Montag A, Huo D, Baron RM, Karrison T, Thirman MJ, Subudhi SK, Chin RK, Felsher DW, Fu YX, McKeithan TW, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci U S A. 2004;101:14198–203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusam S, Vasanwala FH, Dent AL. Transcriptional repressor BCL-6 immortalizes germinal center-like B cells in the absence of p53 function. Oncogene. 2004;23:839–44. doi: 10.1038/sj.onc.1207065. [DOI] [PubMed] [Google Scholar]
- 12.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 13.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 Expression Delineates Heterogeneity in Primary Th2 Cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Margalit O, Amram H, Amariglio N, Simon AJ, Shaklai S, Granot G, Minsky N, Shimoni A, Harmelin A, Givol D, Shohat M, Oren M, et al. BCL6 is regulated by p53 through a response element frequently disrupted in B-cell non-Hodgkin lymphoma. Blood. 2006;107:1599–607. doi: 10.1182/blood-2005-04-1629. [DOI] [PubMed] [Google Scholar]
- 15.Clodi K, Asgary Z, Zhao S, Kliche KO, Cabanillas F, Andreeff M, Younes A. Coexpression of CD40 and CD40 ligand in B-cell lymphoma cells. Br J Haematol. 1998;103:270–5. doi: 10.1046/j.1365-2141.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 16.Pham LV, Tamayo AT, Yoshimura LC, Lo P, Terry N, Reid PS, Ford RJ. A CD40 Signalosome anchored in lipid rafts leads to constitutive activation of NF-kappaB and autonomous cell growth in B cell lymphomas. Immunity. 2002;16:37–50. doi: 10.1016/s1074-7613(01)00258-8. [DOI] [PubMed] [Google Scholar]
- 17.Pham LV, Tamayo AT, Yoshimura LC, Lin-Lee YC, Ford RJ. Constitutive NF-{kappa}B and NFAT activation in aggressive B cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood. 2005 doi: 10.1182/blood-2005-03-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192:1841–8. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 20.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–20. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 22.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–60. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 23.Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu A, Zeleznik-Le N, McKeithan TW. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci U S A. 2002;99:2860–5. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang TT, Dowbenko D, Jackson A, Toney L, Lewin DA, Dent AL, Lasky LA. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J Biol Chem. 2002;277:14255–65. doi: 10.1074/jbc.M110901200. [DOI] [PubMed] [Google Scholar]
- 25.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–14. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 26.Ranuncolo SM, Wang L, Polo JM, Dell’Oso T, Dierov J, Gaymes TJ, Rassool F, Carroll M, Melnick A. BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. J Biol Chem. 2008;283:22565–72. doi: 10.1074/jbc.M803490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polo JM, Ci W, Licht JD, Melnick A. Reversible disruption of BCL6 repression complexes by CD40 signaling in normal and malignant B cells. Blood. 2008;112:644–51. doi: 10.1182/blood-2008-01-131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–92. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]


