Abstract
Abstract The novel H1N1 influenza virus that emerged in humans in Mexico in early 2009 and transmitted efficiently in the human population with global spread has been declared a pandemic strain. Here we review influenza infections in swine since 1918 and the introduction of different avian and human influenza virus genes into swine influenza viruses of North America and Eurasia. These introductions often result in viruses of increased fitness for pigs that occasionally transmit to humans. The novel virus affecting humans is derived from a North American swine influenza virus that has acquired two gene segments [Neuraminidase (NA) and Matrix (M)] from the European swine lineages. This reassortant appears to have increased fitness in humans. The potential for increased virulence in humans and of further reassortment between the novel H1N1 influenza virus and oseltamivir resistant seasonal H1N1 or with highly pathogenic H5N1 influenza stresses the need for urgent pandemic planning.
Keywords: pandemic, H1N1, influenza, swine, zoonoses
Introduction
Influenza A viruses are a genus of the family Orthomyxoviridae. They are RNA viruses with a segmented genome that is comprised of eight negative‐sense, single‐stranded RNA segments. These eight segments encode eleven proteins. The polymerase complex includes the PB2, PB1, and PA proteins as well as the nucleoprotein (NP). There are two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). Matrix protein 1 (M1) is the structural protein of the virus particle while matrix protein 2 (M2) forms an envelope‐spanning proton channel. The genome also encodes two non‐structural proteins (NS1 and NS2) as well as an alternate reading frame protein from PB1 known as PB1‐F2. 1
While their natural reservoir is aquatic birds, influenza A viruses have the ability to infect a number of host species, including swine and humans. Influenza A viruses do exhibit host specificity, but this restriction is not absolute. Most of the time, influenza A viruses in different species evolve along separate paths, but occasionally there is interaction between viruses of different lineages. 1
Because of the segmented nature of the influenza A virus genome, these viruses are able to undergo reassortment if a single cell is concurrently infected with more than one virus. These reassortment events can dramatically change the evolution of influenza A viruses in a certain host. The two most recent human pandemic influenza A viruses were created through the reassortment of human and avian viruses. In 1957, a novel human‐avian H2N2 reassortant virus emerged in the human population. This virus contained the HA, NA, and PB1 from an avian influenza virus, while the remaining gene segments were from the circulating human H1N1. In 1968, a reassortant H3N2 virus emerged that had a novel HA and PB1 from an avian virus and the remaining gene segments from the previously circulating H2N2. 1
Swine play an important role in the ecology of influenza A viruses because they are susceptible to viruses of both the avian and mammalian lineages. The cells of the swine respiratory tract contain receptor sialyloligosaccharides possessing both N‐acetylneuraminic acid‐α2,3‐galactose, which is the preferred receptor for avian influenza viruses and N‐acetylneuraminic acid‐α2,6‐galactose, which is the preferred receptor for mammalian influenza viruses. 2 , 3 This has led to the proposal that swine serve as a “mixing vessel” for influenza viruses of different lineages, providing a place for reassortment and host adaptation to take place. 4
Swine influenza in North America
Reassortment has played a pivotal role in the evolution of influenza A viruses in swine. Influenza was first described as a disease of swine in 1918 5 and the first influenza A virus was isolated from swine in 1930 (Table 1). This original isolate was of the H1N1 subtype and was from the same lineage as the 1918 pandemic virus. This virus entered the human and swine population around the same time and evolved independently in each host. 6 This “classical” swine H1N1 virus continued to circulate as the dominant influenza virus in the North American swine population until 1998. It caused predictable seasonal peaks of disease and was not of great economic concern. 7 While classical H1N1 was the dominant virus, human H3N2 viruses were detected by serology at very low levels, but these viruses never became established in the swine population. 8
Table 1.
Novel influenza A viruses isolated from swine
| Subtype | Origin | Year initally isolated | Country isolated | Reference |
|---|---|---|---|---|
| H1N1 | Swine | 1930 | United States | (6) |
| H3N2 | Human | 1970 | Taiwan | (27) |
| H3N2 | Human | 1977 | Italy | (22) |
| H3N2 | Human | 1977 | China (Hong Kong) | (30) |
| H1N1 | Avian | 1979 | Italy | (23) |
| H1N2 | Swine/human | 1980 | Japan | (55) |
| H3N2 | Swine/human/avian | 1982 | China (Hong Kong) | (56) |
| H3N2 | Human/avian | 1983 | Italy | (24) |
| H1N7 | Human/equine | 1992 | Great Britain | (57) |
| H1N1 | Avian | 1993 | China | (53) |
| H1N2 | Swine/human | 1994 | Great Britain | (25) |
| H3N2 | Human | 1997 | Canada | (58) |
| H3N2 | Swine/human | 1998 | United States | (9) |
| H3N2 | Swine/human/avian | 1998 | United States | (9) |
| H9N2 | Avian | 1998 | China (Hong Kong) | (28) |
| H1N2 | Reassortant swine/swine | 1999 | United States | (13) |
| H4N6 | Avian | 1999 | Canada | (59) |
| H3N2 | Swine/human | 1999 | China (Hong Kong) | (56) |
| H1N1 | Reassortant swine/swine | 2001 | United States | (15) |
| H3N3 | Avian | 2001 | Canada | (20) |
| H1N1 | Avian | 2002 | Canada | (20) |
| H9N2 | Avian/avian | 2003 | China | (37) |
| H1N2 | Human | 2003 | Canada | (12) |
| H1N2 | Reassortant swine/swine | 2004 | China (Hong Kong) | (33) |
| H5N1 | Avian | 2004 | China | (38) |
| H3N1 | Reassortant swine/reassortant swine | 2004 | United States | (17) |
| H1N2 | Reassortant swine/human | 2004 | Canada | (12) |
| H1N1 | Reassortant swine/human | 2005 | United States | (14) |
| H3N8 | Equine | 2005 | China | (60) |
| H1N2 | Reassortant swine/reassortant swine | 2005 | Germany | (26) |
| H1N1 | Human | 2006 | China | (31) |
| H3N1 | Reassortant swine/swine | 2006 | Italy | (61) |
| H2N3 | Reassortant swine/avian | 2006 | United States | (16) |
| H5N1 | Avian | 2006 | Indonesia | (40) |
| H3N1 | Human/swine | 2006 | Korea | (62) |
| H5N2 | Avian | 2008 | Korea | (63) |
| H5N2 | Swine/avian | 2008 | Korea | (63) |
| H1N1 | Reassortant Swine/avian | 2009 | Canada | (64) |
In late 1998, two distinct H3N2 influenza viruses were isolated from swine with severe influenza‐like illness in North Carolina, Minnesota, Iowa, and Texas. The isolate from North Carolina (double reassortant) was a reassortant virus containing HA, NA, and PB1 similar to those of contemporary human H3N2 influenza viruses and M, NP, NS, PA, and PB2 similar to those of the classical H1N1 swine influenza virus lineage. 9 The isolates from Minnesota, Iowa, and Texas (triple reassortant) were even more complex. Like the North Carolina isolate, these viruses contained HA, NA, and PB1 of a contemporary human virus lineage and M, NP, and NS of the classical swine H1N1 lineage. However, these isolates contained PA and PB2 of an avian influenza virus lineage. 9 After the emergence of these two reassortant viruses, the double reassortant did not continue to circulate, but the triple reassortant virus became established in the swine population and continued to circulate and evolve. By early 2000, over 20% of sampled pigs were seropositive for the triple reassortant H3N2. 10 Retrospectively, it was shown that the triple reassortant virus was present in pigs as early as March 1998. 11
Variants of the original H3N2 triple reassortant virus continue to co‐circulate in the North American swine population along with several other reassortant viruses (Figure 1). All of the currently circulating viruses contain a genetically similar constellation of internal genes, but have different surface glycoproteins. Isolated swine influenza A viruses include three distinct H3N2 viruses, each with a human‐like HA from a different year, 10 an H1N2 virus with a human‐like HA, 12 an H1N2 virus with a classical swine‐like HA, 13 an H1N1 virus with a human‐like HA and NA, 14 and an H1N1 virus with a classical swine‐like HA and NA. 15 Other reassortant virus subtypes that have been isolated from pigs since 1998, but have not become established in the swine population include an H3N1 and an H2N3. 16 , 17 Several wholly‐avian viruses, including an H4N6, H3N3, and H1N1 have been isolated from swine herds in Canada, but none have spread beyond the initially infected herds. 18 , 19 , 20
Figure 1.
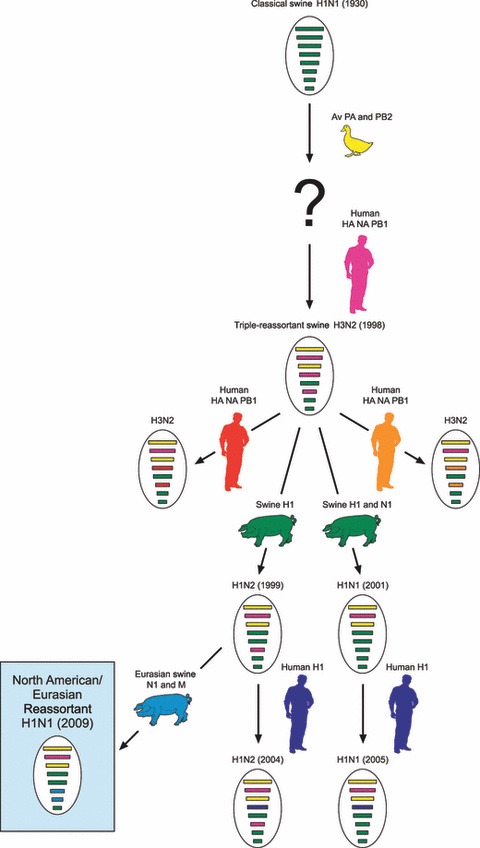
Evolution of swine influenza in North America from 1930‐present. Swine influenza remained relatively stable in the North American swine population from 1930 to 1998. It occasionally crossed over into the human population, but did not spread. After the emergence of the triple reassortant H3N2 viruses, these viruses transmitted to turkeys and continue to circulate in the turkey population. Further reassortment events resulted in two distinct H1N1 reassortants and two distinct H1N2 reassortants. The H1N1 with classical swine H1 and N1 and the H1N2 with human H1 and N2 have been isolated from humans but did not spread. The North American/Eurasian reassortant H1N1 virus is the first swine virus to establish sustained human to human transmission.
Swine influenza in Europe and Asia
The evolution of swine influenza A viruses in Europe and Asia has been distinct from that in North America. Swine influenza was not reported in Europe until 1976, when classical swine H1N1 was detected in pigs in Italy (Table 1). 21 Around the same time, a wholly‐human H3N2 virus was also introduced into the European swine population. 22 In 1979, a wholly avian‐like influenza A virus was isolated from swine in Italy. 23 This avian‐like H1N1 quickly replaced the classical swine H1N1 as the dominant lineage and underwent a reassortment with the human H3N2, resulting in a virus with human‐like HA and NA and avian‐like internal genes. 24 An H1N2 virus that contained a human‐like H1, swine‐like N2, and avian‐like internal genes was isolated from swine in Great Britian in 1994. 25 In 2005, a novel H1N2 that was the result of a reassortment between swine H1N2 and swine H3N2 was isolated in Germany. 26
Most of the European swine influenza viruses also circulate in Asia, but there are several lineages which are only isolated from Asia. Human H3N2 viruses were first isolated from pigs in Taiwan in 1970 27 and continue to circulate in the swine population along with several different reassortant H3N2 viruses. 28 , 29 , 30 Interestingly, viruses with an HA and NA most closely related to the original human H3N2 isolate from 1968 can still be isolated from pigs in China, along with more recent human H3N2 and human H1N1 viruses. 31 In China and Korea, several isolates have been detected that contain gene segments that are genetically similar to those found in the North American swine population. 32 In 2004, an H1N2 virus was isolated in Hong Kong that contained five of the internal genes from the North American triple reassortant swine viruses with a Eurasian lineage H1 and M. 33 In Thailand, several reassortant viruses have been isolated from swine, including an H1N1 that contains the HA and NS from the classical swine H1N1 lineage and the remaining genes from the Eurasian avian‐like swine lineage. An H1N2 reassortant virus with the HA from the classical swine H1N1 lineage and the remaining genes from the Eurasian avian‐like swine lineage was also isolated. 34 , 35 These two reassortant viruses have also been isolated from humans. 36 An avian H9N2 virus was isolated from pigs in Hong Kong in 1998 28 but did not continue to circulate in the pig population of Asia. A separate H9N2 virus reassorted with and obtained the internal genes of an H5N1 virus and both the reassortant and the original virus still circulate. 29 , 37 Avian H5N1 viruses have been isolated sporadically from swine in China and Indonesia, but do not appear to be highly pathogenic in swine and have not become established in the population. 38 , 39 , 40
Influenza zoonoses
The most catastrophic zoonotic influenza event was the H1N1 pandemic of 1918–1919. This virus is proposed to have transmitted directly from birds into humans, although during this time, the disease was also described in the swine population, and it is not known if the virus was transmitted from humans to pigs or from pigs to humans. In the spring of 1918, there was a mild wave of disease, which was followed in the fall by a highly pathogenic wave which killed more than 20 million people worldwide. 41 , 42 , 43 The virus remained in the human population where it evolved separately from the swine H1N1 lineage. It was replaced in the human population by H2N2 in 1957 and was re‐introduced into the human population in 1977. 1
An H1N1 swine influenza virus was isolated from a human in 1974. From 1974 to 2005, there were 43 confirmed cases of transmission of influenza A virus from pigs to humans reported, with six fatalities. Because there are no unique clinical symptoms to differentiate swine influenza from seasonal influenza in humans, this number is probably only a small fraction of the actual cases. Most of these cases were the result of direct exposure to swine or were human‐to‐human transmission within a family cluster. 44 In early 1976, there was an outbreak of severe respiratory disease at Fort Dix, New Jersey. Thirteen soldiers became severely ill, and one died. The cause of the outbreak was determined to be a swine H1N1 virus. Further serological studies showed that over 200 infections occurred, however, within a month, the virus had disappeared from the human population. 45 From December 2005 to February 2009, 11 cases of human infection with swine influenza were reported to the CDC. These infections were all caused by triple reassortant H1N1 or H1N2 swine influenza viruses. 46
In April 2009, two cases of un‐subtypeable influenza A in humans with no known swine contact were reported in southern California. The viruses isolated from these cases were different from the influenza A viruses currently known to be circulating in the North American swine population. The HA and the internal genes were similar to those found in circulating reassortant viruses, but the NA and M were more closely related to the Eurasian avian‐like swine influenza A lineage. 47 The origin of this virus is currently unknown, but it has spread in humans throughout the United States and around the world. As of 24 June 2009, there have been 55 867 reported cases worldwide, with 238 deaths attributed to this virus. 48 The virus was also introduced into the swine population in Canada, but it remains to be seen if it will become established in the swine population there or elsewhere. 49
Conclusions
The successful zoonotic transfer of an influenza A virus containing gene segments of avian, swine and human origin to humans along with consistent human to human transmission on each of the world’s continents fulfills each of the current criteria for a pandemic strain. On 11 June 2009, the WHO raised the pandemic alert from Level 5 to Level 6 and declared the start of an influenza pandemic. 50 At the time of writing the majority of infections in humans were mild but the higher death rate in Mexico and lethal infections in both compromised and uncompromised patients in the United States raises the possibility that the full potential of the virus may not yet have been realized.
The benign nature of influenza in North American swine changed after the acquisition of gene segments from human and avian influenza viruses in 1998 (Figure 1) raising the question of which gene(s) made the virus more active in pigs. The double reassortant that was detected in pigs in 1998 with HA, NA, and PB1 of human influenza origin and M, NP, NS, PA, and PB2 of swine influenza origin was quickly superseded by detection of a triple reassortant that contained gene segments from avian influenza viruses (PA and PB2), swine influenza viruses (M, NP, NS) and human influenza viruses (HA, NA, PB1). 9 It is probable that multiple rounds of reassortment were involved in the genesis of these viruses. Acquisition of this specific internal gene combination provided a high level of fitness for the virus and they spread rapidly through the swine population of North America. 10 Additionally, these viruses had a high propensity to reassort and during the following years generated a range of reassortants that occasionally transmitted to humans but showed little propensity to transmit beyond the index cases. This changed sometime prior to April 2009 when viruses with gene segments (NA, M) from European H1N1 swine‐like influenza viruses were detected in infected humans from the southern United States and subsequently Mexico. 51 , 52 These viruses possessed gene segments similar to those in the H1N1/N2 triple reassortant viruses found in North American swine plus the newly introduced NA and M genes of European swine origin. The likely explanation for the origin of this novel H1N1 influenza virus is that a reassortment event occurred between the North American triple reassortant and the European swine influenza virus. Where this event occurred can only be speculated about. The North American triple reassortant has been reported in Asia where the European swine influenza virus of avian origin has been circulating since 1993. 53 Surveillance of swine influenza viruses in Central and South America has been extremely limited so it is not known if European swine viruses have ever been introduced. Whether reassortment occurred in Asia or America is unknown. Also unknown is the contribution of the NA and M genes of European origin to the increased transmissibility of this virus in humans. At the time of writing 55 867 human cases with 238 deaths have been confirmed. 48 The following are major concerns about this novel pandemic H1N1 virus and the questions that need to be answered:
-
•
Will the virus establish itself in the pig or bird populations of the world?
-
•
Given the propensity of the triple reassortant swine lineage to reassort will it
-
1
acquire the oseltamivir resistance from the seasonal human A/Brisbane/57/2007 (H1N1) like virus?
-
2
reassort with one of the lineages of highly pathogenic H5N1 avian influenza and acquire the high pathogenicity of these viruses while retaining its high human transmissibility?
-
1
-
•
Will the virus (as has been seen with other influenza viruses after zoonotic transfer) show a period of rapid evolution with a high mutation rate at the amino acid level and will this lead to the 2009 H1N1 pandemic virus mimicing the 1918 virus with a mild wave followed by a more pathogenic wave?
-
•
What issues must be resolved prior to the generalized vaccination of humans with the 2009 H1N1 pandemic virus vaccine including
-
1
Safety from the perspective of Guillain–Barre syndrome?
-
2
Immunogenicity of inactivated vaccines and the need for novel adjuvants (MF59, ASO3)?
-
3
The wisdom of using live attenuated vaccines in the face of a pandemic?
-
4
Whether intact inactivated vaccine instead of subunit vaccines would provide greater antigen sparing?
-
1
There are already more questions than answers and it will be a case of being “damned if we do and damned if we don’t” prepare with maximum urgency for a pandemic.
Acknowledgements
This study was supported by Contract HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, and by the American Lebanese Syrian Associated Charities (ALSAC). The authors thank James Knowles with manuscript preparation and Julie Groff for the figure.
References
- 1. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992; 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ito T, Couceiro JN, Kelm S et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 1998; 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virol 1983; 127:361–373. [DOI] [PubMed] [Google Scholar]
- 4. Scholtissek C. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med Principles Pract 1990; 2:65–71. [Google Scholar]
- 5. Koen JS. A practical method for field diagnosis of swine diseases. Am J Vet Med 1919; 14:468–470. [Google Scholar]
- 6. Shope RE. Swine Influenza. III. Filtration experiments and aetiology. J Exp Med 1931; 54:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Easterday BC, Hinshaw VS. Swine Influenza; in Leman AD, Straw BE, Mengeling WL, D’Allaire SD, Taylor DJ. (ed): Diseases of Swine. Ames: Iowa State Press, 1992:349–357. [Google Scholar]
- 8. Chambers TM, Hinshaw VS, Kawaoka Y, Easterday BC, Webster RG. Influenza viral infection of swine in the United States 1988‐1989. Arch Virol 1991; 116:261–265. [DOI] [PubMed] [Google Scholar]
- 9. Zhou NN, Senne DA, Landgraf JS et al. Genetic reassortment of avian, swine, and human influenza A viruses in american pigs. J Virol 1999; 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. Evolution of swine H3N2 influenza viruses in the United States. J Virol 2000; 74:8243–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karasin AI, Schutten MM, Cooper LA et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in north america, 1977‐1999: evidence for wholly human and reassortant virus genotypes [In Process Citation]. Virus Res 2000; 68:71–85. [DOI] [PubMed] [Google Scholar]
- 12. Karasin AI, Carman S, Olsen CW. Identification of human H1N2 and human‐swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J Clin Microbiol 2006; 44:1123–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol 2000; 38:2453–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res 2008; 72:127–154. [DOI] [PubMed] [Google Scholar]
- 15. Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co‐circulate in the United States swine population. Virus Res 2004; 103:67–73. [DOI] [PubMed] [Google Scholar]
- 16. Ma W, Vincent AL, Gramer MR et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 2007; 104:20949–20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma W, Gramer M, Rossow K, Yoon KJ. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J Virol 2006; 80:5092–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karasin AI, Brown IH, Carman S, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol 2000; 74:9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res 2002; 85:199–210. [DOI] [PubMed] [Google Scholar]
- 20. Karasin AI, West K, Carman S, Olsen CW. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol 2004; 42:4349–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nardelli L, Pascucci S, Gualandi GL, Loda P. Outbreaks of classical swine influenza in Italy in 1976. Zentralbl Veterinarmed B 1978; 25:853–857. [DOI] [PubMed] [Google Scholar]
- 22. Castrucci MR, Campitelli L, Ruggieri A et al. Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J Gen Virol 1994; 75(Pt 2):371–379. [DOI] [PubMed] [Google Scholar]
- 23. Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ 1981; 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 24. Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortment between avain and human influenza A viruses in Italian pigs. Virol 1993; 193:503–506. [DOI] [PubMed] [Google Scholar]
- 25. Brown IH, Harris PA, McCauley JW, Alexander DJ. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol 1998; 79(Pt 12):2947–2955. [DOI] [PubMed] [Google Scholar]
- 26. Zell R, Motzke S, Krumbholz A, Wutzler P, Herwig V, Durrwald R. Novel reassortant of swine influenza H1N2 virus in Germany. J Gen Virol 2008; 89(Pt 1):271–276. [DOI] [PubMed] [Google Scholar]
- 27. Kundin WD. Hong Kong A‐2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 1970; 228:857. [DOI] [PubMed] [Google Scholar]
- 28. Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 2001; 75:9679–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu H, Hua RH, Wei TC et al. Isolation and genetic characterization of avian origin H9N2 influenza viruses from pigs in China. Vet Microbiol 2008; 131:82–92. [DOI] [PubMed] [Google Scholar]
- 30. Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science 1977; 196:1454–1455. [DOI] [PubMed] [Google Scholar]
- 31. Yu H, Zhang GH, Hua RH et al. Isolation and genetic analysis of human origin H1N1 and H3N2 influenza viruses from pigs in China. Biochem Biophys Res Commun 2007; 356:91–96. [DOI] [PubMed] [Google Scholar]
- 32. Lee CS, Kang BK, Kim HK et al. Phylogenetic analysis of swine influenza viruses recently isolated in Korea. Virus Genes 2008; 37:168–176. [DOI] [PubMed] [Google Scholar]
- 33. Smith GJ, Vijaykrishna D, Bahl J et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature 2009; 459:1122–1125. [DOI] [PubMed] [Google Scholar]
- 34. Chutinimitkul S, Thippamom N, Damrongwatanapokin S et al. Genetic characterization of H1N1, H1N2 and H3N2 swine influenza virus in Thailand. Arch Virol 2008; 153:1049–1056. [DOI] [PubMed] [Google Scholar]
- 35. Takemae N, Parchariyanon S, Damrongwatanapokin S et al. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respi Viruses 2008; 2:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Komadina N, Roque V, Thawatsupha P et al. Genetic analysis of two influenza A (H1) swine viruses isolated from humans in Thailand and the Philippines. Virus Genes 2007; 35:161–165. [DOI] [PubMed] [Google Scholar]
- 37. Cong YL, Pu J, Liu QF et al. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol 2007; 88(Pt 7):2035–2041. [DOI] [PubMed] [Google Scholar]
- 38. Choi YK, Nguyen TD, Ozaki H et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol 2005; 79:10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu Q, Yang H, Chen W et al. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J Virol 2008; 82:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takano R, Nidom CA, Kiso M et al. A comparison of the pathogenicity of avian and swine H5N1 influenza viruses in Indonesia. Arch Virol 2009; 154:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taubenberger JK, Reid AH, Fanning TG. The 1918 influenza virus: a killer comes into view. Virol 2000; 274:241–245. [DOI] [PubMed] [Google Scholar]
- 42. Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature 2005; 437:889–893. [DOI] [PubMed] [Google Scholar]
- 43. Barry JM, Viboud C, Simonsen L. Cross‐protection between successive waves of the 1918‐1919 influenza pandemic: epidemiological evidence from US Army camps and from Britain. J Infect Dis 2008; 198:1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 2007; 44:1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gaydos JC, Top FH Jr, Hodder RA, Russell PK. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 2006; 12:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shinde V, Bridges CB, Uyeki TM et al. Triple‐reassortant swine influenza A (H1) in humans in the United States, 2005‐2009. N Engl J Med 2009; 360:2616–2625. [DOI] [PubMed] [Google Scholar]
- 47. Novel Swine‐Origin Influenza A (H1N1) Virus Investigation Team . Emergence of a novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 48. Influenza A (H1N1) – update 53. Geneva: World Health Organization, 2009. June 24 [cited 2009 Jun 24];Available from: URL: http://www.who.int/csr/don/2009_06_24/en/index.html. [Google Scholar]
- 49. Influenza A (H1N1) – update 11. Generva: World Health Organization, 2009. May 3 [cited 2009 May 22];Available from: URL: http://www.who.int/csr/don/2009_05_03/en/index.html. [Google Scholar]
- 50. Chen D, Endres RL, Erickson CA et al. Epidermal immunization by a needle‐free powder delivery technology: immunogenicity of influenza vaccine and protection in mice. Nat Med 2000; 6:1187–1190. [DOI] [PubMed] [Google Scholar]
- 51. Centers for Disease Control and Prevention . Swine influenza A (H1N1) infection in two children – Southern California, March–April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:400–402. [PubMed] [Google Scholar]
- 52. Centers for Disease Control and Prevention . Outbreak of swine‐origin influenza A (H1N1) virus infection – Mexico, March–April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:467–470. [PubMed] [Google Scholar]
- 53. Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol 1996; 70:8041–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Influenza A (H1N1) – Update 38. Geneva: World Health Organization, 2009. May 29 [cited 2009 May 25];Available from: URL: http://www.who.int/csr/don/2009_05_25/en/index.html. [Google Scholar]
- 55. Nerome K, Ishida M, Oya A, Oda K. The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol 1982; 62(Pt 1):171–175. [DOI] [PubMed] [Google Scholar]
- 56. Yu H, Hua RH, Zhang Q et al. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol 2008; 46:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brown IH, Alexander DJ, Chakraverty P, Harris PA, Manvell RJ. Isolation of an influenza A virus of unusual subtype (H1N7) from pigs in England, and the subsequent experimental transmission from pig to pig. Vet Microbiol 1994; 39:125–134. [DOI] [PubMed] [Google Scholar]
- 58. Carman S, Stansfield C, Weber J, Bildfell R, Van Dreumel T. H3N2 influenza A virus recovered from a neonatal pig in Ontario‐‐1997. Can Vet J 1999; 40:889–890. [PMC free article] [PubMed] [Google Scholar]
- 59. Karasin AI, Olsen CW, Brown IH, Carman S, Stalker M, Josephson G. H4N6 influenza virus isolated from pigs in Ontario. Can Vet J 2000; 41:938–939. [PMC free article] [PubMed] [Google Scholar]
- 60. Tu J, Zhou H, Jiang T et al. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch Virol 2009; 154:887–890. [DOI] [PubMed] [Google Scholar]
- 61. Moreno A, Barbieri I, Sozzi E et al. Novel swine influenza virus subtype H3N1 in Italy. Vet Microbiol 2009; doi:DOI: 10.1016/j.vetmic.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 62. Shin JY, Song MS, Lee EH et al. Isolation and characterization of novel H3N1 swine influenza viruses from pigs with respiratory diseases in Korea. J Clin Microbiol 2006; 44:3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee JH, Pascua PN, Song MS et al. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J Virol 2009; 83:4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Influenza A (H1N1) – update 11. Geneva: World Health Organization, 2009. May 3 [cited 2009 Jun 1];Available from: URL: http://www.who.int/csr/don/2009_05_03/en/index.html. [Google Scholar]


