Abstract
Morphogenesis of the mammalian secondary palate requires coordination of cell migration, proliferation, differentiation, apoptosis and synthesis of extracellular matrix molecules by numerous signal transduction pathways. Recent evidence suggests a role for members of the Wnt family of secreted cytokines in orofacial development. However, no study has systematically or comprehensively examined the expression of Wnts in embryonic orofacial tissue. We thus conducted a survey of the expression of all known Wnt genes in the developing murine secondary palate. Using an RT-PCR strategy to assay gene expression, 12 of the 19 known members of the Wnt family were found to be expressed in embryonic palatal tissue during key phases of its development. The expression of 5 Wnt family members was found to be temporally regulated. Moreover, these Wnts had unique spatio-temporal patterns of expression which suggested possible roles in palatal ontogeny.
Keywords: Wnt, embryo, orofacial, palate, gene expression
Introduction
Development of the mammalian secondary palate requires the precise coordination of many morphogenetic and cellular processes, the failure of any one of which can result in cleft palate, one of the most prevalent birth defects in humans (Martin et al., 2005). In mice, the palatal shelves originate bilaterally from the oral aspect of the maxillary processes on embryonic day (E) 11.5, growing vertically alongside the tongue on E12.5–E13.5. Subsequent elevation and fusion of the palatal shelves with each other above the tongue on E14.5–E15.5 forms the definitive secondary palate, separating the oral and nasal cavities. Proper development of the palate requires precise spatio-temporal orchestration of cellular apoptosis, proliferation and differentiation. In addition, epithelial to mesenchymal transition (EMT) and apoptosis are fundamental processes critical to morphogenesis in general, and palatal ontogenesis specifically. However, the role of EMT vs. apoptosis in elimination of the medial edge epithelial seam formed when the palatal shelves adhere remains controversial (Fitchett and Hay, 1989; Shuler et al., 1991; Shuler et al., 1992; Nawshad and Hay, 2003; Cuervo and Covarrubias, 2004; Vaziri et al., 2005). Several signaling proteins have been shown to provide regulation of these processes in palatal tissue: TGFβ (Proetzel et al., 1995), BMP (Liu et al., 2005), FGF (Rice et al., 2004), and Notch (Casey et al., 2006). The Wnt family of secreted cytokines have also emerged as functionally important signaling molecules during craniofacial development (Ikeya et al., 1997; Brault et al., 2001; Niemann et al., 2004; Lan et al., 2006; Lee et al., 2008). Currently, 19 genes constitute the vertebrate Wnt family that is divided into at least two distinct classes based upon their downstream activities. Wnts that stimulate the canonical, or β-catenin/TCF/LEF-dependent, pathway include Wnts-1, -3a, and -8, while Wnts-4, -5a and -11 activate a JNK-dependent planar cell polarity pathway (Du et al., 1995). Both pathways involve binding of Wnts to a G-protein coupled plasma membrane receptor, Frizzled (Fz), and require Dishevelled (Dvl) to couple membrane signaling events to mediators of intracellular signaling. Yet a third pathway activated by Wnts is calcium-dependent (Slusarski et al., 1997). Recent studies have suggested that some Wnt family members may play a functional role in orofacial morphogenesis (Brown et al., 2003; Blanton et al., 2004; Juriloff et al., 2006; Lan et al., 2006).
Therefore, to begin to address the significance of Wnts in development of the secondary palate, an evaluation of their expression in this tissue was conducted.
Results
In order to determine the expression of Wnts in the developing secondary palate, RNA was purified from microdissected murine embryonic palatal tissue on embryonic days (E) 12.5, 13.5, and 14.5 and cDNAs prepared for use in semi-quantitative real-time PCR assays Real-time PCR reactions were performed with commercially available probe:primer sets and the signal from each probe:primer set was normalized to that from glyceraldehyde phosphate dehydrogenase (GAPDH). For these experiments, fold-changes of at least 2.0, relative to expression on E12.5, were considered to be significant. Of the 19 Wnt genes tested, 5 underwent significant temporal regulation. The expression of Wnts-2, 10a and 16 was significantly increased on E13.5 when compared to their expression on E12.5, while the expression of Wnts-2, 4, 10a, 10b and 16 was significantly increased on E14.5 when compared to their expression on E12.5 (Table 1). Wnts whose expression was detected but fell below the 2.0 fold-change threshold included Wnts-2b, 5a, 5b, 6, 7b, 9a and 11. Finally, Wnts-1, 3, 3a, 7a, 8a, 8b, and 9b were categorized as being “not detected” because their average threshold values (CT) were >35 and thus below the detection limits of the assay. Therefore, 12 of 19 Wnts were expressed in the developing palate, with 5 exhibiting significant temporal increases in expression.
Table 1.
TEMPORAL EXPRESSION OF WNTs IN MURINE EMBRYONIC SECONDARY PALATAL TISSUE
| Gene | E13.5 | E14.5 |
|---|---|---|
| Wnt-2 | 2.0 ± 0.3 | 3.0 ± 0.5 |
| Wnt-4 | 1.1 ± 0.1 | 2.2 ± 0.3 |
| Wnt-10a | 4.0 ± 0.9 | 5.6 ± 0.4 |
| Wnt-10b | 1.5 ± 0.4 | 3.1 ± 0.6 |
| Wnt-16 | 3.8 ± 0.5 | 3.2 ± 0.5 |
Real-time PCR was performed on cDNAs generated from RNA purified from microdissected murine embryonic palatal tissue on E12.5, E13.5, and E14.5 using primers for the indicated gene. Signals were normalized to that from GAPDH. Fold-change was calculated using the relationship, fold change = 2−ΔΔCt. Data presented represent fold-increases in expression (relative to E12.5) ± standard deviation (n=3–4).
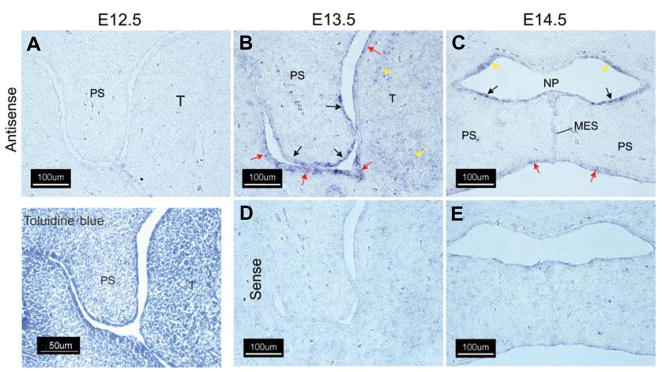
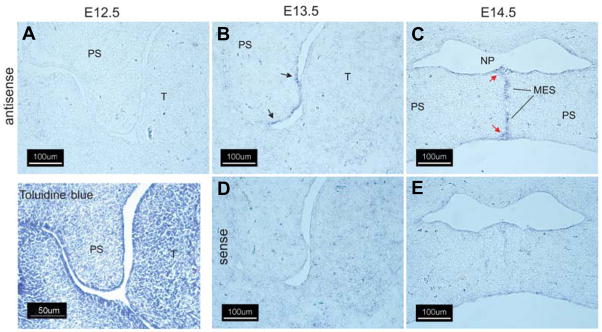
The spatial expression pattern of the Wnts reported in Table 1 was determined by sectional in situ hybridization using specific riboprobes on coronal sections of embryonic mouse heads from E12.5–E14.5 embryos (Figs. 1–5). Wnts that exhibited altered temporal patterns of expression were selected for in situ analysis since these expression patterns may reflect changes in expression in localized cell populations that may in turn provide insight into their developmental function. Wnt-10a underwent one of the largest changes in gene expression on E13.5 and E14.5 when compared to E12.5 (4.0- and 5.6-fold, respectively, Table 1). In situ hybridization with a Wnt-10a-specific riboprobe confirmed its expression in the developing palate (Fig. 1). While Wnt-10a mRNA was not detected in the palatal shelf on E12.5 (Fig. 1A), specific signals were observed in the palatal shelf epithelium on E13.5 (Fig. 1B, black arrows) and in the epithelium of the oronasal cavity and tongue (red arrows). Diffuse staining was also observed in tongue mesenchyme (Fig. 1B, yellow arrows). Following palatal shelf elevation on E14.5, expression of Wnt-10a predominated in the oral and nasal epithelia of the secondary palate (Fig. 1C, red and black arrows, respectively) and in the epithelial lining of the nasopharynx (yellow arrows). No staining was observed when a Wnt-10a sense riboprobe was used (Fig. 1D,E). As with Wnt-10a, Wnt-10b was not detected in sections derived from E12.5 embryos (Fig. 2A). On E13.5, Wnt-10b was expressed in palatal shelf epithelia in a domain that included the future nasal epithelium and the medial edge epithelium (Fig. 2B, black arrows). On E14.5, Wnt-10b was expressed primarily in the medial edge epithelial seam (MES) and epithelial triangles that are present during removal of the MES (red arrows). Strong staining of Wnt-10b mRNA in the MES from palatal shelves of C57BL/6 mouse embryos has also been observed (Dr. Jixiang Ding, personal communication). No staining was observed when a Wnt-10b sense riboprobe was used (Fig. 2D,E).
Fig. 1. Expression of Wnt-10a in the murine secondary palate.
Cryostat sections of fixed, frozen heads from mice on days E12.5–E14.5 were processed for expression of Wnt-10 transcripts by in situ hybridization with either an antisense Wnt-10a-specific riboprobe (A–C) or the sense (negative) control (D,E). An E12.5 coronal section stained with 0.05% (w/v) Toluidine Blue O is included for orientation. No expression of Wnt 10a mRNA was detected on E12.5 (A). On E13.5, expression of Wnt 10a was noted in both the future oral and nasal palatal epithelium and in the medial edge epithelium (B) (black arrows). Expression was also detected in the epithelium of the oral cavity and tongue (B, red arrows). Diffuse expression was observed in tongue mesenchyme on E13.5 (B, yellow arrows). Following shelf elevation and initiation of fusion on E14.5 (C,E), Wnt-10a expression was predominant in palatal nasal epithelium (C, black arrows) in the epithelium lining the nasopharynx (C, yellow arrows) and in the oral epithelium of the palate (C, red arrows). A positive signal was also observed in the medial edge epithelial seam (C, MES,). Abbreviations for all figures are: MES, medial edge seam; NP, nasopharynx; PS, secondary palatal shelf; T, tongue. All panels, 200X magnification.
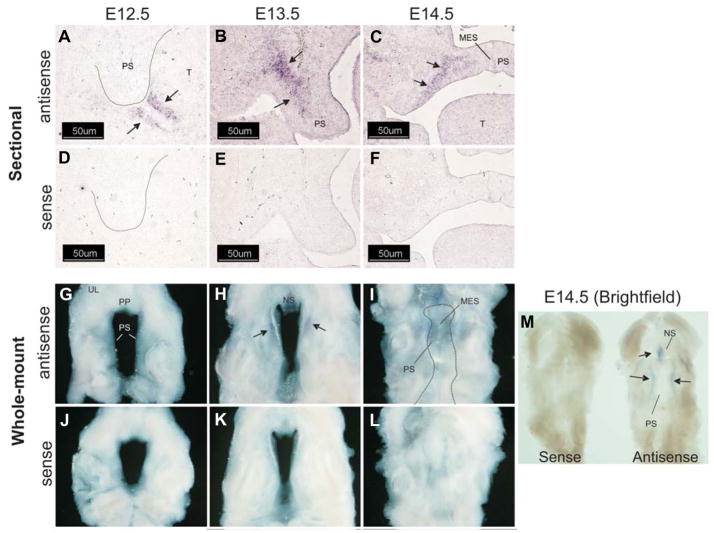
Fig. 5. Whole-mount and sectional in situ hybridization analysis of Wnt-16 expression in the secondary palate.
For sectional in situ hybridization, 12 μm thick coronal frozen sections of embryonic heads on E12.5–E14.5 were prepared and adjacent sections hybridized with an antisense riboprobe (A–D) (for each gestation day) or a control, sense riboprobe (E–H) (for each gestation day). Representative sections were taken from various positions along the anterior-posterior (A–P) axis, as indicated by the corresponding line in panel (I). On E12.5, Wnt-16 was detected in palate mesenchyme along the entire A–P axis, with the expression domain becoming progressively restricted on E13.5 and E14.5. Black arrows indicate specific regions of Wnt-16 expression. These results were confirmed by an independent analysis on intact tissue (I,J) that demonstrated the temporally diminishing domain of Wnt-16 mRNA expression from E12.5 to E14.5. Only a small expression domain remained in the presumptive soft palate on E14.5. For orientation, the palatal shelves are outlined in panel J of each gestation day. All panels, 200X magnification.
Fig. 2. Expression of Wnt-10b in the murine secondary palate.
Cryostat sections of fixed, frozen heads from mice on days E12.5–E14.5 were processed for expression of Wnt-10b transcripts by in situ hybridization with either an antisense Wnt-10b-specific riboprobe (A–C) or the sense (negative) control (D,E). On E12.5, as with Wnt-10a (Fig. 1), there was no detectable expression of Wnt-10b in the palatal shelves (A). On E13.5, expression of Wnt-10b was seen primarily in the epithelium of the palatal shelf (arrows). On E14.5, expression was detected in the disintegrating medial edge seam; compare (C) with (E). Note expression in the epithelial triangles (red arrows). All panels, 200X magnification.
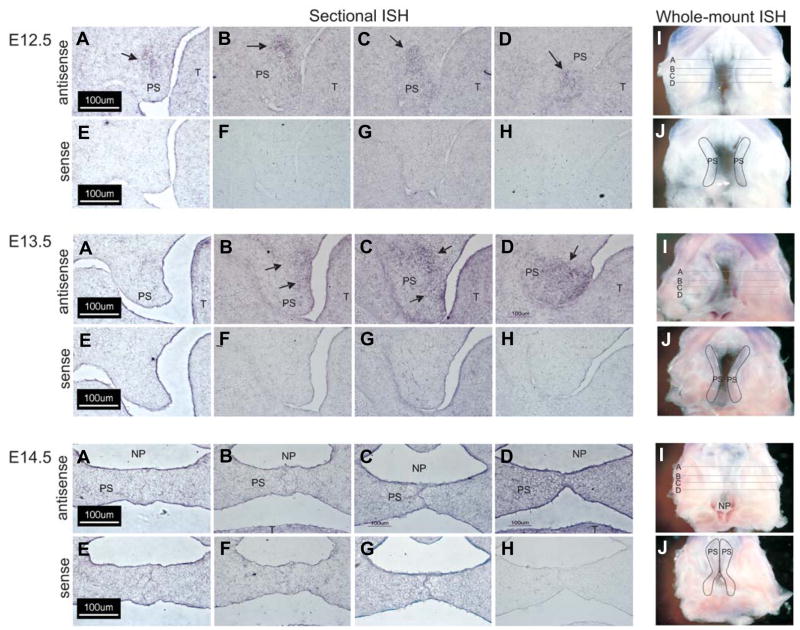
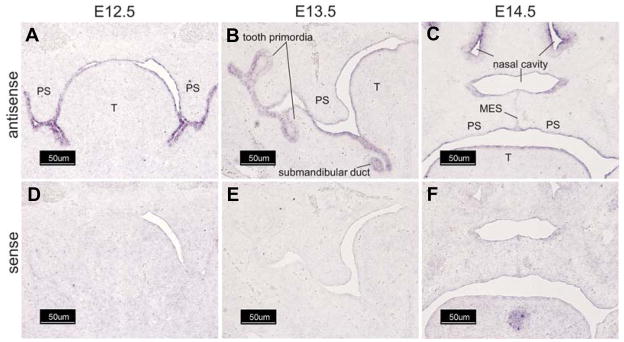
The expression of Wnt-2 in the embryonic palate increased by 2.0- and 3.0-fold on E13.5 and E14.5, respectively (Table 1). In situ hybridization with a Wnt-2 specific riboprobe demonstrated no detectable expression in palatal tissue on E12.5 (Fig. 3A). Wnt-2 expression was detected in a limited domain within the mesenchyme underlying the tongue and oral cavity on E12.5 (Fig. 3A, black arrows). On E13.5 and 14.5, expression of Wnt-2 was detected in the mesenchyme of the palatal shelf (Fig. 3B and 3C, black arrows). On E14.5, expression of Wnt-2 was restricted to a sharply defined domain (Fig 3C, black arrows) medial to the developing maxilla (as assessed by alkaline phosphatase staining, data not shown). In contrast to Wnt-10a and Wnt-10b, there was no evidence of Wnt-2 expression in epithelial cells. Because the expression pattern of Wnt-2 in the secondary palate suggested medial-lateral and antero-posterior differences in staining, we performed whole-mount in situ hybridization with a Wnt-2 riboprobe (Fig. 3G–M). This experiment revealed that Wnt-2 is expressed in a limited domain within the anterior one-third of the secondary palate and in the medial aspect of the developing palatal shelves on E13.5–E14.5. No staining was observed on E12.5 or when a Wnt sense riboprobe was used (Fig. 3D–F and J–L). Wnt-4 was found in the epithelial lining of the entire oral cavity on E12.5 and E13.5, including the palatal shelves and tongue (Fig. 4A and 4B). On E13.5, pronounced expression was also detected in tooth primordia and in the submandibular ducts (Fig. 4B). On E14.5, Wnt-4 expression remained conspicuous in epithelia lining the oral and nasal cavities as well as the palatal medial edge epithelial seam (Fig. 4C). No staining was observed when a Wnt-4 sense riboprobe was used (Fig. 4D–F).
Fig. 3. Wnt-2 is expressed in palatal shelf mesenchyme.
Cryostat sections of fixed, frozen heads from mice on days E12.5–E14.5 were processed for expression of Wnt-2 transcripts by in situ hybridization with either an antisense Wnt-2-specific riboprobe (A–C) or the sense (negative) control (D–F). No detectable expression of Wnt-2 was observed in E12.5 palates (A,D) (palatal shelves outlined in black). Wnt-2 transcripts were, however, detected in the mesenchyme subjacent to the epithelium forming the junction between the oral cavity and tongue (arrows). On E13.5 and 14.5, expression of Wnt-2 was detected in the mesenchyme of the palatal shelf (B,C) (arrows) and extending laterally into the maxilla on E13.5 but not on E14.5. No signal was obtained with a sense Wnt-2 riboprobe (D–F). Panels (A,B,D,E) 200X magnification, panels (C,F), 100X magnification. Bar is 50 μm. Whole-mount in situ hybridization was also performed (G–M). No Wnt-2 mRNA was detected on E12.5 (G) but on E13.5 (H) a domain lateral to the medial edge of the palatal shelves and restricted to the anterior one-third was observed (arrows). On E14.5, the Wnt-2 mRNA expression domain was still confined to the anterior palate with signals also observed in the nasal septum (M) (arrows). Because the signal on E14.5 samples was difficult to discern on darkfield images (I), a brightfield image is also included (M) that clearly demonstrates expression with the antisense riboprobe. No signals were obtained when a Wnt-2 sense riboprobe was used (J–L and M). UL, upper lip; PP, primary palate; PS, palatal shelf; NS, nasal septum; MES, medial edge seam. The fused secondary palate is outlined in panel I.
Fig. 4. Wnt-4 is expressed in multiple epithelial layers.
Cryostat sections of fixed, frozen heads from mice on days E12.5–E14.5 were processed for expression of Wnt-4 transcripts by in situ hybridization with either an antisense Wnt-4-specific riboprobe (A–C) or the sense (negative) control (D–F). Wnt-4 was markedly expressed in the epithelia lining the oral cavity, tongue, and palatal shelves on E12.5 (A). On E13.5, further staining was found in tooth primordia and in the submandibular duct, as well as in the palatal shelf epithelium. On E14.5, expression in the secondary palate was found in epithelia of the nasal cavity, tongue, and palatal shelf (oral and nasal surfaces). On E14.5 expression of Wnt-4 was also found in tooth primordia and in the submandibular and sublingual ducts (not shown). All panels, 100X magnification. Bar is 50 μm.
Wnt-16 was up-regulated by 3.8- and 3.2-fold on E13.5 and E14.5, respectively (Table 1). The expression of Wnt-16 mRNA along the anterior-posterior (A–P) axis of the secondary palate was analyzed by both sectional and whole-mount in situ hybridization. Wnt-16 expression was seen to be differentially expressed along the palatal A–P axis (Fig. 5). On E12.5, Wnt-16 expression was detected in palate mesenchyme from each section analyzed (E12.5, Fig. 5A–D, black arrows) with no apparent difference in expression along the A–P palatal axis. These results were confirmed by whole-mount in situ hybridization (Fig. 5I–J). On E13.5, expression of Wnt-16 mRNA was absent in the anterior palatal process (E13.5, Fig. 5A). Black arrows indicate expression in the posterior two-thirds of the palatal processes (Fig. 5B–D). These results were also confirmed by whole-mount in situ hybridization (Fig. 5I–J). On E14.5, only diffuse expression of Wnt-16 was detected in the most posterior aspect of the palatal processes (Fig. 5, E14.5, D). Whole-mount in situ hybridization again confirmed that expression of Wnt-16 was confined to a region of the palate adjacent to the posterior nasopharynx (np). No staining was observed when a Wnt-16a sense riboprobe was used (Fig. 5E–H, J). These results allow the conjecture that Wnt-16 may be important for antero-posterior patterning of the palatal process.
Discussion
In order to begin to elucidate the role of the Wnt family of cytokines in palate development, expression of each of the 19 known vertebrate Wnt genes in the developing murine embryonic palate was determined. Of the 19 Wnts tested, 12 were found to be expressed in embryonic palatal tissue. Seven of these (Wnts-2b, -5a, -5b, -6, -7b, -9a and -11) were constitutively expressed in the embryonic palate during E12.5–E14.5. Five others were significantly up-regulated from E12.5 to E14.5 (listed in Table 1). Although the palatal shelves first appear on ~E11.5, they are rudimentary and the experiments reported here used E12.5 as the baseline for expression. Thus, any temporal alterations in expression reflect morphogenetic changes that occur subsequent to inductive signals from the maxilla. For these experiments, a relatively high threshold was set (2-fold change) for changes in expression to be considered as significant since thresholds less than this proved unreliable and difficult to reproduce. While Wnts whose expression fell below the 2.0-fold change threshold may prove to have a role in orofacial developmental, for the purposes of the present study we nevertheless chose to focus on those (Wnts-2, -4, -10a, -10b, and -16) exhibiting significant temporal changes in expression. Each of these Wnts displayed distinct tissue-specific patterns of expression. Wnts-4, -10a, and -10b were found to be expressed primarily in epithelial tissue, whereas Wnts-2 and -16 were expressed in specific mesenchymal domains within the developing secondary palate.
Epithelial-specific expression of Wnts -4, -10a and -10b suggests a role in the establishment or maintenance of epithelial polarity. The Wnt-mediated non-canonical, planar cell polarity (PCP) pathway is crucial in the establishment of epithelial cell morphology (Dollar et al., 2005). Of the Wnts identified in the embryonic oral epithelium, however, only Wnt-4 has been demonstrated to activate the PCP pathway (Lyons et al., 2004). The more restricted domain of expression for Wnt-10b within the future medial edge epithelial (MEE) cells on E13.5 and in the disintegrating medial edge seam on E14.5 suggests that Wnt-10b may play a role in palatal fusion. In addition to Wnt-10b and Wnt-10a, a number of genes essential for MEE differentiation (e.g. TGFβ3) are also expressed in these cells and functional interaction among these genes and/or the proteins they encode, is likely. Indeed, we have previously demonstrated that the signaling pathways activated by Wnt and TGFβ functionally interact in cultured palate cells (Warner et al., 2005). In addition, TGFβ has also been shown to induce the expression of a number of Wnt genes, such as Wnts-2, -4, -5a, and -10a in a human cell line (Zhou et al., 2004).
Enhanced expression of Wnts-2 and -16 in palate mesenchyme on E13.5 and E14.5 (Table 1) suggests a possible role for these Wnts in cell proliferation and/or extracellular matrix synthesis, two processes crucial for growth and reorientation of the shelves during this period (Morris-Wiman and Brinkley, 1992; Sasaki et al., 2004). Further, the expression of Wnt-2 in a well-defined domain in the anterior palate and adjacent to the developing maxilla (Fig. 3C) may indicate that Wnt-2 functions to define or limit the medial growth of the maxilla. Consistent with this idea is the observation that alkaline phosphatase staining and Wnt-2 expression are mutually exclusive on E13.5 and E14.5 in the developing palate (data not shown). Wnt-16 expression was progressively restricted to the most posterior aspect of the secondary palate, suggesting a possible role in anterior-posterior patterning of the palate. One must be careful, however, in speculating protein function based upon mRNA expression patterns since Wnts are secreted and may act at some distance from their cells of origin. Indeed, there may be extensive interaction between palatal mesenchyme and the overlying epithelium (Rice et al., 2004).
Few studies have examined the expression or activity of Wnts in the developing palate. Mice with mutations in the Wnt-9b gene (linked to the A/WySn mouse phenotype) (Juriloff et al., 2006) exhibit a cleft palate. The absence of Wnt-9b expression in the developing secondary palate (Lan et al., 2006) indicates that the cleft phenotype in these mice is secondary to other defect(s). Consistent with these data is our failure to detect expression of Wnt-9b by RT-PCR during any stage of palate development. Wnt-3 mutations have also been linked to the human syndrome, tetra-amelia, which includes cleft palate among a myriad of other defects (Niemann et al., 2004). We also failed to detect Wnt-3 on any day of palate development. Because tetra-amelia is a complex disorder with many defects, it is possible that the cleft seen in these patients is also secondary to other defect(s). Wnt-6 and Wnt-10a have been linked to human cases of non-syndromic cleft palate, although it is not clear if the defect is primary or secondary to other defects (Beaty et al., 2006). Most recently, knockdown of Wnt-11 was reported to alter palatal shelf fusion by perturbing apoptosis of the medial edge seam cells (Lee et al., 2008).
Functional GSK-3β, a key component of the canonical Wnt signaling pathway, appears to be necessary for proper fusion of the palatal shelves in mice (Liu et al., 2007). We attempted to identify the cells responding to canonical Wnt signals using transgenic embryos harboring the β-galactosidase gene under control of the TCF/β-catenin-inducible promoter (TOPGAL; DasGupta and Fuchs, 1999). In our hands, these embryos were found to have highly variable expression of β-galactosidase on E13.5–E15.5 and thus proved unreliable as a read-out for canonical Wnt activation, at least in the secondary palate (data not shown). Interestingly, we failed to detect expression of any of the Wnts that define the canonical pathway (Wnts-1, -3a, and 8), even though we have previously shown that stimulation of palate mesenchymal cells in vitro with Wnt-3a activates the canonical/β-catenin pathway (Warner et al., 2005). We did, however, find expression of the founding members of the non-canonical class (Wnts-4, -5a, and -11) suggesting a possible role for non-canonical Wnt signaling in palatal ontogeny. Although the biological function(s) of many Wnts are presently unknown, it is becoming increasingly clear that many act through more than one signaling pathway to elicit numerous cellular responses that are cell- and tissue-dependent (Wallingford et al., 2000; Civenni et al., 2003; Kishida et al., 2004; Tao et al., 2005).
Recent studies have demonstrated that several Wnt members are differentially expressed and temporally modulated in embryonic palatal tissue (Mukhopadhyay et al., 2004). Changes in expression for Wnts-4, and –10b (Mukhopadhyay et al., 2004) are consistent with data presented in the current study. While the biological function(s) of many Wnts are presently unknown, Wnt-5a may play an important role in palatal development. Expression of Wnt-5a in the mouse secondary palate was shown to be ubiquitous, and Wnt5a−/− knockout mice develop a cleft palate (Xiao et al., 2005). However, these mice also exhibit other craniofacial abnormalities, including tongue defects, which may indirectly contribute to the palatal defect.
With the demonstration of expression of many Wnts in the developing secondary palate, studies can now be designed to explore the role of specific Wnts in orofacial development, dissect the contributions of the disparate signaling pathways activated by Wnts (e.g. canonical/β-catenin and non-canonical pathways), and examine integration with other signaling pathways. These studies provide the necessary foundation from which to build a model for Wnt signaling during orofacial development.
Materials and Methods
Animals
ICR mice (Harlan, Indianapolis, IN) were maintained at a temperature of 22…C with a 12 hour light cycle and provided access to appropriate nutrition ad libitum. To obtain embryos of a defined gestational age, mature males and females were housed overnight and the presence of a vaginal plug the following morning was taken as evidence of copulation and was designated embryonic day 0.5 (E0.5). Pregnant mice were euthanized using inhaled carbon dioxide on E12.4–E14.5 of gestation. Embryos were removed from pregnant dams for microdissection of palate tissue for gene expression studies or the entire head was processed for in situ hybridization as detailed below.
RNA isolation and RT-PCR
RNA was purified using the RNeasy kit (Qiagen, Valencia, CA) and cDNAs were synthesized from total RNA using the SuperScript first strand cDNA synthesis system (Gibco Invitrogen Corp. Gaithersburg, MD). The quantity and purity of extracted total RNA was assessed by spectrophotometric UV absorbance at 260/280 nm. An amount equivalent to 10–20 ng of the initial input of total RNA was used as the template in real-time PCR assays with probe:primer pairs purchased from Applied Biosystems (Foster City, CA). QRT-PCR analysis was performed using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). All data were normalized to the amplification signal from GAPDH, which does not display changes in expression during the time frame examined (Greene et al., 2003). Fold-change values were determined according to the relationship: fold-change = 2−ΔΔCt, where Ct is the threshold value for real-time PCR amplification detection, ΔCt is the difference in Ct for the same probe:primer pair (e.g. Wnt 2) on E13.5 or E14.5 vs. E12.5, and Δ ΔCt = ΔCt, sample minus ΔCt, GAPDH (Livak and Schmittgen, 2001). Each probe primer set was used on cDNAs prepared from 3–4 independent sets of RNA derived from E12.5–E14.5 embryos. Threshold (CT) values greater than 35 were considered “not detected” because values within this range correspond to expression levels representative of single copy transcripts and are not reliable in fold-change calculations (Applied Biosystems). A 2.0-fold level of change was considered significant in these experiments.
Riboprobe synthesis
For sectional in situ hybridization, pBluescript SK(−) plasmid (Stratagene, La Jolla, CA) containing either mouse Wnt-10a or Wnt 10-b (a generous gift from Dr. Gregory Shackleford, Stanford University) was linearized with the appropriate restriction enzyme to generate a 1200 nucleotide or 1300 nucleotide riboprobe antisense or sense riboprobe, respectively. cDNAs for Wnt-2 and Wnt-4 were provided by Dr. Meng Sheng Qiu, University of Louisville, and were linearized by restriction digestion to obtain riboprobes of 600 nucleotides or 1000 nucleotides in length, respectively. A Wnt-16 cDNA vector was constructed using an RT-PCR-based approach to amplify a 600 nucleotide riboprobe. Riboprobes were prepared with the DIG RNA labeling kit according to the manufacturer’s instructions (Roche Diagnostics, Indianapolis, IN) and purified by repeated LiCl precipitation. RNA yields were measured using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and RNA quality and integrity determined by agarose gel electrophoresis.
In situ hybridization
For sectional in situ hybridization, heads from E12.5–E14.5 mouse embryos were removed and fixed by overnight incubation in 4% paraformaldehyde/PBS at 4°C. Heads were then infused with 30% (w/v) sucrose overnight at 4°C, followed by a second overnight incubation at 4°C in a 1:1 mixture of 30% (w/v) sucrose and Tissue Tek O.C.T. Compound (Sakura Finetek, Torrance, CA). Heads were then frozen in dry ice/ethanol and stored at −80°C. Coronal sections were cut to a thickness of 12 μm in a cryostat and placed onto RNase-free glass slides (Fisher Scientific, Pittsburgh, PA) and dried overnight at room temperature. Sections were then fixed with 4% paraformaldehyde/PBS, bleached with hydrogen peroxide (6% v/v), digested with 1 μg/ml proteinase K for 15 min, and postfixed with 0.2% glutaraldehyde/4% paraformaldehyde. Sections were incubated overnight at 65°C with the appropriate antisense or sense riboprobe (1 μg/ml). Following washes, specific hybridization was detected with an alkaline phosphatase-labeled anti-digoxigenin antibody (0.375 units/ml, Roche Diagnostics) followed by color development with 375 μg/ml nitro blue tetrazolium chloride, 475 μg/ml 5-Bromo-4-chloro-3-indolyl phosphate (added from a 50X stock solution, Roche Diagnostics) and 2 mM tetramisole (Sigma Chemical Company, St. Louis, MO) and stored in the dark for 3–4 days at room temperature. The reaction was terminated by the addition of PBT, pH 5.5 and sections mounted under Mowiol 4–88 (EMD Biosciences, Inc., La Jolla, CA) or Crystal/Mount (Biomeda Corp., Foster City, CA). Sections were visualized using a Nikon Eclipse E600 microscope and photographs taken with a Nikon DXM1200F digital camera and processed with Nikon ACT-1 v. 2.62 software (Nikon, Inc. Melville, NY). Whole-mount in situ hybridization was performed on dissected E12.5–E14.5 maxillary/palate tissue essentially as described above except that the tissue was digested with 10 μg/ml proteinase K for 15 min (E12.5), 20 min (E13.5), or 25 min (E14.5), antibody washes performed overnight, and color development allowed to proceed for a maximum of 2 hours. Tissue was cleared in 80% glycerol and visualized and photographed with a Nikon SMZ1500 stereomicroscope equipped with a Nikon SMZ1500 stereomicroscope equipped with a Nikon DXM1200F digital camera controlled by ACT-1 v. 2.62 software (Nikon, Inc.).
Acknowledgments
The authors thank Dr. Gregory M. Shackleford (Stanford University) for the Wnt-10a and Wnt-10b cDNAs, Dr. Meng Sheng Qiu (University of Louisville) for Wnt-2 and Wnt-4 cDNAs, and Dr. Jixiang Ding and Ms. Qun Li for providing TOPGAL transgenic mice and for helpful discussions. This work was supported in part by NIH grants DE12363 (to M.M.P.), DE05550, HD053509 and P20 RR017702 from the COBRE Program of the National Center for Research Resources (all to R.M.G.), and the Commonwealth of Kentucky Research Challenge Trust Fund.
Abbreviations used in this paper
- Dvl
Dishevelled
- Fz
Frizzled
- GSK-3β
glycogen synthase kinase 3β
- LEF
lymphoid enhancer factor
- MES
medial edge seam
- PBS
phosphate buffered saline
- RT-PCR
reverse-transcription-polymerase chain reaction
- TCF
T-cell factor
- TGFβ
transforming growth factor β
References
- BEATY TH, HETMANSKI JB, FALLIN MD, PARK JW, SULL JW, MCINTOSH I, LIANG KY, VANDERKOLK CA, REDETT RJ, BOYADJIEV SA, et al. Analysis of candidate genes on chromosome 2 in oral cleft case-parent trios from three populations. Hum Genet. 2006;120:501–518. doi: 10.1007/s00439-006-0235-9. [DOI] [PubMed] [Google Scholar]
- BLANTON SH, BERTIN T, SERNA ME, STAL S, MULLIKEN JB, HECHT JT. Association of chromosomal regions 3p21.2, 10p13, and 16p13.3 with nonsyndromic cleft lip and palate. Am J Med Genet A. 2004;125:23–27. doi: 10.1002/ajmg.a.20426. [DOI] [PubMed] [Google Scholar]
- BRAULT V, MOORE R, KUTSCH S, ISHIBASHI M, ROWITCH DH, MCMAHON AP, SOMMER L, BOUSSADIA O, KEMLER R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- BROWN NL, KNOTT L, HALLIGAN E, YARRAM SJ, MANSELL JP, SANDY JR. Microarray analysis of murine palatogenesis: temporal expression of genes during normal palate development. Dev Growth Differ. 2003;45:153–165. doi: 10.1034/j.1600-0854.2004.00686.x. [DOI] [PubMed] [Google Scholar]
- CASEY LM, LAN Y, CHO ES, MALTBY KM, GRIDLEY T, JIANG R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIVENNI G, HOLBRO T, HYNES NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO Rep. 2003;4:166–171. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUERVO R, COVARRUBIAS L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development. 2004;131:14–24. doi: 10.1242/dev.00907. [DOI] [PubMed] [Google Scholar]
- DASGUPTA R, FUCHS E. Multiple roles for activated LEF/TCF transcription factor complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- DOLLAR GL, WEBER U, MLODZIK M, SOKOL SY. Regulation of Lethal giant larvae by Dishevelled. Nature. 2005;437:1376–1380. doi: 10.1038/nature04116. [DOI] [PubMed] [Google Scholar]
- DU SJ, PURCELL SM, CHRISTIAN JL, MCGREW LL, MOON RT. Identification of distinct classes and functiional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FITCHETT JE, HAY ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev Biol. 1989;131:455–474. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- GREENE RM, NUGENT P, MUKHOPADHYAY P, WARNER DR, PISANO MM. Intracellular dynamics of Smad-mediated TGFbeta signaling. J Cell Physiol. 2003;197:261–271. doi: 10.1002/jcp.10355. [DOI] [PubMed] [Google Scholar]
- IKEYA M, LEE SM, JOHNSON JE, MCMAHON AP, TAKADA S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- JURILOFF DM, HARRIS MJ, MCMAHON AP, CARROLL TJ, LIDRAL AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- KISHIDA S, YAMAMOTO H, KIKUCHI A. Wnt-3a and Dvl induce neurite retraction by activating Rho-associated kinase. Mol Cell Biol. 2004;24:4487–4501. doi: 10.1128/MCB.24.10.4487-4501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAN Y, RYAN RC, ZHANG Z, BULLARD SA, BUSH JO, MALTBY KM, LIDRAL AC, JIANG R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JM, KIM JY, CHO KW, LEE MJ, KWAK S, CAI J, JUNG HS. Wnt11/Fgfr1b cross-talk modulates the fate of cells in palate development. Dev Biol. 2008;314:341–350. doi: 10.1016/j.ydbio.2007.11.033. [DOI] [PubMed] [Google Scholar]
- LIU KJ, ARRON JR, STANKUNAS K, CRABTREE GR, LONGAKER MT. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- LIU W, SUN X, BRAUT A, MISHINA Y, BEHRINGER RR, MINA M, MARTIN JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- LIVAK KJ, SCHMITTGEN TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- LYONS JP, MUELLER UW, JI H, EVERETT C, FANG X, HSIEH JC, BARTH AM, MCCREA PD. Wnt-4 activates the canonical beta-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/beta-catenin activity in kidney epithelial cells. Exp Cell Res. 2004;298:369–387. doi: 10.1016/j.yexcr.2004.04.036. [DOI] [PubMed] [Google Scholar]
- MARTIN JA, HAMILTON BE, SUTTON PD, VENTURA SJ, MENACKER F, MUNSON ML. Births: final data for 2003. Natl Vital Stat Rep. 2005;54:1–116. [PubMed] [Google Scholar]
- MORRIS-WIMAN J, BRINKLEY L. An extracellular matrix infrastructure provides support for murine secondary palatal shelf remodelling. Anat Rec. 1992;234:575–586. doi: 10.1002/ar.1092340413. [DOI] [PubMed] [Google Scholar]
- MUKHOPADHYAY P, GREENE RM, ZACHARIAS W, WEINRICH MC, SINGH S, YOUNG WW, JR, PISANO MM. Developmental gene expression profiling of mammalian, fetal orofacial tissue. Birth Defects Res A Clin Mol Teratol. 2004;70:912–926. doi: 10.1002/bdra.20095. [DOI] [PubMed] [Google Scholar]
- NAWSHAD A, HAY ED. TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J Cell Biol. 2003;163:1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIEMANN S, ZHAO C, PASCU F, STAHL U, AULEPP U, NISWANDER L, WEBER JL, MULLER U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am J Hum Genet. 2004;74:558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROETZEL G, PAWLOWSKI SA, WILES MV, YIN M, BOIVIN GP, HOWLES PN, DING J, FERGUSON MW, DOETSCHMAN T. Transforming growth factor-β 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICE R, SPENCER-DENE B, CONNOR EC, GRITLI-LINDE A, MCMAHON AP, DICKSON C, THESLEFF I, RICE DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASAKI Y, TANAKA S, HAMACHI T, TAYA Y. Deficient cell proliferation in palatal shelf mesenchyme of CL/Fr mouse embryos. J Dent Res. 2004;83:797–801. doi: 10.1177/154405910408301012. [DOI] [PubMed] [Google Scholar]
- SHULER CF, GUO Y, MAJUMDER A, LUO RY. Molecular and morphologic changes during the epithelial-mesenchymal transformation of palatal shelf medial edge epithelium in vitro. Int J Dev Biol. 1991;35:463–472. [PubMed] [Google Scholar]
- SHULER CF, HALPERN DE, GUO Y, SANK AC. Medial edge epithelium fate traced by cell lineage analysis during epithelial-mesenchymal transformation in vivo. Dev Biol. 1992;154:318–330. doi: 10.1016/0012-1606(92)90071-n. [DOI] [PubMed] [Google Scholar]
- SLUSARSKI DC, YANG-SNYDER J, BUSA WB, MOON RT. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- TAO Q, YOKOTA C, PUCK H, KOFRON M, BIRSOY B, YAN D, ASASHIMA M, WYLIE CC, LIN X, HEASMAN J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- VAZIRI SF, HALLBERG K, HARFE BD, MCMAHON AP, LINDE A, GRITLI-LINDE A. Fate-mapping of the epithelial seam during palatal fusion rules out epithelial-mesenchymal transformation. Dev Biol. 2005;285:490–495. doi: 10.1016/j.ydbio.2005.07.027. [DOI] [PubMed] [Google Scholar]
- WALLINGFORD JB, ROWNING BA, VOGELI KM, ROTHBACHER U, FRASER SE, HARLAND RM. Dishevelled controls cell polarity during Xenopus gastrulation. Nature. 2000;405:81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- WARNER DR, GREENE RM, PISANO MM. Cross-talk between the TGFbeta and Wnt signaling pathways in murine embryonic maxillary mesenchymal cells. FEBS Lett. 2005;579:3539–3546. doi: 10.1016/j.febslet.2005.05.024. [DOI] [PubMed] [Google Scholar]
- XIAO J, ZHU EX, NAGATSUKA H, GUNDUZ M, LI C, MINOO P, NAGAI N. Wnt5a Gene Plays a Role in Mouse Embryonic Orofacial Development. J Hard Tissue Biology. 2005;14:355–356. [Google Scholar]
- ZHOU S, EID K, GLOWACKI J. Cooperation between TGF-beta and Wnt pathways during chondrocyte and adipocyte differentiation of human marrow stromal cells. J Bone Miner Res. 2004;19:463–470. doi: 10.1359/JBMR.0301239. [DOI] [PubMed] [Google Scholar]