Abstract
Personalized treatment using stem, modified or genetically engineered, cells is becoming a reality in the field of medicine, in which allogenic or autologous cells can be used for treatment and possibly for early diagnosis of diseases. Hematopoietic, stromal and organ specific stem cells are under evaluation for cell-based therapies for cardiac, neurological, autoimmune and other disorders. Cytotoxic or genetically altered T-cells are under clinical trial for the treatment of hematopoietic or other malignant diseases. Before using stem cells in clinical trials, translational research in experimental animal models are essential, with a critical emphasis on developing noninvasive methods for tracking the temporal and spatial homing of these cells to target tissues. Moreover, it is necessary to determine the transplanted cell’s engraftment efficiency and functional capability. Various in vivo imaging modalities are in use to track the movement and incorporation of administered cells. Tagging cells with reporter genes, fluorescent dyes or different contrast agents transforms them into cellular probes or imaging agents. Recent reports have shown that magnetically labeled cells can be used as cellular magnetic resonance imaging (MRI) probes, demonstrating the cell trafficking to target tissues. In this review, we will discuss the methods to transform cells into probes for in vivo imaging, along with their advantages and disadvantages as well as the future clinical applicability of cellular imaging method and corresponding imaging modality.
Keywords: Cellular Magnetic Resonance (CMRI), Stem cells, Cell Tracking, SPION, Magnetic Cell Labeling
1. Introduction
Despite the uncertainty of the clinical outcome, stem cells are increasingly being used to treat cardiovascular, neurological and other diseases (1–8). Genetically modified cells are considered for the use in the treatment of genetic disorders or in the treatment malignant tumors (9–13). Cytotoxic T-cells (CTLs) or engineered T-cells are in the process of clinical trials for the treatment of hematopoietic or other malignant diseases (14, 15). However, there is no Food and Drug Administration (FDA) approved imaging modality or imaging contrast agent available that can be used for the long term monitoring the migration of the administered cells, in vivo.
Recently, extensive animal investigations have used different imaging modalities to track the migration and follow up of administered cells. Usually, investigators manipulate cells ex vivo either by incorporating different exogenous imaging-contrast agents or by transfecting different reporter genes (16–23). Cells carrying contrast agents can be then detected by optical, magnetic resonance imaging (MRI) or nuclear medicine imaging methods. Cells carrying reporter genes are suitable for optical or nuclear medicine imaging techniques, such as single photon emission computed tomography (SPECT) or positron emission tomography (PET). In all cases introduction of exogenous agents in allogenic or autologous cells is restricted by the FDA for use in humans (16, 24). In this review we will discuss the recent advancements in translational research that utilizes in vivo imaging techniques to track the migration and homing of administered cells and the use of different contrast agents to tag cells. We will also address the advantages and disadvantages of genetic modulation of cells for the purpose of tracking and functional modification, for both therapeutic and diagnostic purposes for use in clinical trials.
2. Cells as probes for imaging modalities
2a. Optical and fluorescent imaging
There is growing interest in the field of optical imaging for establishing the efficacy of engineered stem cells for therapeutic applications. One of the major ways to ex vivo label cells is the use of reporter gene systems. This approach became crucial for cellular/molecular imaging and is based on using cells that were genetically engineered ex vivo. The gene of interest is chosen based on the imaging modality to be used, physiological events that are to be monitored or therapeutic goals to be achieved. However, the criteria for designing reporter gene systems are usually established based on the combination of imaging and therapeutic goals. The reporter gene of interest encodes the protein, that when expressed, interacts with a specific imaging probe and the level of probe accumulation is proportional to the reporter gene expression levels. Currently, a need exists for an unambiguous, non-invasive identification of delivered cells and ideally, that corresponds to expression of previously silent markers of differentiation. The implementation of vector constructs that equip the cell with genetically encoded imaging probes is needed. The attributes of such constructs will be addressed in this section.
Tissue- Specific Opportunities
Promoter driven tissue specific expression of a transgene invites a variety of options for addressing the identification of an optimal promoter for robust gene expression during proliferation and differentiation (25), with the subsequent potential for delineating cells throughout the course of their development (26). Likewise, tissue specificity of the expression vector provides the capacity for investigating individual factors instrumental in the rise of a given cellular lineage (27) and subsequent visualization of the induction of such events (28). Progress reports are also possible from the cells destinations, for example reporter genes responsive to hypoxia (29) or other cellular stresses (30, 31) triggered by microenvironment. Ultimately, vectors incorporating the ascribed specifications can also be equipped for execution of therapeutic mechanisms such as that of oncolytic retrovirus, herpes vector gene transfer (32, 33) or other targeted suicide gene delivery (34, 35).
Transduction Methods
Reliable and cytocompatible labeling of stem cells in which preservation of proliferation and multi-lineage differentiation potential is necessary, requires realization of mechanistic cross-over between transcriptional regulation of the transgene and the host cell. Currently, lentiviral and non-integrating lentiviral vector (36) based transduction has become increasingly popular owing to its highly efficient, long-term incorporation, with efficacy now extending to include stem cells (37, 38). Successful transduction in dividing and non-dividing cells is also possible with replication deficient adenovirus, however potential toxicity (39) and relatively more limited efficiency of transgene expression has been reported (40). Retroviral transduction is limited because it requires actively dividing cells and there is a potential (41) tendency for insertional mutagenesis or generation of replication competent virus (42). There is the benefit of minimization of gene silencing through viral transduction. However, there is also the propensity for gene silencing noted in cell lines carrying a cytomegalovirus (CMV) promoter independent of transduction method used (43–45). Nonviral gene delivery generally has a lower transduction efficiency and while circumventing the safety concerns of viral methods, it presents additional complications by way of modifying intracellular pathway (46) or stimulating of the immune or anti-inflammatory responses by cationic transfection agents (47, 48) or poor degradability and cytotoxicity (49).
Luciferase and β-galactosidase Provided Feedback
Bioluminescence imaging (BLI) can not be translated to the clinic. The technique is particularly relevant for experimental studies with the goal of clinical transition of cell-based therapy. The establishment of preliminary groundwork has been noted recently in a number of cell tracking studies (50). Furthermore, as an imaging modality BLI has limited spatial resolution capacity (2–3mm) and therefore depth penetration (1cm) with essentially no background (maximal signal to noise ratio) and can be used in a fairly high throughput manner requiring minimal post processing of the data. BLI data reflects viable, metabolically active, cell-generated activity only, and will not provide false positives results that are particularly beneficial for cell trafficking studies. Among the genetically expressed photoproteins used as bioluminescent reporters, that of the sea pansy, Renilla reformis, (475nm em. peak, substrate coelenterazine) and firefly, photinus, (560 nm em. peak, substrate luciferin) are the most popular bioluminescent reporters for small animal imaging. These are often used for in vivo sequential differential imaging, providing fast, convenient and noninvasive measurements to be obtained before, during and after treatments (22, 51, 52).
At present, the overall trend within BLI and in vivo optical imaging is the active search and development of substrates having “red shifted” fluorogenic properties that take advantage of far-red (600–650nm) and near-infra-red (650–900nm) spectral windows in order to minimize the absorption and scattering of photons from tissue. In the interest of resolving signal from deeper within tissues, the search for luciferin and luciferin-like substrates that are more thermostable and maintain high overall photon yield while emitting a majority of photons in the red portion of the emission spectra (above 600nm) is currently underway (53, 54). Other variations of luciferase are being considered including click beetle red (CBRed) and click beetle green (CBGr68 and CBGr99) [56–59]. There is also an active search to produce stable, red-shifted mutants of Renilla (55). Thermostable red and green firefly luciferase mutants are also being optimized for dual-color bioluminescent reporter assays potentially capable of monitoring two distinct activities at 37°C (56). Similarly, novel red and green-emitting luciferases of railroad worms (Phrixothrix) are being developed for multiple gene expression in mammalian cells (57). However, the practicality of such reporters for BLI in live animals has not been reported.
There is increasing interest in expanding the use of beta galactosidase activity for immunohistochemical application toward that of non-invasive, in vivo detection methods. Recent studies have used β-Gal-derived cleavage of a far-red smart fluorogenic substrate (DDAOG) for in vitro and in vivo 2D-fluorescence reflectance imaging (58, 59). However, accurate quantitation of this application may warrant three-dimensional tomographic imaging and potentially a more red-shifted substrate molecule. Direct application of “sequential reporter-enzyme luminescence,” (SRL) toward imaging of β-galactosidase activity in live mice (60) has been reported, as Fluc-generated luminescence is derived through activation of caged galactoside-luciferin conjugate, lugal (61). Unlike standard Fluc assessment, this SLR approach does permit detection of signal outside of living cells as does that of another model proposing a specific blood assay for circulating luciferase (62). Either approach may potentially be applicable in monitoring circulating cell viability in vivo.
Fluorescent Feedback
Organic fluorophores such as rhodamine, fluorescein, DAPI, PKH26 and alexa488, are among the most commercially available, inexpensive, widely and easily used for a variety of straightforward shorter term labeling applications in cell and developmental biology [68, 69]. However, these fluorphores are often subjected to photobleaching and/or quenching and may be sensitive to changes in pH and chemical degradation. Moreover, organic dyes hold little promise for the current calling of long term labeling for cell tracking strategies. Alternatively, genetically encoded fluorescent proteins such as green fluorescent protein (GFP) have widespread application in cell labeling and potentially has far better photostability and overall luminescence time than organic dyes (25) (29, 32, 33). Even genetically encoded fluorescent proteins like GFP are subject to the limitations of generally broad emission spectra capable of generating false positive results and an overlap of emission spectra with tissue autofluorescence as well as absorption and limited resolution to a few millimeters. Enhanced red-shifted versions of fluorescent proteins such as DsRed proved capable of giving several orders of magnitude higher signal intensity in vivo, as compared to bioluminescence, however the large background autofluorescence severely reduces signal-to-noise ratio (63). Potential improvements in brightness and photostability of in vivo fluorescence imaging are underway with subsequent generations of monomeric red fluorescent protein (mRFP1) (64), of the ‘mFruits’ such as tdTomato and TagRFP-T (65, 66). In addition methods for optimizing expression of individual components of multimodality fusion vectors are in progress, such as a thermostable variant of firefly luciferase joined with mRFP and herpes simplex virus 1 thymidine kinase gene (tk) that demonstrated superior expression from all three reporter proteins (65).
With the widespread implementation of fluorescence and bioluminescence based applications essential in extracting functional non-invasive disease state information, an overwhelming need exists for more accurate methods for quantification of transgene expression (63, 67–70). Many critical parameters such as the tissue-to-detector geometry, auto-fluorescence, tissue optical properties, absorption and scattering remain unaccounted for in current state of data analyses. As a result, these largely account for artifacts that can potentially present in raw fluorescence data, thus comprising accurate quantification (21, 71). Quantitation using ratios accounting for such parameters that exist in 2D fluorescence imaging data are beginning to be developed (21). Moreover, accurate quantification may only be possible when measurements are properly controlled and signals are normalized. Other methods for obtaining more accurate quantitation of the detected fluorescence include use of blue-shifted excitation filters to subtract out tissue autofluorescence (63) and the application of an array of fluorescent filters accompanying spectral unmixing algorithms (72). Ultimately, the capacity for actual collection and reconstruction of tomographic data will need to become mainstream, in order to pave the way for possible clinical transition.
2b. Nuclear medicine imaging
Reporter gene strategies for transplanted cells
Gene reporter systems that are currently in use in nuclear medicine cellular imaging can be classified into three groups: (a) genes encoding for cell surface receptors that specifically bind the probe (such as dopamine D2 receptor), (b) genes encoding for membrane associated transporters that transport the probe across the cell membrane (such as sodium iodide symporter- NIS) and (c) genes encoding for enzymes that biochemically modify the probe (such as thymidine kinase – tk). Regardless of the mechanism, this specific interaction between reporter gene product and the administered probe generates a signal that can be detected by imaging modalities such as MRI, PET, SPECT or optical imaging.
Reporter gene approaches have many advantages over direct and indirect cell labeling methods. Stable transfection of cells ensure for long term expression of the reporter gene that does not dilute out in proliferating cells. Furthermore, over time accumulated divided cells can generate increased signal that can be detected with repeated imaging. In addition, the signals detected prove the in vivo presence of viable cells. Reporter gene approaches have great potential in gaining insights in particular mechanisms of stem cell based therapies. For example, by employing tissue specific promoters in driving the transcription of reporter gene, one can monitor the state of cell differentiation. However, as reporter gene imaging approaches continue to develop, the concerns with regard to immunogenicity and long term cell specific expression still need to be overcome.
One of the most widely used reporter genes for PET imaging is wild-type herpes simplex virus type 1 thymidine kinase (HSV1-tk) and it’s HSV1-sr39tk mutant. This enzyme efficiently phosphorylates purine and pyrimidine analogs and has been very successfully used with radio labeled reporter probes such as 124I-2′-fluoro-2′-deoxy-1-β-D-β-arabinofuranosyl-5-iodouracil (FIAU), 18F-2′-fluoro-2′-deoxy-1-β-D-β-arabinofuranosyl-5-ethyluracil (FEAU) and 18F-9-(4- 18F-fluoro-3-hydroxymethyl-butyl)guanine (FHBG) (73-76). One of the advantages of enzymatic reporter gene systems, such as –tk enzyme, is the signal amplification that occurs as a result of imaging probe trapping and accumulation. This signal amplification is generally not generated by receptor and transporter based reporters. However, the major limitation for successful translation of HSV-tk reporter gene into clinical setting is the immune reaction that the viral protein elicits in humans (77). Although several studies described the use of human derived reporter genes for somatostatin receptor (78, 79), norepinephrine transporter (80) and sodium iodide symporter (81, 82) in various applications, these findings did not eliminate the need for human derived HSV-tk equivalent. In an elegant recent study, Ponomarev et al. reported the use of human mitochondrial thymidine kinase type 2 (hTK2) as PET reporter gene (83). By eliminating nuclear localization signal, the retrovirus-mediated expression of this kinase was targeted to cytosol where it efficiently phosphorylated [18F] FEAU and [124I] FIAU. In addition, this gene carried the role of a suicide gene when used in combination with anticancer nucleoside analogs, such as d-arabinofuranosyl-cytosine. Besides the use in anticancer therapeutic strategies, introduction of suicide reporter genes into stem cells may serve as a potential safety mechanism against possible cellular oncogenic transformation; an attractive approach that warrants further investigations. In a very recent human trial Yaghoubi et al. (84) reported successful utilization of genetically modified CD8+ cytolytic T-cells carrying IL-13 zetakine and HSV1-tk genes in a case of glioblastoma multiforme. The authors detected the distribution of cytolytic T-cells in the tumor as well as other parts of the body by PET scanning using 18F-FHBG (Figure 1).
Figure 1.
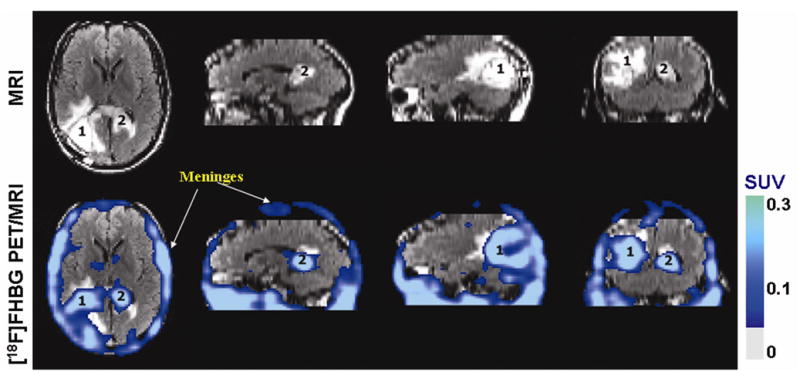
MRI and PET over MRI superimposed brain images of the patient who had been infused autologous cytolytic T-cells expressing IL13 zetakine and HSV1-tk genes. Images were acquired approximately two hours after [18F]FHBG injection. MRI images show tumors with associated edema in the left parieto-occipital region (1), which was partially resected, as well as in the center near corpus callosum (2) of the brain. The infused cells had localized at the site of tumor 1 and also trafficked to tumor 2. [18F]FHBG activity at both sites is higher than the brain background. Background [18F]FHBG activity is low within the central nervous system due to cells’ inability to cross the blood brain barrier, however, activity can be observed in the meninges. The tumor 1 to meninges and tumor 2 to meninges [18F]FHBG activity ratios in this patient was 1.75 and 1.57, respectively. Whereas the average resected tumor site to meninges and intact tumor site to meninges [18F]FHBG activity ratio in control patients was 0.86 and 0.44, respectively. SUV = standard uptake value. (Courtesy of Drs. Shahriar S Yaghoubi and Sanjiv S Gambhir, Department of Radiology, Stanford University, CA)
Cellular imaging based on the use of reporter genes strongly depends on stable, persistent and long term expression of the desired protein. In particular, monitoring the long term fate and trafficking of stem cells can not be accomplished without securing the long term expression of the reporter genes. This long term expression is usually achieved by utilizing viral expression systems and the most widely used are adeno- and lenti- virus vectors. Although adenovirus constructs ensure the strong expression of reporter genes, they may lead to the leaky expression of immunogenic adenoviral proteins that could lead to host immune response (85). In addition, long term expression that would carry on to the daughter cells in proliferating population is hindered due to the episomal gene expression (the reporter gene is not integrated into the cell chromatin). On the other hand, lentivirus based vectors exhibited many advantages over the adenovirus system. Lentiviral vectors stably integrate into the host cell chromatin that enable long term expression in dividing and non dividing cells (86), are not prone to gene silencing (87) and in small animals do not elicit immunogenic reaction (88). Therefore, lentivirus-based vectors appeared more suitable for most of the molecular imaging applications and have been successfully used by many groups (74, 88–90). The robust expression of reporter genes that are currently in use is usually achieved by utilizing strong viral promoters, such as human cytomegalovirus (CMV) promoter. However, the most common drawback of the CMV promoter, when establishing stably transfected mammalian cell lines, is gene silencing. This phenomenon was attributed to epigenetic mechanisms such as DNA methylation (91) and in vitro and in vivo studies by Krishnan et al. demonstrated that in embryonic rat cardiomyoblast, CMV silencing was completely reversed by treatment with 5-azacytidine (43). To circumvent this problem, Love et al. utilized lenti-virus based triple fusion reporter (firefly-luciferase, monomeric red fluorescent protein and HSV1-sr39tk) whose expression was driven by a modified myeloproliferative sarcoma virus promoter (mnd) (92). This strong promoter drove the continuous expression of the triple-fusion reporter in implanted human mesenchymal stem cells for more than 3 months. Recent efforts were focused on developing reporter gene constructs using mammalian promoters such as Elongation factor-1α (EF-1α) and ubiquitine C. Further investigations will be needed to delineate the optimal promoters that would efficiently drive the expression of the reporter gene of choice, while avoiding epigenetic silencing.
Another attractive reporter gene commonly used with SPECT imaging is sodium iodide symporter (NIS). Since it’s cloning in 1996 (93) human NIS gene has been widely used in imaging applications in conjunction with 99mTc-pertechnetate or 124I and in anticancer therapy with 131I and 188Re. By active transport via transgenically encoded and expressed NIS channel, cells can take up radioactive probe and subsequently be monitored by gamma camera or SPECT scanners. Availability of human reporter gene has been a major advantage in exploiting NIS gene as an imaging and therapeutic tool. Various studies utilized viral vectors to stably express NIS in cells (94–98). However, when tracking the NIS expressing cells in whole body imaging applications, due to the presence of endogenous NIS regions like thyroid, stomach and bladder, may result in background signals.
Reporter gene approach has also been used with MR imaging, where investigators used genes that facilitate iron uptake (99–101). Genove et al. utilized adeno viral vector to express a metalloprotein from the ferritin family as the cell sequesters endogenous iron from the organism without the need for an exogenous contrast agent. Cells that endogenously generated superparamagnetic forms of iron oxide nanoparticles were detected by MRI in in vitro and in vivo settings. However, further studies are needed to translate this novel approach into the clinical application.
Currently, many studies utilize the combination of two or more reporter genes that would enable the use of different imaging modalities to overcome the drawbacks associated with a single reporter gene and/or associated detection system. One of the combinatorial approaches in molecular imaging is the use of fusion reporter genes containing fluorescent and PET reporter genes that provide information with high resolution and in tomographic manner (102). However, the insufficient sensitivity that these construct provided led to the novel constructs that included triple fusion genes composed of bioluminescent, fluorescent and PET reporter genes. Ray et al. evaluated the activity, in vitro and in vivo, of variants triple fusion reporter constructs that encoded for luciferase, red fluorescent protein and HSV–tk. By using thermostable firefly luciferase lacking the peroxisome localization sequence they increased the enzyme activity and bioluminescence and thus the sensitivity for the optical aspect of imaging (65). This improved triple fusion vector enabled higher sensitivity detection of less number of cells. The multimodality approach in using fusion reporter genes in molecular imaging is continuously evolving and various construct has been currently used by many groups (74, 103). Hwang et al. constructed a dual membrane protein reporter system consisting of hNIS and D2R (linked with an internal ribosomal entry site (IRES)) in an attempt to overcome the shortcomings of each reporter gene and to enable the simultaneous use of designated receptors for therapeutic and imaging purposes (104). This system resulted in expression attenuation of the gene downstream of IRES as well as in competitive effect of two over-expressed membrane associated receptors, demonstrating the difficulties associated with optimizing all the components of the efficient multi-reporter gene system.
Further studies are needed to generate the efficient multi-reporter gene that would enable the use of multimodal molecular imagining for trafficking of less number of cells with greater sensitivity and higher spatial resolution. These studies will need to focus on designing constructs that would eliminate localization sequences of reporter genes to increase cytoplasmic localization and therefore possible increase in the activity of intracellular proteins; designing construct that would enable optimal expression of all the encoded genes and once expressed, reporter genes would not interfere with each other or with cellular function.
2c. Multimodal imaging
Investigators have been working to develop new types of contrast agents that can be detected by two different imaging modalities, which could be complementary to each other. Other advantages of bimodal imaging agents would be to determine the status of administered cells. For example, cells labeled with MRI contrast agent alone can be tracked by MRI, however this approach does not provide information on the functional status of the administered cells. If a bimodal contrast agent was used to transfect genes into the cells, the expression of the gene product can be detected by another complementary imaging modalities such as optical imager or nuclear medicine techniques indicative of the functional status of the cells. Bimodal contrast agents can also be applied in PET and bioluminescence imaging (89). Giesel et al. (105) were able to label mesenchymal stem cells (MSC) using a bifunctional gadoflurine M-Cy3.5 for both MRI and optical imaging. Gadoflurine M-Cy3.5 is designed with a hydrophilic tail that allows the agent to be inserted in the cell wall and then internalized into cytosol. Intracerebral implantation of 106 gadoflurine M-Cy3.5 labeled MSC allowed for clear visualization of cells in the rat brain on T1 weighted imaging at clinical relevant 1.5 Tesla that could be confirmed by fluorescent microscopy. Brekke et al. (106) used bimodal gadolinium rhodamine dextran (GRID) agent to label neural stem cells (NSC) to determine the labeling efficiency and the toxicity of the agent and observed significant loss of viability and proliferative capacity of the cells. Like gadoflurine M-Cy3.5, GRID labeled cells can be tracked by MRI (in vivo) and fluorescent microscopy (ex vivo samples). However, these agents may not indicate real functional status of the administered cells.
A better approach would be to make transgenic cells that carry reporter genes for different imaging modalities or transgenic cells can be labeled with MRI contrast agents before administration. Love et al (92) have reported long term follow up of administered transgenic MSC that carried reporter genes for PET and bioluminescence imaging. For the most of the bioluminescence imaging, administration of exogenous substrate is necessary which may not be permitted for future clinical use. Recently we have used magnetically labeled transgenic endothelial progenitor cells (EPC) to determine the migration, incorporation and expression of gene product in a mouse model of breast cancer. MRI determined cell migration and incorporation into the tumor and the functional status of the incorporated cells (gene expression) was determined by SPECT imaging (Figure 2). We used FDA approved agents, ferumoxides and protamine sulfate, to magnetically label cells and human sodium iodide symporter (hNIS) gene to determine the functional status of the incorporated cells.
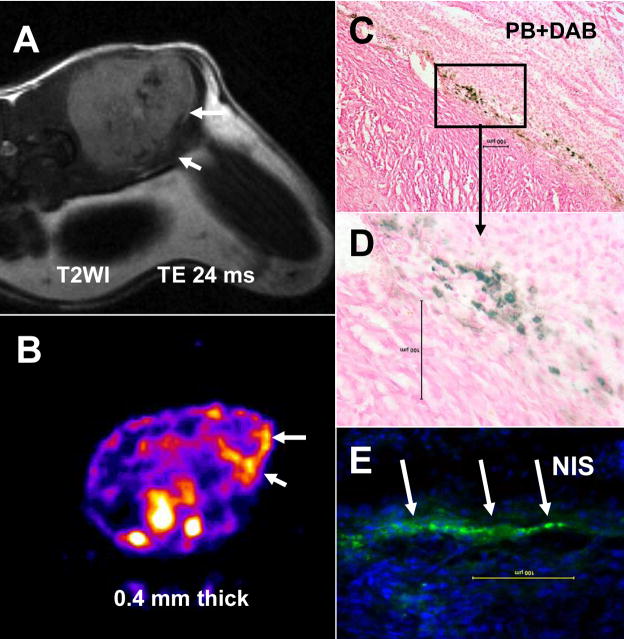
Figure 2.
Tracking of magnetically labeled transgenic EPCs by MRI and SPECT studies. (A) T2WI obtained by a clinical 3T MRI system with TE of 24 ms, 1 mm thick, 256×256 matrices and 3.6 cm FOV. White arrows indicate accumulated iron labeled cells (low signal intensity areas) that carry and express hNIS genes at the site of incorporation, which was detected by Tc-99m SPECT study (B). Presence of iron labeled cells is confirmed by DAB enhanced Prussian blue staining (C, D) and presence of hNIS is confirmed by immunohistochemistry using anti hNIS antibody and FITC tagged secondary antibodies (arrows) (E). Magnetically labeled transgenic EPCs acted as both MRI probes and gene carrier systems.
2d. Magnetic resonance imaging
2d.1. Superparamagnetic Agents
Superparamagnetic iron oxide nanoparticles (SPION) are family of MRI contrast agents that are presently being used to efficiently label cells for cellular imaging. There are various methods used to prepare SPION, resulting in a wide range of physiochemical differences including core size (e.g., ultrasmall (U)SPIO), shape, mono or oligocrystalline composition, and outer coating that may alter the ability to use these agents to label cells. There are FDA approved and FDA non-approved SPIONs are available in the market. One of the advantages of using SPIONs to label cells is that they are biodegradable and can be utilized by the cells in iron metabolism pathways (107, 108). However, these SPIONs need to be modified for efficient labeling of cells. Our group has developed a technique to make ferumoxides (FDA approved agent) transfection agents complexes to facilitate cellular uptake by endocytosis (109–113). Very recently, instead of commonly used cellular transfection agents, such as lipofectamin, we introduced the use of protamine sulfate (FDA approved agent) to generate ferumoxides-protamine sulfate (FePro) complexes for efficient labeling of different mammalian cells, including stem cells and T-lymphocytes (16). These labeled cells have been used in different animal models and tracked by both, high strength and clinical strength MRI systems (114–116). One of the advantages of labeling cells using FDA approved agents is the possibility of clinical trial without facing major toxicity issues related to contrast and transfection agents. Due to the susceptible effect SPION, labeled cells can easily be detected by MRI, compared to the cells labeled with gadolinium or T1- based MRI contrast agents.
Besides transfection agents mediated labeling of cells with SPION through facilitated endocytosis, various other modifications and methods were employed to label cells to be used as cellular probes for MRI. Investigators have modified the surface charge of the nanoparticles by coating it with cationic materials or modified the surface of the coating by attaching membrane penetrable peptides. The types of coatings include dextran and modified cross-linked dextran, dendrimers, starches, citrate and viral particles, and are usually attached through electrostatic interactions with the surface of the iron oxide crystal core contributing to the hydrodynamic size and zeta potential of the SPION (117). The zeta potential or the average potential difference, expressed in millivolts, exists between the surface of the (U)SPION immersed in distilled water and the bulk of the liquid. The SPIONs have been characterized as either carrying positive or negative zeta potential that determines the contrast agent’s ability to interact with cell/plasma membrane. Dextran-coated SPIO nanoparticles such as ferumoxides, ferucarbotran or ferumoxtran-10 are clinically approved MR contrast agents for use as hepatic imaging agents or have been used in clinical trials as blood pool agent or for lymphangiography (118–122) and are also being used to label cells. There are also experimental (U)SPIONs that have been used for labeling cells. The cationic coated USPIONs, carboxypropyl trimethyl ammonium (WSIO) and citrate (VSOP C184) were designed so that they would attach to the negative surface charge of plasma membranes through electrostatic interactions and then get incorporated into endosomes of macrophages (123).
Modified SPIONs for labeling cells to use as probes for cellular MRI
Physico-chemical modifications have been tried by different groups to facilitate cellular uptake of SPIONs, especially by non-phagocytic cells. Bulte et al. have used generation 4.5 polyamindoamine (PAMAM) dendrimer as a coating of SPIONs that resulted in the synthesis of magnetodendrimers (MD-100) (124), which were used to label oligodendroglial progenitors derived neural stem cells (NSC). The labeled cells were transplanted into the ventricles of neonatal dysmyelinated Long Evans Shaker rats and the migration of labeled cells into the brain parenchyma could be observed by CMRI up to 42 days following implantation. Josephson et al. (125) modified dextran coating of USPIONs by cross-linking the dextran strands (CLIO) and then covalently attaching HIV-1 Tat proteins to the surface that has allowed for efficient and effective labeling of non-phagocytic cells presumably through macropinocytosis. Using MR imaging, homing of CLIO-Tat labeled lymphocytes could be visualized in the liver and spleen in normal mice (126). CLIO-Tat labeled T-cells have been used in adoptive transfer in autoimmune diabetes mouse model and labeled cells have been shown to selectively home to specific antigens in B16 melanoma in mouse model by in vivo MRI (127–129). The monoclonal antibody (OX-26) to the rat transferrin receptor was covalently attached to USPIO nanoparticles (MION-46L) and used to label rat progenitor oligodendrocytes (CG-4). Labeled rat CG-4 cells were directly implanted into spinal cords of myelin deficient rats and ex vivo MR images obtained on day 10–14 days after implantation, demonstrated excellent correlation between the hypointense regions and blooming artifacts caused by the presence of labeled cells and the degree of myelination in the spinal cord detected on immuno-histochemistry (130). Ahrens et al. also labeled dendritic cells by biotinylating anti-CD-11 MoAb in conjunction with strepavidin attached to dextran coated SPIONs. Instead of using peptide, dendrimers or antibodies, investigators have used hemagglutinatin virus of Japan (HVJ) envelope to encapsulate SPIONs to label microglial cells in culture (131–133). The HVJ SPIO labeled cells were intra-cardially injected and clusters of cells could be seen within 1 day following transplantation in the brains of mice.
Micron sized iron oxide commercially available particles or beads (MPIO) are also being used to label cells for cellular MRI studies in experimental models. These agents are from 0.3 to >5 microns in size and contain greater than 60% of magnetite in a polymer coating that can include a fluorescent marker that allows for dual detection of labeled cells by MRI and fluorescent microscopy. MPIOs have been used to track macrophage infiltration in transplantation rejection, to monitor single cell migration in tissues and to locate implanted stem cells in an area of myocardial infarction (134–138) Recently, Shapiro et al. (134) demonstrated uptake of very large MPIO of 5.8 microns in size in cultured hepatocytes and has been able to visualize single cells at 7 Tesla on T2* weighted images. Heyn et al. (139) have shown that following IC injection of enhanced green fluorescent protein (EGFP) transfected 231BR breast cancer cells labeled with MPIO in mice, the number of hypointense regions detected on a balance steady state gradient echo image (e.g., FIESTA) decreased with time.
Mechanical methods for labeling cells to use as probes for cellular MRI
Mechanical approaches such as the gene gun or electroporation have been used to effectively introduce MRI contrast agents into cells. The gene gun fires nanoparticles or magnetic beads directly into cells in culture, driving the particles through the cell membrane or directly into the nucleus. However, it is unknown what the long-term effects are on functional, metabolic and differential capabilities of the cell (140). Moreover this technique for labeling cells has its own limitations with respect to the efficiency, potential tissue damages created by the impact of the particles and small area of coverage (141). Since less traumatic methods to label cells with MR SPIONs are available, it is unlikely that the gene gun approach be used in the future.
Magnetofection is a technique that utilizes strong magnetic force to introduce SPION or desired genome attached with magnetic nanoparticles within the cells (142–147). This technique delivers nanoparticles directly to the cytoplasm and it is effective for DNA transfection. However, direct delivery to the cell cytoplasm may be a deterrent for magnetic cell labeling because of possible cytotoxicity following the release of iron into the cytoplasm or nucleus. This technique is useful for rapid labeling only in adherent cells. It’s applicability in labeling suspension cells or cell with small cytoplasm to nuclear ratio (such as T-cells and hematopoietic stem cells) has not been verified. Moreover, details on toxicity and nuclear uptake have not been described yet.
Transfection agents for labeling cells to use as probes for cellular MRI
In 2002, we combined commercially available SPION (e.g., ferumoxtran and ferumoxides) with commonly available polycationic transfection agents to effectively label cells. Different commercially available transfection agents have been tried with varying results (109–112, 148–151). However, most of the commercially available transfection agents are toxic to the cells at relatively low doses and importantly, these transfection agents are not FDA approved and can not be used clinically. By mixing two FDA approved agents, ferumoxides (Feridex IV, Berlex, NJ) and protamine sulfate together a complex is generated that efficiently and effectively labels stem cells (16, 111, 112, 152–154). Ferumoxides are dextran-coated colloidal iron oxide nanoparticles that magnetically saturate at low fields and have an extremely high NMR T2 relativity. Changes in R2 (R2=1/T2) are linear with respect to iron concentration. Protamine sulfate is an FDA-approved drug containing >60% arginine and is used for the treatment of heparin anticoagulation overdose. Cells are labeled with the ferumoxides-protamine sulfate (FePro) complex via macropinocytosis and can be imaged at clinically relevant MRI fields using standard imaging techniques. The concentration of iron in cells is dependent on nuclear-cytoplasm ratio, the iron concentration in the nano or micron sized particles, iron content in media, incubation times and method of endocytosis of the particles (16, 110, 111, 134) (Figure 3). Unlabeled stem cells usually contain less that 0.1 picograms of iron per cell whereas the labeled cells grown in suspension (i.e., hematopoietic stem cells, T-cells) contain 1–5 picograms iron per cells. Cells that adhere to culture dish (i.e., mesenchymal stem cells, human cervical cancer cells, macrophages) can take up from 5 to >20 picograms iron per cell (16, 111, 112).
Figure 3.
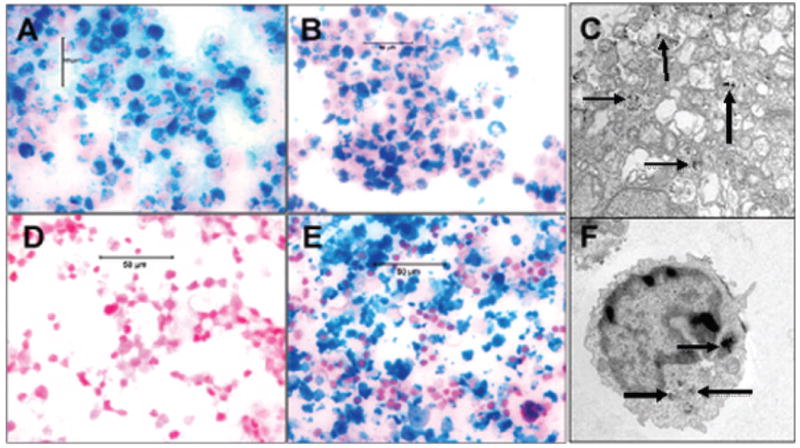
Prussian blue staining and transmission electron micrography (TEM) of cells. Labeling of cells with ferumoxides-protamine sulfate complexes using our new technique of 4 hours incubation. (A) immature dendritc cells, (b) U251 human glioma cells and corresponding TEM (C). Note the black particles within endosomes (arrows). (D) unlabeled cytotoxic T-cells, (E) labeled cytotoxic T-cells and corresponding TEM (F). Note the black particles within the endosomes (arrows).
2d.2. Paramagnetic Agents
Both gadolinium and manganese based nanoparticles are being utilized to labeled cells for in vivo tracking by MRI. Both mechanical as well as simple incubation methods are used to facilitate the uptake of the particles buy cells.
Direct injection of high concentrations of gadolinium chelates into Xenopus laevis egg enabled tracking of the labeled cell proliferation and migration during development, using MRI and optical imaging (155). However, this approach is not practical or efficient method for labeling mammalian cells with MR contrast agents. Electroporation has recently been used to label cells with gadolinium chelates and SPIO nanoparticles (156, 157). There is relatively little experience using this approach with MRI contrast agents to label cells and it is unclear as to the long-term effects on cell viability when using this method. Electroporation is commonly used to introduce DNA into the cell genome and well known to be associated with cell stress due to chemical imbalances and efflux or influx of chemicals from within the cell and surrounding media, altering the cells viability and survival. The type, size and number of cells, media conditions, the magnitude and duration of the electric pulse, the handling of cells post electroporation may all be the factors that influence cell viability and survival following electroporation with MR contrast agent. It has been shown that significant amount of cell lysis and death occurred during electroporation and following labeling with contrast agents (158). Recently, it has been reported that the magnetic labeling of embryonic stem cells by electroporation resulted in significant decrease in the percentage of viable cells compared to labeling cells with transfection agents complexed to ferumoxides (150). In addition, by labeling ESC by electroporation method, the ability of these to differentiate into cardiac progenitor cells was inhibited, therefore indicating that this method may not be clinically useful approach (150).
Electroporation method has also been used to label rat glioma cells using manganese oxide (MnO) nanoparticles. Gilad et al. have shown effective labeling and tracking of labeled cells in rat brain after implantation, using a 9.4 Tesla animal imaging system (159). However, the MnO labeled cells were not clearly visualized after 3 days of implantation. Shapiro et al. also used MnCO3, and MnO3 nanoparticles to label cells by simple incubation, however, in vivo tracking has not been reported by this group yet (160). Similarly Sotak et al. have reported effective labeling of murine hepatocytes using Mn-III-transferrin but in vivo tracking has not been tried yet (161). Investigators are actively working on making engineered nanoparticles containing different metals that can elicit both, T1 and T2 effects (162).
3. Iron metabolism in SPION labeled cells and limitations of SPION labeling
Labeling cells with ferumoxides does not alter the viability and functional capability of cells or the differential capacity of stem cells (16, 163). Ferumoxides-protamine sulfate labeled embryonic, mesenchymal, hematopoietic and neural stem cells showed similar rates of differentiation to different lineages, compared to control unlabeled cells (16, 150, 163–165).
In addition, studies have shown no significant changes in reactive oxygen species production in SPION labeled cells (110, 166) (167). In general, when concentrations of SPIONs exceed more than 100 μg/ml, some toxicity may be observed depending on the type of cells used for labeling (112). No significant reduction in viability of mesenchymal stromal cells (MSCs) was observed after incubation with SPION at the concentrations of up to 250 μg Fe/ml (168). SPION labeled MSCs maintained their multipotent capability in vitro. In the presence of specific factors labeled MSCs differentiated along adipogenic, chondrogenic, and osteogenic lineages (163) (166) (169), In vivo in their ability to differentiate to bone and hematopoieitc supporting stroma was also preserved, when transplanted in to the flanks of nude mice along with a carrier such as hydroxyapaptitie (Edyta Pawelczyk, personal communication). Recently, Farrell et al. also demonstrated chondrogenic differentiation of SPION labeled MSCs in a mouse model (169). However, the authors also observed morphological differences in the appearance of implanted scaffolds between labeled and unlabeled cells following chondrogenic differentiation. The cause of the alterations is not known, however the contributing role of the scaffold itself could not be excluded. Number of reports showed that other types of stem and progenitor cells including hematopoetic, neural stem cells or neural progenitors also maintained their differentiation capacity after labeling with SPIONs (124, 132, 163).
Following internalization, iron oxide particles remain for an extended period of time in the endosomes of slowly dividing cells or may circulate back to the extracellular space in rapidly dividing cells (16, 110). In some cases, intracellular SPIONs are transferred from early to late endosomes, followed by fusion with lysosomes (170–172). A recent study by Arbab et al. showed that a lysosomal pH of 4.5 could dissolve iron oxide particles over 3–5 days (173). This observation suggested that free iron could be released from iron oxide core of SPIONs into the cytoplasm and made available to participate in the cellular metabolic pathways (173). Since the maintenance of proper levels of “labile iron” is of crucial importance to living cells, Pawelczyk et al. in a study published two years ago determined whether SPION labeling affected cellular iron homeostasis (166). The authors have shown that ferumoxides (dextran coated SPIONs with the particle size 80 – 150 nm) –protamine sulfate labeled cells can detect and safely handle the intracellular iron load that can be 10 – 100 higher compared to unlabeled cells. In response to iron oxide particles loading into the endosomes, cells increased gene and protein expression levels of ferritin, a major iron-storage protein and at the same time transiently decreased gene and protein expression of transferrin receptor 1(TfR-1) that is involved in iron transport across the cell membrane (166). TfR-1 levels returned to control levels one week post-labeling with SPIONs, while for ferritin levels required two weeks in rapidly dividing cells and more than a month in slowly dividing cells.
Monocytes’ and macrophages’ intrinsic ability to phagocytose a large amount of SPIONs without the aid of transfection agents makes them interesting cells for studying using cellular MRI. Labeling macrophages by simple incubation with a contrast agent makes them good markers of inflammatory status in human studies in vivo. Labeled macrophages can be visualized in inflammatory lesions of stroke, brain tumors or artherosclerosis (174–176). As an antigen presenting cells and professional phagocytes, macrophages are also involved in the removal of cellular debris in the areas of inflammation followed by induction of appropriate immune responses. Siglienti et al. reported recently that SPION (Resovist®, carboxydextran coated SPION, 62 nm mean particle size) internalization by macrophages (on average 4.33±0.61 pg iron/cell) modulates in vitro, their cytokine profile towards an anti-inflammatory or more immunosuppressive phenotype by increasing interleukin (IL)-10 and reducing tumor necrosis factor (TNF)-α production (177). The authors have shown that during SPION-labeled macrophage interaction with T cells, IL-12p40 was inhibited. The results obtained by Sigilienti’s group with regard to proinflammatory cytokine production are most probably due to the use of rodent peritoneal macrophages. Stein et al (178)have shown that the capacity of rodent peritoneal macrophage population to release high levels of TNF-α strongly depends on the process of recruitment of peritoneal macrophages as well as on the subsequent stimuli. Peritoneal rat and mouse macrophages isolated upon thioglycollate stimulation can release high levels of TNF-α in response to LPS. On the other hand, resident peritoneal macrophages isolated from non stimulated animals (similar that are used in study by Sigilienti’s group) release only small amounts of TNF-α. At present, it is not known whether local cytokine concentrations in inflammatory lesions in vivo would be affected by the infiltration of iron oxide labeled macrophages. However, Muller et al examined in detail the safety and lack of proinflammatory activity in human macrophages labeled with a smaller iron oxide particles, Ferumoxtran-10, a dextran -coated ultrasmall SPIONs with a particle size 20 to 40 nm (179). This study showed that short term (48 hour) or long term (2 weeks) incubation with Ferumoxtran-10 at doses up to a 1mg/ml had no effect on baseline or stimulated production of pro-inflammatory cytokines such as IL-12, IL-6, TNF-α, IL-1β, superoxide anion production nor interfered with Fc-receptor mediated phagocytosis. Furthermore, extremely high intracellular concentrations of Ferumoxtran-10 of 10 mg/ml resulted in only 20–30% reduction in viability across various incubation times. In the other study, however, ferumoxides induced significant apoptosis in human monocytes after 4 hours at concentrations of 0.5 mg/ml and above (180). In a very recent publication we have shown no modulatory effects of SPION on the cytokine production by macrophage like THP-1 cells (181).
One of the key questions that still remain in cellular MRI is the fate of the SPION during cellular degradation. Up to 80 % of cells may die during direct implantation of stem cells into tissue due to trauma, ischemia or apoptosis (182–186). Subsequently, these stem cells or released SPIONs could be taken up by host macrophages and thus confounding the interpretations of MRI and histological results. Amsalem et al. and Terrovitis et al. (185, 186) recently investigated the relationship between the signal detected on MRI and the survival and engraftment of SPION- labeled mesenchymal stromal cells in acute myocardial infarction model in rats. Both groups were able to detect enhanced MRI signal 3 to 4 weeks post-transplantation, however they were not able to detect on histology sections many originally transplanted iron oxide labeled cells, but instead they detected resident macrophages with phagocytosed SPIONs (185, 186). These studies may indicate some limitations in the use of cellular MRI to monitor stem cell transplantation in cardiac cell therapy. Further research is required to address cell viability, labeling strategies and more advanced MRI techniques for the cardiac cell therapy field.
There are also reports that demonstrated that SPIONs and other exogenous or endogenous labels can be, in vivo, transferred from the labeled cells to tissue macrophages following direct transplantation of labeled stem cells. Lepore et al. showed that following implantation of ferumoxides labeled lineage-restricted neural precursors into an intact spinal cord, some of tissue macrophages contained iron taken up from grafted cells (187). In a different model, Brekke et al. demonstrated the in vivo transfer of gadolinium rhodamine dextran label from neural stem cells to macrophages in gliomas in rat brain (188). Despite all the reports acknowledging the possibility of misinterpreting the results from studies with labeled transplanted cells, none have quantified the number of macrophages taking up the exogenous label in the region of interest. Pawelczyk et al. have developed an in vitro model of localized inflammation using a Boyden Chamber (BC) where they quantified the number of SPION or bromodeoxyuridine (BrdU) labeled cells being engulfed by activated macrophages (189). Ferumoxide-Protamine sulfate or BrdU labeled MSCs were loaded into the matrigel with various ratios of activated macrophages in upper wells of the BC, while the lower wells contained the chemokines that allowed for selective migration of macrophages. After 24 and 96 hours of incubation, macrophages in the lower and upper wells were harvested and analyzed by flow cytometry with anti-CD68 and anti-dextran antibodies. The flow cytometric analysis of activated macrophages from lower or upper wells revealed from 10 to 20 % dextran or up to 10 % BrdU positive macrophages after 96 hours of incubation. Transfer of iron to activated macrophages was less than 10% of the total iron load in labeled cells, indicating that the hypointense regions observed on in vivo cellular MRI in the area of magnetically labeled cells would be considered minimal. This study provides the first report on the quantification of the label being transferred to macrophages and warrants the validation of transplanted labeled cells with staining for bystander cell markers.
4. Quantitation of administered cells by new MRI technique and analysis
Because of the potent contrast effects and inherent lack of cell toxicity, most of the magnetic resonance labels currently used in cell tracking are SPIONs of various sizes. Detection of iron labeled cells has been accomplished through T1, T2 and T2* weighted imaging (124, 130, 190–192). The NMR relaxation characteristics differ substantially when compartmentalized within cells compared with when they are within regions of freely diffusible water (193). As a result, T2* weighted gradient echo acquisitions provide the greatest sensitivity to the presence of intracellular SPION (193). The susceptibility effect on the SPION label extends well outside the volume occupied by the cell, and this extension augments its delectability. T2* weighted measures, however, are sensitive to background field inhomogeneities induced by imperfect shimming, blood, and endogenous ferritin deposits and thus have poorer specificity for iron particles. Conversely, T2 and T1 weighted spin echo acquisitions can be 2 ~3 orders of magnitude less sensitive to iron labeled cells, respectively, than T2* measurements (193). Balanced steady state free precession (b-SSFP) sequence (also known as FIESTA or True-FISP) imaging method has been shown to provide similar sensitivity as gradient echo imaging and a spin echo like insensitivity to background magnetic field inhomogeneities (194, 195).
T2 or T2 * based imaging methods depict iron labeled cells as pronounced local signal voids or hypointense regions. Differentiation between the signal loss caused by the intracellular nanoparticles and native low signals, for example those from artifacts or metals such as calcium, is challenging. Furthermore, the detection of labeled cells is limited by partial volume effects, in which signal void detection is dependent on the resolution of the image. If the signal void or hypointense voxels created by the agent is too small, it could be at the detection limits of MRI. To overcome these limitations, various positive contrast methods have been developed. Contrast enhancement by selecting the off resonance tissues caused by iron labeled cells was reported by Cunningham et al (196). Stuber et al. used inversion recovery on-resonance water suppression (IRON) to pre-saturate on-resonant water generating voxels with hyperintensities from off-resonance regions near SPIO labeled cells(197). Zurikya and Hu used a diffusion-mediated off-resonance saturation method to obtain images with positive contrast (198). Alternatively, Seppenwoolde et al. achieved positive contrast by dephasing the background signal with a slice gradient, while in the region near the paramagnetic marker the signal was conserved because the induced dipole field compensated for the dephasing gradient (199). A similar approach has been used for imaging of iron labeled cells and is known as gradient echo acquisition for superparamagnetic particles (GRASP) (200). Positive contrast images can also be derived from the magnetic field map by applying different post-processing techniques (201, 202). Bakker et al. exploited the echo-shift by applying a shifted reconstruction window in k-space (203). A susceptibility gradient mapping (SGM) technique has been developed that calculates the positive contrast images from a regular complex gradient echo dataset (204). The SGM method generates a parameter map of the 3D susceptibility gradient vector for every voxel by computing the echo-shifts in all three dimensions. A comparison of positive contrast images is shown in Figure 4. A common drawback of the positive contrast method is that, whereas signals can be quantified very efficiently, other important features of MRI, such as the detection of anatomical details, can be represented very poorly.
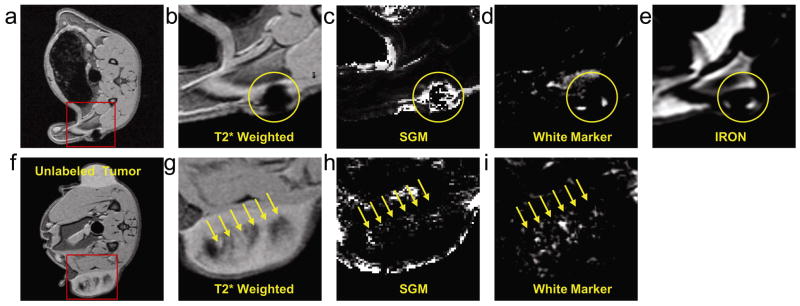
Figure 4.
Top row: Images were acquired when the SPIO labeled tumor was approximately 5 mm in diameter, representing highly concentrated SPIO labeled tumor cells (yellow circle). An axial slice of the rat (a) and zoom views of the labeled tumor with T2* weighted (b), SGM (c), White Marker (d) and IRON (e) techniques. Bottom row: Images were acquired when the SPIO labeled tumor was approximately 20 mm in diameter, representing diluted SPIO nanoparticles (yellow arrow). An axial slice of the rat (f) and zoom views of the labeled tumor with T2* weighted (g), SGM (h), White Marker (i) techniques. The IRON technique failed to generate positive contrast images of the diluted SPIO nanoparticles.
The detection threshold for SPION labeled cells is affected by a number of factors, including field strength, SNR, pulse sequence, acquisition parameters etc. Heyn et al. estimated that femptomolar quantities of SPIONs could be detected under typical micro-imaging conditions with b-SSFP sequence (194). Verdijk et al. concluded that 1000 cells/mm3 could be detected in patients treated with SPION labeled therapeutic cells (205) while Dahnke and Schaeffter predicted the detection limit of 120 cells/mm3 in the brain and 385 cells/mm3 in the liver on a 3T whole body MR scanner (206). Recent studies have demonstrated the feasibility to detect a small number of cells or even single cells. Hoehn et al. demonstrated in vivo detection of 500 cells implanted in the rat brain at 7T (207). Single SPIO labeled cells were observed in vitro studies at high field strength (208), and later at 1.5T (209).
To date, the majority of the cellular MR imaging studies performed a qualitative assessment of the hypo- or hyperintensities observed in tissues containing SPIO labeled cells. Quantitative measurement of cellular migration may allow monitoring the effectiveness of stem cell delivery and therefore the optimization of the therapy. Because R2 relaxation rate is sensitive to both iron concentration and distribution of the nanoparticles (193, 210), it is not suitable for the use in quantitation of iron oxide concentration by itself. A simple linear relationship, however, exists between the iron concentration and R2* change in vitro, for cell in suspensions, where the magnetic material is distributed in clusters(193). Using a multiple readout gradient echo pulse sequence, T2* relaxation times can be determined for the labeled cells in tissues, therefore coming one step closer toward quantitation of SPION distributions. Bos et al. demonstrated that R2* increase in the liver was of the right order corresponding to the number of MSCs injected in the portal vein in a rat model (211). Using a standard calibration curve, quantitative prediction of the number of labeled cells in a given region was therefore obtained within the brain of transplanted EAE mice (212).
However, R2* based quantification of the number of cells in vivo remains complicated, especially in longitudinal studies. First, the T2* relaxation rate is not only influenced by SPION in labeled cells, but also by macroscopic susceptibilities that arise from air-tissue interface. These susceptibility artifacts lead to overestimated relaxation rates or obscure low concentrations of numbers of labeled cells. Several methods have been proposed to correct for the macroscopic magnetic susceptibility influence such as increasing the spatial resolution (213), altering the slice selection gradient (214), or utilizing main field inhomogeneities correction to compensate for magnetic field susceptibilities from tissues that do not contain magnetically labeled cells (206). Second, quantitation of iron labeled cells in vivo can be complicated by the existence of free iron. It is difficult to completely separate extracellular iron in the microenvironment from the labeled cells. Free iron could also be found at injected sites where hemorrhage and labeled dead cells are often present (210). Because intracellular SPIOs have much longer T2 than nanoparticles freely suspended in the extracellular space, measuring both T2 and T2* relaxation times could reduce the interference from this iron pool and lead to a more accurate quantification of the number of intracellular SPION (215). Finally, it should be noted that MRI quantitation of cells labeled with SPION is an indirect technique. As such, signal change is due to the amount of SPION and not the number of cells. As cells proliferate and the iron is divided between daughter cells, the total iron content and the signal from each cell decreases. Furthermore, the iron from cells undergoing apoptosis or cell lysis can be internalized by resident tissue macrophages, resulting in signal falsely attributable to cells (186, 189).
5. Cells as both, cellular and therapeutic probes
In a clinical scenario, cell based therapy will be directed to two different approaches; 1) repair of damaged tissues using either genetically engineered cell or unmodified cells, 2) antitumor approaches using transgenic cells that carry suicidal gene and/or genes that can release cytotoxic cytokines upon activation or activated cells (such as Cytotoxic T-cells) directed against tumors. However, in both approaches investigators need to know the migration and accumulation status of the administered cells along with the functional improvement of the target organs/tissues. Moreover, due to the small number of administered cells compared to the total cells needed for the repair of the tissues, cells need to be modified so that endogenous cells can migrate to the site of interest. Recently reported clinical trials using stem cells in myocardial infarction were unable to track the administered cells and functional improvement was assessed by clinical signs and symptoms (1, 4).
Due to its unique property to migrate to the pathological lesions, stem cells are considered a unique choice to be the delivery vehicle for therapeutic genes to the tumors, especially for glioma (216–218). Rat neural stem cells (NSCs) expressing the cytosine deaminase gene, injected at a site distant from the primary tumor exhibit extensive migration and stable expression of the gene, indicating persistent ability to destroy tumor cells locally as well as distantly from the main tumor mass or metastatic foci (12). Mouse neural progenitor cells transfected with retroviral IL-4 injected into the brains of mice with glioblastoma exhibited migration, engraftment and destruction of tumor coincident with improved mouse survival rate. Glioma-bearing mice treated with murine NSCs producing IL-12 resulted in prolonged survival compared with controls, and transplanted cells demonstrated strong tropism for disseminated glioma. These effects were also associated with enhanced T-cell infiltration in tumor microsatellites, as well as long-term tumor immunity (219). Mesenchymal stem cells, pluripotent bone marrow stromal cells, were also used to carry genes to glioma and considered as an effective delivery vehicles (216, 220–222). Schichor et al. have pointed out that cells should have the following criteria to be used as gene delivery vehicles for glioma; 1) cells should be available from each glioma patient to create an autologous system without immune response; 2) within human brain parenchyma, cells should exhibit active motility directed toward glioma tissues (220). Ferrari et al. have shown the migration and incorporation of HSV-tk transfected mouse EPCs in subcutaneous tumor in a mouse model; however, the investigators did not show the incorporation of the transfected cells by in vivo imaging (223). It is utmost necessary to device a way to track the migration and homing of these transgenic cells to their site of interaction. We have investigated the feasibility of using transgenic cells as therapeutic and diagnostic probes in breast cancer animal model using magnetically labeled transgenic (hNIS) endothelial progenitor cells. MRI was used to track the migration of cells to the site of tumor and Tc-99m SPECT was utilized to determine the genetic expression (functional capability to carry suicidal gene) of these cell to the sites of tumor (Figure 2).
Genetically modified T-cells or cytotoxic T-cells are being considered for the treatment of hematologic as well as non-hematologic solid malignant tumors (14, 15, 224, 225). Tumor immunology has long been a focus of cell-based vaccine therapy research and as mentioned earlier, dendritic, as well as T cells, are considered the best candidates for developing such therapies. Dendritic cell-based vaccination therapy against recurrent glioma, that is in clinical trials (226, 227), utilized patient’s dendritic cells that were pulsed ex vivo with a glioma cell-lysate collected from the same patient, and it was shown that autologous administration of these tumor cell-lysate-pulsed dendritic cells initiated immunogenic activity against glioma cells, delaying tumor recurrence and/or decreasing the recurrence rate (226–229). Animal studies also showed an increased number of cytotoxic T lymphocytes (CTL), compared to control or pre-vaccination levels, in experimental glioma that utilized cell-lysate-pulsed dendritic cell therapy, indicating in vivo sensitization of T-cells to glioma presumably due to the administered primed dendritic cells (230–232). Studies describing the accumulation of dendritic cells and CTLs at the site of tumors indicated the initiation of tumor based immune reaction (233, 234). In addition, in vivo effectiveness of CTLs that were sensitized in vitro by dendritic cells was demonstrated in rat glioma model, as reported by Mercahnt et al. (235). Gliosarcoma 9L cell line was also shown to initiate an immunogenic reaction when transplanted peripherally or intracerebrally into syngeneic rat (236, 237). Animal experiments performed by our group also demonstrated initiation of cellular immunity in syngeneic fisher rats (114). It is also important to determine the migration and accumulation of these sensitized T-cells to the sites of tumor. Kircher et al. (Can Res 2003) have shown the migration and accumulation of sensitized T-cells to the implanted tumor by cellular MRI (128). Our group also reported the use of sensitized splenocytes (sensitized T-cells) to detect tumor (rat glioma) by cellular MRI (114). We are currently utilizing in vitro technique to make cytotoxic T-cells against glioma and other tumors. These cells are labeled with ferumoxides according to our established labeling technique (16) and used as diagnostic probes to determine the tumor and its metastasis (Figure 5). These genetically modified or sensitized T-cells can be used as therapeutic and diagnostic probes. For therapeutic purpose approximately 10–20% of the cells should be magnetically labeled for detection by cellular MRI.
Figure 5.
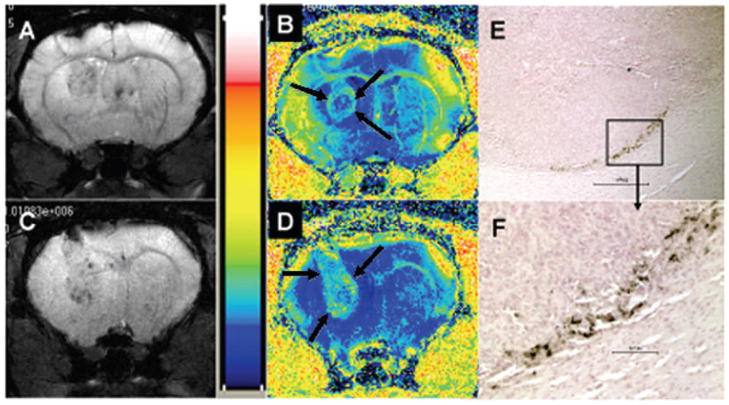
Gradient echo images and corresponding R2* maps of rat brain tumors created with U251 human glioma cells that received magnetically labeled non-sensitized human T-cells (A, B) and magnetically labeled sensitized T-cells (C, D). T-cells were sensitized by glioma cells lysate primed mature dendritic cells. E and F showed accumulated sensitized T-cells in the tumor (C, D) delineated by DAB enhanced Prussian blue staining. Note the increased R2* values in the tumor that received sensitized T-cells compared to surrounding brain tissues (D, arrows). The R2* values seen in tumor that received non-sensitized cells are not increased compared to surrounding brain tissues (B, arrows).
6. Investigations those are essential to apply for IND in FDA for cellular probes
Translation to the clinic
There have been four reports in the literature on using MRI to monitor the migration of magnetically labeled cells that are in early phase clinical trials. De Vries et al. has reported the use of magnetically labeled dendritic cells on a phase I clinical trial. The magnetically labeled dendritic cells were transformed from monocytes that were labeled with ferumoxides for two days and incubated in appropriately conditioned media (238). The labeled dendritic cells were transplanted directly into lymph nodes of patients with melanoma and the migration of these cells was serially monitored by MRI through adjacent lymph nodes. This was the first clinical trial to demonstrate the clinical utility of labeling cells with ferumoxides for monitoring cellular vaccine therapy. These investigators were able to delineate if labeled cells were actually implanted into lymph nodes or surrounding subcutaneous fat. In addition, the authors indicated that serial MRI demonstrated that ferumoxides labeled dendritic cells were cleared from the subcutaneous fat by 30 days following injection (personnel communication L de Vries). The authors indicated that MRI could detect approximately 2000 ferumoxides labeled dendritic cells per voxel (i.e, imaging volume element), and therefore with improvements in MRI techniques the detection of fewer numbers of labeled cells within a region of interest should be possible.
Zhu et al. (239) reported a study in small group of patients who had suffered traumatic brain injury with open head trauma. Cells were extracted from these patients, placed in culture and labeled with ferumoxides. In this early phase trial, patients received intracerebral injections of ferumoxides labeled or unlabeled “neural stem cells” around the area of injured brain based on T2 weighted images. Approximately 50,000 cells were implanted at each site in the brain with up to 10 implantations done per patient (personnel communication J Zhu). Serial MRIs over 21 days demonstrated the migration of ferumoxides labeled neural stem cells from injections sites into white and gray matter that was not observed in patients receiving unlabeled cells. The authors did not report any neurological complications as a result of implanting cells into patients with brain trauma. The authors recently presented that the hypointense areas were visible in the brains of patients with brain trauma for approximately 10 weeks on T2* weighted MRI following implantation of ferumoxides labeled neural stem cells (personnel communication J Zhu).
In 2008, Toso C et al (240) reported four diabetic patients who received ferucarbotran labeled cadaveric islet via the catheter into the portal vein. These authors injected between 30,000 and 300,000 islets in the patients with approximately a 90% viability of the transplanted labeled cells. Only three of the four patients injected with ferucarbotran labeled islets were evaluated on MRI at 1.5 Tesla and the islets were detected as hypointense regions in the periphery of the liver for approximately 6 weeks following the infusion of cells. These results indicate that it may be possible to monitor magnetically labeled islet cell transplantations in diabetic patients and suggests that this approach may be useful in monitoring transplantation rejection.
The fourth study was published in 2007 (241) and it involved a cohort of spinal cord injury patients in South America that received CD34+ autologous hematopoietic stem cells labeled with SPION containing beads, normally used for magnetic cell sorting (i.e., Dyna beads). Approximately 700,000 labeled cells were injected via lumbar puncture into the cerebral spinal fluid of patients and MRI was performed in the area of spinal cord injury. Hypointense regions were observed around the area of spinal cord lesion on sagital T2*w imaging. However, the authors reported no clinical improvement in these patients.
It is important to note that all four studies were performed outside the United States and informed consent was obtained in each patient on intramural research board approved protocols. However, no government oversight (i.e., similar to Food and Drug Administration) was required and no investigative new drug submission was needed to carry out these studies.
The translation of novel MRI contrast agents from experimental studies to the clinical applications will require a significant amount of preparation and perseverance by the investigators in order to successfully evaluate the agent in phase I clinical trial. It is very important that investigators establish an early dialogue with the appropriate regulatory agencies, such as the food and drug administration (FDA), to discuss the preclinical experimental studies that would be needed for the investigative new drug (IND) submission. It is also recommended that investigators familiarize themselves with the guidelines that the FDA has published on the web site http://www.fda.gov/cber/guidelines.htm and http://www.fda.gov/cber/rules.htm for cellular therapies. For certain cellular therapies, preclinical studies may be required for completion of IND and should reflect the proposed clinical indications as closely as possible. Moreover, the investigator will need to provide the evidence to support therapeutic rational, evaluate number of cells to be used, number of animals needed for statistically valid results and appropriate monitoring of animals receiving transplanted cells for site specific toxicities.
Unfortunately, the FDA guidelines on how to evaluate magnetically labeling of cells with novel SPIONs for cellular MRI are not ready at this time. Magnetically labeling of cell products do not fall under the auspices of the FDA approved exploratory or Phase 0 trials even though pharmaceutical grade agents are being used for cell labeling. An IND will need to be prepared by the investigator in order to use the agent clinically and the agents used for labeling cells need to be manufactured either with clinical grade products or made with good laboratory practices (GLP) (115, 242). The FDA will also require certification and possible reformulation of the novel contrast agents made in a GLP facility to complete the chemistry manufacturing and control (CMC) section of the IND. The CMC section would need to be provided by the investigator in order to use their agent to magnetically label cells in an early phase clinical trial.
Research groups planning on using a novel agents to label cells will need to demonstrate the following: 1) The novel MRI contrast agents are not toxic to the cells in culture or in experimental animals (i.e., mice and rats) with a large therapeutic window and lethal dose of 50% of the level for the product alone; 2) The novel agent does not alter cell proliferation and viability, differentiation capacity or result in an increase in reactive oxygen species or apoptosis of the labeled cells compared to controls; and 3) the composition of the agent is standardized and can be manufactured in a reproducible manner. In addition for labeling of stem cells, the investigators will need to demonstrate that the contrast agent does not alter characteristics and potency of the stem cells (243). It is important to note that these concepts such as the ability to self replicate or form colony forming units when cells are placed in culture and to differentiate or support niches may be difficult to define for each stem cell to be labeled.
In order to use the novel agent in clinical trials the investigator will need to demonstrate the following: 1) production of the contrast agent to be used to label cells should be done using compounds that are approved for clinical use or an exemption will be required; 2) cell labeling with the novel MR contrast agent can be performed using good manufacturing practices (GMP) in an approved facility; 3) the GMP facility would have approved standard operating procedures for handling, process and evaluating stem cells or other cells and be able to perform labeling on large scale; 4) the GMP facility has appropriate quality assurance and standard operating procedure for linking stem cells to patient who will receive the transplantation; 5) labeling of cells does not result in a significant cells loss; 6) the phenotype of the cells are unaltered as result of the labeling; and 8) there are no toxins or infections agents present in resulting product that would be released to the subjects in a clinical trial.
Preclinical studies performed in experimental disease models will probably include the infusion of magnetically labeled and unlabeled cells along with sham controls and assessment of toxicity. Serum chemistries and complete blood count evaluations will probably be required. MRI studies on the experimental animals can be performed using either clinical scanners or higher field strength scanners that are routinely used for MRI in rodents. Unfortunately, qualitative assessment of cellular MRI alone to determine the presence or extent of migration of the fewest numbers of magnetically labeled cells is inadequate and will probably require quantitative image analysis approaches to reveal the presence of labeled cells that are sparsely mixed with host cells through-out the target tissue. In order to use quantitative MRI approaches (i.e., T1, T2, or T2* relaxation properties) to determine the presence of labeled cells in tissues, there will be need to improve the hardware stability and reproducibility in order to perform serial MRI studies and track magnetically labeled cells over time. MRI hardware instabilities and inhomogeneities may all contribute to inaccuracies in quantitative relaxation rate measurements of tissue over time. In order to qualitatively improve MRI sensitivity of magnetically labeled cells in tissues, smaller voxels will be required to limit partial volume effects and it may be necessary to use 7T MRI to increase signal to noise, and sensitivity to changes in magnetic susceptibility. Susceptibility weighted imaging approaches (244) may also be useful in localizing magnetically labeled cells within target tissue.
Pathological imaging correlation of the target tissue should be performed; however pathological examination of major organs will be required to determine if the presence of labeled cells resulted in inflammatory or pathological changes. Of note, prior to the study it will be important to demonstrate that the immuno-histochemical techniques that will be used to assess the presence of magnetically labeled cells are able to identify the cells from surrounding host tissue. Disease specific indications for specific magnetically labeled cells will need to be developed along with documentation submitted to FDA with an institution review board approved clinical protocols. The tracking of magnetically labeled cells will involve a multidisciplinary team approach working together to label cells and monitor the patients during the early phase clinical trials.
Acknowledgments
This work was performed in part from support by the intramural research program in the Clinical Center at the National Institutes of Health and the following extramural supports; NS058589, CA129801, CA122031.
Abbreviations
- CTLs
cytotoxic T-cells
- FDA
Food and Drug Administration
- SPECT
single photon emission computed tomography
- PET
positron emission tomography
- CMV
cytomegalovirus
- PEI
polyethylenimine
- BLI
bioluminescence imaging
- GFP
green fluorescent protein
- mRFP1
monomeric red fluorescent protein
- ttk
1 thymidine kinase gene
- NIS
sodium iodide symporter
- FIAU
124I-2′-fluoro-2′-deoxy-1-β-D-β-arabinofuranosyl-5-iodouracil
- FEAU
18F-2′-fluoro-2′-deoxy-1-β-D-β-arabinofuranosyl-5-ethyluracil
- FHBG
18F-9-(4- 18F-fluoro-3-hydroxymethyl-butyl)guanine
- NSC
neural stem cells
- MSC
mesenchymal stem cell
- EPC
endothelial progenitor cells
- SPION
superparamagnetic iron oxide nanoparticles
- HVJ
hemagglutinatin virus of Japan
- MION
micron sized iron oxide
- EGFP
enhanced green fluorescent protein
- IRON
inversion recovery on-resonance water suppression
- b-SSFP
balanced steady state free precession sequence (also known as FIESTA or True-FISP)
- SGM
susceptibility gradient mapping
- GRASP
gradient echo acquisition for superparamagnetic particles
- IND
investigative new drug
References
- 1.Chachques JC, Trainini JC, Lago N, Masoli OH, Barisani JL, Cortes-Morichetti M, et al. Myocardial assistance by grafting a new bioartificial upgraded myocardium (MAGNUM clinical trial): one year follow-up. Cell Transplant. 2007;16(9):927–34. doi: 10.3727/096368907783338217. [DOI] [PubMed] [Google Scholar]
- 2.Malafronte C, Achilli F. Stem cells mobilization in acute myocardial infarction (stem-AMI trial): preliminary data of a perspective, randomized, single blind trial. Minerva Cardioangiol. 2007 Dec;55(6):721–31. [PubMed] [Google Scholar]
- 3.Cleland JG, Coletta AP, Abdellah AT, Cullington D, Clark AL, Rigby AS. Clinical trials update from the American Heart Association 2007: CORONA, RethinQ, MASCOT, AF-CHF, HART, MASTER, POISE and stem cell therapy. Eur J Heart Fail. 2008 Jan;10(1):102–8. doi: 10.1016/j.ejheart.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Lunde K, Solheim S, Aakhus S, Arnesen H, Moum T, Abdelnoor M, et al. Exercise capacity and quality of life after intracoronary injection of autologous mononuclear bone marrow cells in acute myocardial infarction: results from the Autologous Stem cell Transplantation in Acute Myocardial Infarction (ASTAMI) randomized controlled trial. Am Heart J. 2007 Oct;154(4):710 e1–8. doi: 10.1016/j.ahj.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Ripa RS, Haack-Sorensen M, Wang Y, Jorgensen E, Mortensen S, Bindslev L, et al. Bone marrow derived mesenchymal cell mobilization by granulocyte-colony stimulating factor after acute myocardial infarction: results from the Stem Cells in Myocardial Infarction (STEMMI) trial. Circulation. 2007 Sep 11;116(11 Suppl):I24–30. doi: 10.1161/CIRCULATIONAHA.106.678649. [DOI] [PubMed] [Google Scholar]
- 6.Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002 Aug;30(4):215–22. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 7.Escolar ML, Poe MD, Provenzale JM, Richards KC, Allison J, Wood S, et al. Transplantation of umbilical-cord blood in babies with infantile Krabbe’s disease. N Engl J Med. 2005 May 19;352(20):2069–81. doi: 10.1056/NEJMoa042604. [DOI] [PubMed] [Google Scholar]
- 8.Yoon SH, Shim YS, Park YH, Chung JK, Nam JH, Kim MO, et al. Complete spinal cord injury treatment using autologous bone marrow cell transplantation and bone marrow stimulation with granulocyte macrophage-colony stimulating factor: Phase I/II clinical trial. Stem Cells. 2007 Aug;25(8):2066–73. doi: 10.1634/stemcells.2006-0807. [DOI] [PubMed] [Google Scholar]
- 9.Benedetti S, Pirola B, Pollo B, Magrassi L, Bruzzone MG, Rigamonti D, et al. Gene therapy of experimental brain tumors using neural progenitor cells. Nat Med. 2000;6(4):447–50. doi: 10.1038/74710. [DOI] [PubMed] [Google Scholar]
- 10.Benedetti S, Di Meco F, Cirenei N, Bruzzone MG, Pollo B, Florio N, et al. IL-4 gene transfer for the treatment of experimental gliomas. Adv Exp Med Biol. 1998;451:315–21. doi: 10.1007/978-1-4615-5357-1_49. [DOI] [PubMed] [Google Scholar]
- 11.Arap W, Pasqualini R. Engineered embryonic endothelial progenitor cells as therapeutic Trojan horses.[comment] Cancer Cell. 2004;5(5):406–8. doi: 10.1016/s1535-6108(04)00121-7. [DOI] [PubMed] [Google Scholar]
- 12.Aboody KS, Najbauer J, Schmidt NO, Yang W, Wu JK, Zhuge Y, et al. Targeting of melanoma brain metastases using engineered neural stem/progenitor cells. Neuro-oncol. 2006;8(2):119–26. doi: 10.1215/15228517-2005-012. Epub 2006 Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Palma M, Venneri MA, Roca C, Naldini L. Targeting exogenous genes to tumor angiogenesis by transplantation of genetically modified hematopoietic stem cells. Nat Med. 2003 Jun;9(6):789–95. doi: 10.1038/nm871. [DOI] [PubMed] [Google Scholar]
- 14.Sadelain M, Riviere I, Brentjens R. Targeting tumours with genetically enhanced T lymphocytes. Nat Rev Cancer. 2003 Jan;3(1):35–45. doi: 10.1038/nrc971. [DOI] [PubMed] [Google Scholar]
- 15.Brentjens RJ, Santos E, Nikhamin Y, Yeh R, Matsushita M, La Perle K, et al. Genetically targeted T cells eradicate systemic acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2007 Sep 15;13(18 Pt 1):5426–35. doi: 10.1158/1078-0432.CCR-07-0674. [DOI] [PubMed] [Google Scholar]
- 16.Arbab AS, Yocum GT, Kalish H, Jordan EK, Anderson SA, Khakoo AY, et al. Efficient magnetic cell labeling with protamine sulfate complexed to ferumoxides for cellular MRI.[see comment] Blood. 2004;104(4):1217–23. doi: 10.1182/blood-2004-02-0655. [DOI] [PubMed] [Google Scholar]
- 17.Baumjohann D, Hess A, Budinsky L, Brune K, Schuler G, Lutz MB. In vivo magnetic resonance imaging of dendritic cell migration into the draining lymph nodes of mice. Eur J Immunol. 2006 Sep;36(9):2544–55. doi: 10.1002/eji.200535742. [DOI] [PubMed] [Google Scholar]
- 18.Kamaly N, Kalber T, Ahmad A, Oliver MH, So PW, Herlihy AH, et al. Bimodal paramagnetic and fluorescent liposomes for cellular and tumor magnetic resonance imaging. Bioconjug Chem. 2008 Jan;19(1):118–29. doi: 10.1021/bc7001715. [DOI] [PubMed] [Google Scholar]
- 19.Aime S, Barge A, Cabella C, Crich SG, Gianolio E. Targeting cells with MR imaging probes based on paramagnetic Gd(III) chelates. Curr Pharm Biotechnol. 2004 Dec;5(6):509–18. doi: 10.2174/1389201043376580. [DOI] [PubMed] [Google Scholar]
- 20.Anderson SA, Lee KK, Frank JA. Gadolinium-fullerenol as a paramagnetic contrast agent for cellular imaging. Invest Radiol. 2006;41(3):332–8. doi: 10.1097/01.rli.0000192420.94038.9e. [DOI] [PubMed] [Google Scholar]
- 21.Bogaards A, Sterenborg HJ, Trachtenberg J, Wilson BC, Lilge L. In vivo quantification of fluorescent molecular markers in real-time by ratio imaging for diagnostic screening and image-guided surgery. Lasers Surg Med. 2007 Aug;39(7):605–13. doi: 10.1002/lsm.20525. [DOI] [PubMed] [Google Scholar]
- 22.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh AY, Lin SA, Cao F, Cao Y, van der Bogt KE, Chu P, et al. Molecular imaging of bone marrow mononuclear cell homing and engraftment in ischemic myocardium. Stem Cells. 2007 Oct;25(10):2677–84. doi: 10.1634/stemcells.2007-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrucci JT, Stark DD. Iron oxide-enhanced MR imaging of the liver and spleen: review of the first 5 years. AJR American Journal of Roentgenology. 1990;155(5):943–50. doi: 10.2214/ajr.155.5.2120963. [DOI] [PubMed] [Google Scholar]
- 25.Chan KK, Wu SM, Nissom PM, Oh SK, Choo AB. Generation of high-level stable transgene expressing human embryonic stem cell lines using Chinese hamster elongation factor-1 alpha promoter system. Stem Cells Dev. 2008 Aug;17(4):825–36. doi: 10.1089/scd.2007.0233. [DOI] [PubMed] [Google Scholar]
- 26.Ovchinnikov DA, van Zuylen WJ, DeBats CE, Alexander KA, Kellie S, Hume DA. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J Leukoc Biol. 2008 Feb;83(2):430–3. doi: 10.1189/jlb.0807585. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt K, Schinke T, Haberland M, Priemel M, Schilling AF, Mueldner C, et al. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005 Mar 14;168(6):899–910. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang do W, Kang JH, Jeong JM, Chung JK, Lee MC, Kim S, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. Eur J Nucl Med Mol Imaging. 2008 Jan;35(1):135–45. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- 29.Brader P, Riedl CC, Woo Y, Ponomarev V, Zanzonico P, Wen B, et al. Imaging of hypoxia-driven gene expression in an orthotopic liver tumor model. Mol Cancer Ther. 2007 Nov;6(11):2900–8. doi: 10.1158/1535-7163.MCT-07-0432. [DOI] [PubMed] [Google Scholar]
- 30.Che J, Doubrovin M, Serganova I, Ageyeva L, Beresten T, Finn R, et al. HSP70-inducible hNIS-IRES-eGFP reporter imaging: response to heat shock. Mol Imaging. 2007 Nov–Dec;6(6):404–16. [PubMed] [Google Scholar]
- 31.Xie X, Cao F, Sheikh AY, Li Z, Connolly AJ, Pei X, et al. Genetic modification of embryonic stem cells with VEGF enhances cell survival and improves cardiac function. Cloning Stem Cells. 2007 Winter;9(4):549–63. doi: 10.1089/clo.2007.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aboody-Guterman KS, Pechan PA, Rainov NG, Sena-Esteves M, Jacobs A, Snyder EY, et al. Green fluorescent protein as a reporter for retrovirus and helper virus-free HSV-1 amplicon vector-mediated gene transfer into neural cells in culture and in vivo. Neuroreport. 1997 Dec 1;8(17):3801–8. doi: 10.1097/00001756-199712010-00029. [DOI] [PubMed] [Google Scholar]
- 33.Eisenberg DP, Adusumilli PS, Hendershott KJ, Chung S, Yu Z, Chan MK, et al. Real-time intraoperative detection of breast cancer axillary lymph node metastases using a green fluorescent protein-expressing herpes virus. Ann Surg. 2006 Jun;243(6):824–30. doi: 10.1097/01.sla.0000219738.56896.c0. discussion 30–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foloppe J, Kintz J, Futin N, Findeli A, Cordier P, Schlesinger Y, et al. Targeted delivery of a suicide gene to human colorectal tumors by a conditionally replicating vaccinia virus. Gene Ther. 2008 May 15; doi: 10.1038/gt.2008.82. [DOI] [PubMed] [Google Scholar]
- 35.Kirn DH, Wang Y, Le Boeuf F, Bell J, Thorne SH. Targeting of interferon-beta to produce a specific, multi-mechanistic oncolytic vaccinia virus. PLoS Med. 2007 Dec;4(12):e353. doi: 10.1371/journal.pmed.0040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007 Nov;15(11):1947–54. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- 37.Okada S, Ishii K, Yamane J, Iwanami A, Ikegami T, Katoh H, et al. In vivo imaging of engrafted neural stem cells: its application in evaluating the optimal timing of transplantation for spinal cord injury. Faseb J. 2005 Nov;19(13):1839–41. doi: 10.1096/fj.05-4082fje. [DOI] [PubMed] [Google Scholar]
- 38.Consiglio A, Gritti A, Dolcetta D, Follenzi A, Bordignon C, Gage FH, et al. Robust in vivo gene transfer into adult mammalian neural stem cells by lentiviral vectors. Proc Natl Acad Sci U S A. 2004 Oct 12;101(41):14835–40. doi: 10.1073/pnas.0404180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Chirmule N, Gao GP, Qian R, Croyle M, Joshi B, et al. Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages. Mol Ther. 2001 May;3(5 Pt 1):697–707. doi: 10.1006/mthe.2001.0329. [DOI] [PubMed] [Google Scholar]
- 40.Sung RS, Qin L, Bromberg JS. TNFalpha and IFNgamma induced by innate anti-adenoviral immune responses inhibit adenovirus-mediated transgene expression. Mol Ther. 2001 May;3(5 Pt 1):757–67. doi: 10.1006/mthe.2001.0318. [DOI] [PubMed] [Google Scholar]
- 41.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994 Jan;68(1):510–6. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi Y, Hahm SH, Lee KH. Retroviral gene therapy: safety issues and possible solutions. Curr Gene Ther. 2005 Feb;5(1):25–35. doi: 10.2174/1566523052997514. [DOI] [PubMed] [Google Scholar]
- 43.Krishnan M, Park JM, Cao F, Wang D, Paulmurugan R, Tseng JR, et al. Effects of epigenetic modulation on reporter gene expression: implications for stem cell imaging. Faseb J. 2006 Jan;20(1):106–8. doi: 10.1096/fj.05-4551fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaetano C, Catalano A, Palumbo R, Illi B, Orlando G, Ventoruzzo G, et al. Transcriptionally active drugs improve adenovirus vector performance in vitro and in vivo. Gene Ther. 2000 Oct;7(19):1624–30. doi: 10.1038/sj.gt.3301296. [DOI] [PubMed] [Google Scholar]
- 45.Rosenqvist N, Hard Af Segerstad C, Samuelsson C, Johansen J, Lundberg C. Activation of silenced transgene expression in neural precursor cell lines by inhibitors of histone deacetylation. J Gene Med. 2002 May–Jun;4(3):248–57. doi: 10.1002/jgm.268. [DOI] [PubMed] [Google Scholar]
- 46.Wei J, Yan W, Li X, Chang WC, Tai HH. Activation of thromboxane receptor alpha induces expression of cyclooxygenase-2 through multiple signaling pathways in A549 human lung adenocarcinoma cells. Biochem Pharmacol. 2007 Sep 1;74(5):787–800. doi: 10.1016/j.bcp.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leon-Ponte M, Kirchhof MG, Sun T, Stephens T, Singh B, Sandhu S, et al. Polycationic lipids inhibit the pro-inflammatory response to LPS. Immunol Lett. 2005 Jan 15;96(1):73–83. doi: 10.1016/j.imlet.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 48.Vangasseri DP, Cui Z, Chen W, Hokey DA, Falo LD, Jr, Huang L. Immunostimulation of dendritic cells by cationic liposomes. Mol Membr Biol. 2006 Sep–Oct;23(5):385–95. doi: 10.1080/09687860600790537. [DOI] [PubMed] [Google Scholar]
- 49.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999 Aug;16(8):1273–9. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 50.Beilhack A, Schulz S, Baker J, Beilhack GF, Nishimura R, Baker EM, et al. Prevention of acute graft-versus-host disease by blocking T-cell entry to secondary lymphoid organs. Blood. 2008 Mar 1;111(5):2919–28. doi: 10.1182/blood-2007-09-112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah K, Tang Y, Breakefield X, Weissleder R. Real-time imaging of TRAIL-induced apoptosis of glioma tumors in vivo. Oncogene. 2003;22(44):6865–72. doi: 10.1038/sj.onc.1206748. [DOI] [PubMed] [Google Scholar]
- 52.Shah K, Hingtgen S, Kasmieh R, Figueiredo JL, Garcia-Garcia E, Martinez-Serrano A, et al. Bimodal viral vectors and in vivo imaging reveal the fate of human neural stem cells in experimental glioma model. J Neurosci. 2008 Apr 23;28(17):4406–13. doi: 10.1523/JNEUROSCI.0296-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kajiyama N, Nakano E. Enhancement of thermostability of firefly luciferase from Luciola lateralis by a single amino acid substitution. Biosci Biotechnol Biochem. 1994 Jun;58(6):1170–1. doi: 10.1271/bbb.58.1170. [DOI] [PubMed] [Google Scholar]
- 54.Branchini BR, Ablamsky DM, Murtiashaw MH, Uzasci L, Fraga H, Southworth TL. Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2007 Feb 15;361(2):253–62. doi: 10.1016/j.ab.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 55.Venisnik KM, Olafsen T, Loening AM, Iyer M, Gambhir SS, Wu AM. Bifunctional antibody-Renilla luciferase fusion protein for in vivo optical detection of tumors. Protein Eng Des Sel. 2006 Oct;19(10):453–60. doi: 10.1093/protein/gzl030. [DOI] [PubMed] [Google Scholar]
- 56.Michelini E, Cevenini L, Mezzanotte L, Ablamsky D, Southworth T, Branchini BR, et al. Combining intracellular and secreted bioluminescent reporter proteins for multicolor cell-based assays. Photochem Photobiol Sci. 2008 Feb;7(2):212–7. doi: 10.1039/b714251j. [DOI] [PubMed] [Google Scholar]
- 57.Nakajima Y, Kimura T, Suzuki C, Ohmiya Y. Improved expression of novel red- and green-emitting luciferases of Phrixothrix railroad worms in mammalian cells. Biosci Biotechnol Biochem. 2004 Apr;68(4):948–51. doi: 10.1271/bbb.68.948. [DOI] [PubMed] [Google Scholar]
- 58.Tung CH, Zeng Q, Shah K, Kim DE, Schellingerhout D, Weissleder R. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Research. 2004;64(5):1579–83. doi: 10.1158/0008-5472.can-03-3226. [DOI] [PubMed] [Google Scholar]
- 59.Josserand V, Texier-Nogues I, Huber P, Favrot MC, Coll JL. Non-invasive in vivo optical imaging of the lacZ and luc gene expression in mice. Gene Ther. 2007 Nov;14(22):1587–93. doi: 10.1038/sj.gt.3303028. [DOI] [PubMed] [Google Scholar]
- 60.Wehrman TS, von Degenfeld G, Krutzik PO, Nolan GP, Blau HM. Luminescent imaging of beta-galactosidase activity in living subjects using sequential reporter-enzyme luminescence. Nat Methods. 2006 Apr;3(4):295–301. doi: 10.1038/nmeth868. [DOI] [PubMed] [Google Scholar]
- 61.Miska W, Geiger R. Synthesis and characterization of luciferin derivatives for use in bioluminescence enhanced enzyme immunoassays. New ultrasensitive detection systems for enzyme immunoassays, I. J Clin Chem Clin Biochem. 1987 Jan;25(1):23–30. doi: 10.1515/cclm.1987.25.1.23. [DOI] [PubMed] [Google Scholar]
- 62.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008 Feb;5(2):171–3. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004 Jan;3(1):9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 64.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):7877–82. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007 Apr 1;67(7):3085–93. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 66.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008 Jun;5(6):545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dehghani H, Davis SC, Jiang S, Pogue BW, Paulsen KD, Patterson MS. Spectrally resolved bioluminescence optical tomography. Opt Lett. 2006 Feb 1;31(3):365–7. doi: 10.1364/ol.31.000365. [DOI] [PubMed] [Google Scholar]
- 68.Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, et al. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Phys Med Biol. 2005 Dec 7;50(23):5421–41. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- 69.Gade TP, Koutcher JA, Spees WM, Beattie BJ, Ponomarev V, Doubrovin M, et al. Imaging transgene activity in vivo. Cancer Res. 2008 Apr 15;68(8):2878–84. doi: 10.1158/0008-5472.CAN-07-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baba S, Cho SY, Ye Z, Cheng L, Engles JM, Wahl RL. How reproducible is bioluminescent imaging of tumor cell growth? Single time point versus the dynamic measurement approach. Mol Imaging. 2007 Sep–Oct;6(5):315–22. [PubMed] [Google Scholar]
- 71.Mansfield JR, Gossage KW, Hoyt CC, Levenson RM. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J Biomed Opt. 2005 Jul–Aug;10(4):41207. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- 72.Farkas DL, Du C, Fisher GW, Lau C, Niu W, Wachman ES, et al. Non-invasive image acquisition and advanced processing in optical bioimaging. Comput Med Imaging Graph. 1998 Mar–Apr;22(2):89–102. doi: 10.1016/s0895-6111(98)00011-1. [DOI] [PubMed] [Google Scholar]
- 73.Buursma AR, Rutgers V, Hospers GA, Mulder NH, Vaalburg W, de Vries EF. 18F-FEAU as a radiotracer for herpes simplex virus thymidine kinase gene expression: in-vitro comparison with other PET tracers. Nucl Med Commun. 2006 Jan;27(1):25–30. doi: 10.1097/01.mnm.0000186609.12895.20. [DOI] [PubMed] [Google Scholar]
- 74.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006 Feb 21;113(7):1005–14. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tjuvajev JG, Doubrovin M, Akhurst T, Cai S, Balatoni J, Alauddin MM, et al. Comparison of radiolabeled nucleoside probes (FIAU, FHBG, and FHPG) for PET imaging of HSV1-tk gene expression. J Nucl Med. 2002 Aug;43(8):1072–83. [PubMed] [Google Scholar]
- 76.Yaghoubi SS, Barrio JR, Namavari M, Satyamurthy N, Phelps ME, Herschman HR, et al. Imaging progress of herpes simplex virus type 1 thymidine kinase suicide gene therapy in living subjects with positron emission tomography. Cancer Gene Ther. 2005 Mar;12(3):329–39. doi: 10.1038/sj.cgt.7700795. [DOI] [PubMed] [Google Scholar]
- 77.Berger C, Flowers ME, Warren EH, Riddell SR. Analysis of transgene-specific immune responses that limit the in vivo persistence of adoptively transferred HSV-TK-modified donor T cells after allogeneic hematopoietic cell transplantation. Blood. 2006 Mar 15;107(6):2294–302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rogers BE, Parry JJ, Andrews R, Cordopatis P, Nock BA, Maina T. MicroPET imaging of gene transfer with a somatostatin receptor-based reporter gene and (94m)Tc-Demotate 1. J Nucl Med. 2005 Nov;46(11):1889–97. [PubMed] [Google Scholar]
- 79.Zinn KR, Buchsbaum DJ, Chaudhuri TR, Mountz JM, Grizzle WE, Rogers BE. Noninvasive monitoring of gene transfer using a reporter receptor imaged with a high-affinity peptide radiolabeled with 99mTc or 188Re. J Nucl Med. 2000 May;41(5):887–95. [PubMed] [Google Scholar]
- 80.Doubrovin MM, Doubrovina ES, Zanzonico P, Sadelain M, Larson SM, O’Reilly RJ. In vivo imaging and quantitation of adoptively transferred human antigen-specific T cells transduced to express a human norepinephrine transporter gene. Cancer Res. 2007 Dec 15;67(24):11959–69. doi: 10.1158/0008-5472.CAN-07-1250. [DOI] [PubMed] [Google Scholar]
- 81.Che J, Doubrovin M, Serganova I, Ageyeva L, Zanzonico P, Blasberg R. hNIS-IRES-eGFP dual reporter gene imaging. Mol Imaging. 2005 Apr–Jun;4(2):128–36. doi: 10.1162/15353500200504193. [DOI] [PubMed] [Google Scholar]
- 82.Groot-Wassink T, Aboagye EO, Glaser M, Lemoine NR, Vassaux G. Adenovirus biodistribution and noninvasive imaging of gene expression in vivo by positron emission tomography using human sodium/iodide symporter as reporter gene. Hum Gene Ther. 2002 Sep 20;13(14):1723–35. doi: 10.1089/104303402760293565. [DOI] [PubMed] [Google Scholar]
- 83.Ponomarev V, Doubrovin M, Shavrin A, Serganova I, Beresten T, Ageyeva L, et al. A human-derived reporter gene for noninvasive imaging in humans: mitochondrial thymidine kinase type 2. J Nucl Med. 2007 May;48(5):819–26. doi: 10.2967/jnumed.106.036962. [DOI] [PubMed] [Google Scholar]
- 84.Yaghoubi SS, Jensen MC, Satyamurthy N, Budhiraja S, Paik D, Czernin J, et al. Noninvasive detection of therapeutic cytolytic T cells with (18)F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2008 Nov 18; doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y, Nunes FA, Berencsi K, Furth EE, Gonczol E, Wilson JM. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4407–11. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Y, Ramezani A, Lewis R, Hawley RG, Thomson JA. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 2003;21(1):111–7. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 87.Pfeifer A, Ikawa M, Dayn Y, Verma IM. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc Natl Acad Sci U S A. 2002 Feb 19;99(4):2140–5. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toelen J, Deroose CM, Gijsbers R, Reumers V, Sbragia LN, Vets S, et al. Fetal gene transfer with lentiviral vectors: long-term in vivo follow-up evaluation in a rat model. Am J Obstet Gynecol. 2007 Apr;196(4):352 e1–6. doi: 10.1016/j.ajog.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 89.Deroose CM, De A, Loening AM, Chow PL, Ray P, Chatziioannou AF, et al. Multimodality imaging of tumor xenografts and metastases in mice with combined small-animal PET, small-animal CT, and bioluminescence imaging. J Nucl Med. 2007 Feb;48(2):295–303. [PMC free article] [PubMed] [Google Scholar]
- 90.Sander WE, Metzger ME, Morizono K, Bonifacino A, Penzak SR, Xie YM, et al. Noninvasive molecular imaging to detect transgene expression of lentiviral vector in nonhuman primates. J Nucl Med. 2006 Jul;47(7):1212–9. [PubMed] [Google Scholar]
- 91.Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004 Apr;6(4):395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- 92.Love Z, Wang F, Dennis J, Awadallah A, Salem N, Lin Y, et al. Imaging of mesenchymal stem cell transplant by bioluminescence and PET. J Nucl Med. 2007 Dec;48(12):2011–20. doi: 10.2967/jnumed.107.043166. [DOI] [PubMed] [Google Scholar]
- 93.Smanik PA, Liu Q, Furminger TL, Ryu K, Xing S, Mazzaferri EL, et al. Cloning of the human sodium lodide symporter. Biochem Biophys Res Commun. 1996;226(2):339–45. doi: 10.1006/bbrc.1996.1358. [DOI] [PubMed] [Google Scholar]
- 94.Bouchentouf M, Benabdallah BF, Dumont M, Rousseau J, Jobin L, Tremblay JP. Real-time imaging of myoblast transplantation using the human sodium iodide symporter. Biotechniques. 2005 Jun;38(6):937–42. doi: 10.2144/05386IT01. [DOI] [PubMed] [Google Scholar]
- 95.Miyagawa M, Beyer M, Wagner B, Anton M, Spitzweg C, Gansbacher B, et al. Cardiac reporter gene imaging using the human sodium/iodide symporter gene. Cardiovasc Res. 2005;65(1):195–202. doi: 10.1016/j.cardiores.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 96.Dwyer RM, Bergert ER, O’Connor MK, Gendler SJ, Morris JC. Sodium iodide symporter-mediated radioiodide imaging and therapy of ovarian tumor xenografts in mice. Gene Ther. 2006;13(1):60–6. doi: 10.1038/sj.gt.3302599. [DOI] [PubMed] [Google Scholar]
- 97.Lee WW, Moon DH, Park SY, Jin J, Kim SJ, Lee H. Imaging of adenovirus-mediated expression of human sodium iodide symporter gene by 99mTcO4 scintigraphy in mice. Nucl Med Biol. 2004;31(1):31–40. doi: 10.1016/s0969-8051(03)00100-8. [DOI] [PubMed] [Google Scholar]
- 98.Niu G, Anderson RD, Madsen MT, Graham MM, Oberley LW, Domann FE. Dual-expressing adenoviral vectors encoding the sodium iodide symporter for use in noninvasive radiological imaging of therapeutic gene transfer. Nucl Med Biol. 2006;33(3):391–8. doi: 10.1016/j.nucmedbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 99.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–17. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, et al. MRI detection of transcriptional regulation of gene expression in transgenic mice. Nat Med. 2007;13(4):498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- 101.Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11:450–4. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 102.Loimas S, Wahlfors J, Janne J. Herpes simplex virus thymidine kinase-green fluorescent protein fusion gene: new tool for gene transfer studies and gene therapy. Biotechniques. 1998 Apr;24(4):614–8. doi: 10.2144/98244st01. [DOI] [PubMed] [Google Scholar]
- 103.Tai JH, Nguyen B, Wells RG, Kovacs MS, McGirr R, Prato FS, et al. Imaging of gene expression in live pancreatic islet cell lines using dual-isotope SPECT. J Nucl Med. 2008 Jan;49(1):94–102. doi: 10.2967/jnumed.107.043430. [DOI] [PubMed] [Google Scholar]
- 104.Hwang do W, Kang JH, Chang YS, Jeong JM, Chung JK, Lee MC, et al. Development of a dual membrane protein reporter system using sodium iodide symporter and mutant dopamine D2 receptor transgenes. J Nucl Med. 2007 Apr;48(4):588–95. doi: 10.2967/jnumed.106.036533. [DOI] [PubMed] [Google Scholar]
- 105.Giesel FL, Stroick M, Griebe M, Troster H, von der Lieth CW, Requardt M, et al. Gadofluorine m uptake in stem cells as a new magnetic resonance imaging tracking method: an in vitro and in vivo study. Invest Radiol. 2006 Dec;41(12):868–73. doi: 10.1097/01.rli.0000246147.44835.4c. [DOI] [PubMed] [Google Scholar]
- 106.Brekke C, Morgan SC, Lowe AS, Meade TJ, Price J, Williams SC, et al. The in vitro effects of a bimodal contrast agent on cellular functions and relaxometry. NMR Biomed. 2007 Apr;20(2):77–89. doi: 10.1002/nbm.1077. [DOI] [PubMed] [Google Scholar]
- 107.Weissleder R, Stark DD, Engelstad BL, Bacon BR, Compton CC, White DL, et al. Superparamagnetic iron oxide: pharmacokinetics and toxicity. AJR American Journal of Roentgenology. 1989;152(1):167–73. doi: 10.2214/ajr.152.1.167. [DOI] [PubMed] [Google Scholar]
- 108.Pouliquen D, Le Jeune JJ, Perdrisot R, Ermias A, Jallet P. Iron oxide nanoparticles for use as an MRI contrast agent: pharmacokinetics and metabolism. Magn Reson Imaging. 1991;9(3):275–83. doi: 10.1016/0730-725x(91)90412-f. [DOI] [PubMed] [Google Scholar]
- 109.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228(2):480–7. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 110.Arbab AS, Bashaw LA, Miller BR, Jordan EK, Lewis BK, Kalish H, et al. Characterization of biophysical and metabolic properties of cells labeled with superparamagnetic iron oxide nanoparticles and transfection agent for cellular MR imaging.[see comment] Radiology. 2003;229(3):838–46. doi: 10.1148/radiol.2293021215. [DOI] [PubMed] [Google Scholar]
- 111.Arbab AS, Yocum GT, Wilson LB, Parwana A, Jordan EK, Kalish H, et al. Comparison of transfection agents in forming complexes with ferumoxides, cell labeling efficiency, and cellular viability. Molecular Imaging: Official Journal of the Society for Molecular Imaging. 2004;3(1):24–32. doi: 10.1162/15353500200403190. [DOI] [PubMed] [Google Scholar]
- 112.Arbab AS, Bashaw LA, Miller BR, Jordan EK, Bulte JW, Frank JA. Intracytoplasmic tagging of cells with ferumoxides and transfection agent for cellular magnetic resonance imaging after cell transplantation: methods and techniques. Transplantation. 2003;76(7):1123–30. doi: 10.1097/01.TP.0000089237.39220.83. [DOI] [PubMed] [Google Scholar]
- 113.Arbab AS, Jordan EK, Wilson LB, Yocum GT, Lewis BK, Frank JA. In vivo trafficking and targeted delivery of magnetically labeled stem cells. Human Gene Therapy. 2004;15(4):351–60. doi: 10.1089/104303404322959506. [DOI] [PubMed] [Google Scholar]
- 114.Arbab AS, Rad AM, Iskander AS, Jafari-Khouzani K, Brown SL, Churchman JL, et al. Magnetically-labeled sensitized splenocytes to identify glioma by MRI: a preliminary study. Magn Reson Med. 2007 Sep;58(3):519–26. doi: 10.1002/mrm.21343. [DOI] [PubMed] [Google Scholar]
- 115.Arbab AS, Frank JA. Cellular MRI and its role in stem cell therapy. Regen Med. 2008 Mar;3(2):199–215. doi: 10.2217/17460751.3.2.199. [DOI] [PubMed] [Google Scholar]
- 116.Arbab AS, Pandit SD, Anderson SA, Yocum GT, Bur M, Frenkel V, et al. Magnetic resonance imaging and confocal microscopy studies of magnetically labeled endothelial progenitor cells trafficking to sites of tumor angiogenesis. Stem Cells. 2006 Mar;24(3):671–8. doi: 10.1634/stemcells.2005-0017. [DOI] [PubMed] [Google Scholar]
- 117.Jung CW, Jacobs P. Physical and chemical properties of superparamagnetic iron oxide MR contrast agents: ferumoxides, ferumoxtran, ferumoxsil. Magn Reson Imaging. 1995;13(5):661–74. doi: 10.1016/0730-725x(95)00024-b. [DOI] [PubMed] [Google Scholar]
- 118.Bluemke D, Weber TM, Rubin D, de Lange EE, Semelka R, Redvanly RD, Chezmar J, Outwater E, Carlos R, Saini S, Holland GA, Mammone JF, Brown JJ, Milestone B, Javitt MC, Jacobs P. Hepatic MR imaging with ferumoxides: multicenter study of safety and effectiveness of direct injection protocol. Radiology. 2003 Aug;228(2):457–64. doi: 10.1148/radiol.2282012061. [DOI] [PubMed] [Google Scholar]
- 119.Harisinghani M, Barentsz J, Hahn PF, Deserno WM, Tabatabaei S, van de Kaa CH, de la Rosette J, Weissleder R. Noninvasive detection of clinically occult lymph-node metastases in prostate cancer. N Engl J Med. 2003 Sept 4;343(25):2491–9. doi: 10.1056/NEJMoa022749. [DOI] [PubMed] [Google Scholar]
- 120.Harisinghani MG, Saini S, Weissleder R, Halpern EF, Schima W, Rubin DL, et al. Differentiation of liver hemangiomas from metastases and hepatocellular carcinoma at MR imaging enhanced with blood-pool contrast agent Code-7227. Radiology. 1997;202(3):687–91. doi: 10.1148/radiology.202.3.9051017. [DOI] [PubMed] [Google Scholar]
- 121.Harisinghani M, Saini S, Weissleder R, Hahn PF, Yantiss RK, Tempany C, Wood BJ, Mueller PR. MR lymphangiography using ultrasmall superparamagnetic iron oxide in patients with primary abdominal and pelvic malignancies: radiographic-pathologic correlation. AJR Am J Roentgenol. 1999 May;172(5):1347–51. doi: 10.2214/ajr.172.5.10227514. [DOI] [PubMed] [Google Scholar]
- 122.Mack MG, Balzer JO, Straub R, Eichler K, Vogl TJ. Superparamagnetic iron oxide-enhanced MR imaging of head and neck lymph nodes. Radiology. 2002;222(1):239–44. doi: 10.1148/radiol.2221010225. [DOI] [PubMed] [Google Scholar]
- 123.Fleige G, Seeberger F, Laux D, Kresse M, Taupitz M, Pilgrimm H, et al. In vitro characterization of two different ultrasmall iron oxide particles for magnetic resonance cell tracking. Invest Radiol. 2002 Sep;37(9):482–8. doi: 10.1097/00004424-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 124.Bulte JW, Douglas T, Witwer B, Zhang SC, Strable E, Lewis BK, et al. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nature Biotechnology. 2001;19(12):1141–7. doi: 10.1038/nbt1201-1141. [DOI] [PubMed] [Google Scholar]
- 125.Josephson L, Tung CH, Moore A, Weissleder R. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconjug Chem. 1999;10(2):186–91. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 126.Dodd CH, Hsu HC, Chu WJ, Yang P, Zhang HG, Mountz JD, Jr, et al. Normal T-cell response and in vivo magnetic resonance imaging of T cells loaded with HIV transactivator-peptide-derived superparamagnetic nanoparticles. Journal of Immunological Methods. 2001;256(1–2):89–105. doi: 10.1016/s0022-1759(01)00433-1. [DOI] [PubMed] [Google Scholar]
- 127.Moore A, Sun PZ, Cory D, Hogemann D, Weissleder R, Lipes MA. MRI of insulitis in autoimmune diabetes. Magn Reson Med. 2002 Apr;47(4):751–8. doi: 10.1002/mrm.10110. [DOI] [PubMed] [Google Scholar]
- 128.Kircher MF, Allport JR, Graves EE, Love V, Josephson L, Lichtman AH, et al. In vivo high resolution three-dimensional imaging of antigen-specific cytotoxic T-lymphocyte trafficking to tumors. Cancer Res. 2003 Oct 15;63(20):6838–46. [PubMed] [Google Scholar]
- 129.Moore A, Grimm J, Han B, Santamaria P. Tracking the recruitment of diabetogenic CD8+ T-cells to the pancreas in real time. Diabetes. 2004 Jun;53(6):1459–66. doi: 10.2337/diabetes.53.6.1459. [DOI] [PubMed] [Google Scholar]
- 130.Bulte JW, Zhang S, van Gelderen P, Herynek V, Jordan EK, Duncan ID, et al. Neurotransplantation of magnetically labeled oligodendrocyte progenitors: magnetic resonance tracking of cell migration and myelination. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):15256–61. doi: 10.1073/pnas.96.26.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Toyoda K, Tooyama I, Kato M, Sato H, Morikawa S, Hisa Y, et al. Effective magnetic labeling of transplanted cells with HVJ-E for magnetic resonance imaging. Neuroreport. 2004 Mar 22;15(4):589–93. doi: 10.1097/00001756-200403220-00004. [DOI] [PubMed] [Google Scholar]
- 132.Miyoshi S, Flexman JA, Cross DJ, Maravilla KR, Kim Y, Anzai Y, Oshima J, Minoshima S. Transfection of Neuroprogenitor Cells with Iron Nanoparticles for Magnetic Resonance Imaging Tracking: Cell Viability, Differentiation, and Intracellular Localization. Mol Imaging iol. 2005 Aug;4(1):10. doi: 10.1007/s11307-005-0008-1. [DOI] [PubMed] [Google Scholar]
- 133.Song Y, Morikawa S, Morita M, Inubushi T, Takada T, Torii R, et al. Magnetic resonance imaging using hemagglutinating virus of Japan-envelope vector successfully detects localization of intra-cardially administered microglia in normal mouse brain. Neurosci Lett. 2006 Feb 27;395(1):42–5. doi: 10.1016/j.neulet.2005.10.074. [DOI] [PubMed] [Google Scholar]
- 134.Shapiro EM, Skrtic S, Koretsky AP. Sizing it up: cellular MRI using micron-sized iron oxide particles. Magn Reson Med. 2005 Feb;53(2):329–38. doi: 10.1002/mrm.20342. [DOI] [PubMed] [Google Scholar]
- 135.Hinds KA, Hill JM, Shapiro EM, Laukkanen MO, Silva AC, Combs CA, et al. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003 Aug 1;102(3):867–72. doi: 10.1182/blood-2002-12-3669. [DOI] [PubMed] [Google Scholar]
- 136.Hill JM, Dick AJ, Raman VK, Thompson RB, Yu ZX, Hinds KA, et al. Serial cardiac magnetic resonance imaging of injected mesenchymal stem cells. Circulation. 2003 Aug 26;108(8):1009–14. doi: 10.1161/01.CIR.0000084537.66419.7A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ho C, Hitchens TK. A non-invasive approach to detecting organ rejection by MRI: monitoring the accumulation of immune cells at the transplanted organ. Curr Pharm Biotechnol. 2004 Dec;5(6):551–66. doi: 10.2174/1389201043376535. [DOI] [PubMed] [Google Scholar]
- 138.Wu YL, Ye Q, Foley LM, Hitchens TK, Sato K, Williams JB, et al. In situ labeling of immune cells with iron oxide particles: an approach to detect organ rejection by cellular MRI. Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1852–7. doi: 10.1073/pnas.0507198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006 Nov;56(5):1001–10. doi: 10.1002/mrm.21029. [DOI] [PubMed] [Google Scholar]
- 140.Zhang RL, Zhang L, Zhang ZG, Morris D, Jiang Q, Wang L, et al. Migration and differentiation of adult rat subventricular zone progenitor cells transplanted into the adult rat striatum. Neuroscience. 2003;116(2):373–82. doi: 10.1016/s0306-4522(02)00696-6. [DOI] [PubMed] [Google Scholar]
- 141.Davidson JM, Krieg T, Eming SA. Particle-mediated gene therapy of wounds. Wound Repair Regen. 2000 Nov–Dec;8(6):452–9. doi: 10.1046/j.1524-475x.2000.00452.x. [DOI] [PubMed] [Google Scholar]
- 142.Bhattarai SR, Kim SY, Jang KY, Lee KC, Yi HK, Lee DY, et al. Laboratory formulated magnetic nanoparticles for enhancement of viral gene expression in suspension cell line. J Virol Methods. 2008 Feb;147(2):213–8. doi: 10.1016/j.jviromet.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 143.Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kruger A, et al. Magnetofection: enhancing and targeting gene delivery by magnetic force in vitro and in vivo. Gene Ther. 2002 Jan;9(2):102–9. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- 144.Plank C, Scherer F, Schillinger U, Bergemann C, Anton M. Magnetofection: enhancing and targeting gene delivery with superparamagnetic nanoparticles and magnetic fields. J Liposome Res. 2003 Feb;13(1):29–32. doi: 10.1081/lpr-120017486. [DOI] [PubMed] [Google Scholar]
- 145.Soeren W. Gersting USJLPNCRCPDRJR. Gene delivery to respiratory epithelial cells by magnetofection. The Journal of Gene Medicine. 2004;6(8):913–22. doi: 10.1002/jgm.569. [DOI] [PubMed] [Google Scholar]
- 146.Stephanie Huth JLSWGCRCPUWJR. Insights into the mechanism of magnetofection using PEI-based magnetofectins for gene transfer. The Journal of Gene Medicine. 2004;6(8):923–36. doi: 10.1002/jgm.577. [DOI] [PubMed] [Google Scholar]
- 147.Mykhaylyk O, Antequera YS, Vlaskou D, Plank C. Generation of magnetic nonviral gene transfer agents and magnetofection in vitro. Nat Protocols. 2007;2(10):2391–411. doi: 10.1038/nprot.2007.352. [DOI] [PubMed] [Google Scholar]
- 148.Bulte JW, Arbab AS, Douglas T, Frank JA. Preparation of magnetically labeled cells for cell tracking by magnetic resonance imaging. Methods in Enzymology. 2004;386:275–99. doi: 10.1016/S0076-6879(04)86013-0. [DOI] [PubMed] [Google Scholar]
- 149.Frank JA, Zywicke H, Jordan EK, Mitchell J, Lewis BK, Miller B, et al. Magnetic intracellular labeling of mammalian cells by combining (FDA-approved) superparamagnetic iron oxide MR contrast agents and commonly used transfection agents. Acad Radiol. 2002 Aug;9(Suppl 2):S484–7. doi: 10.1016/s1076-6332(03)80271-4. [DOI] [PubMed] [Google Scholar]
- 150.Suzuki Y, Zhang S, Kundu P, Yeung AC, Robbins RC, Yang PC. In vitro comparison of the biological effects of three transfection methods for magnetically labeling mouse embryonic stem cells with ferumoxides. Magn Reson Med. 2007 Jun;57(6):1173–9. doi: 10.1002/mrm.21219. [DOI] [PubMed] [Google Scholar]
- 151.Neri M, Maderna C, Cavazzin C, Deidda-Vigoriti V, Politi LS, Scotti G, et al. Efficient In Vitro Labeling Of Human Neural Precursor Cells With Superparamagnetic Iron Oxide Particles: Relevance For In Vivo Cell Tracking. Stem Cells. 2007 Nov 15; doi: 10.1634/stemcells.2007-0251. [DOI] [PubMed] [Google Scholar]
- 152.Montet-Abou K, Montet X, Weissleder R, Josephson L. Transfection agent induced nanoparticle cell loading. Mol Imaging. 2005 Jul–Sep;4(3):165–71. doi: 10.1162/15353500200505100. [DOI] [PubMed] [Google Scholar]
- 153.Reynolds F, Weissleder R, Josephson L. Protamine as an efficient membrane-translocating peptide. Bioconjug Chem. 2005 Sep–Oct;16(5):1240–5. doi: 10.1021/bc0501451. [DOI] [PubMed] [Google Scholar]
- 154.Wu YJ, Muldoon LL, Varallyay C, Markwardt S, Jones RE, Neuwelt EA. In Vivo Leukocyte Labeling with Intravenous Ferumoxides/Protamine Sulfate Complex and in vitro Characterization for Cellular Magnetic Resonance Imaging. Am J Physiol Cell Physiol. 2007 September 26;2007:00215.2007. doi: 10.1152/ajpcell.00215.2007. [DOI] [PubMed] [Google Scholar]
- 155.Jacobs RE, Fraser SE. Magnetic resonance microscopy of embryonic cell lineages and movements. Science. 1994 Feb 4;263(5147):681–4. doi: 10.1126/science.7508143. [DOI] [PubMed] [Google Scholar]
- 156.Crich SG, Lanzardo S, Barge A, Aime S. Visualization through magnetic resonance imaging of DNA internalized following “in vivo” electroporation. Molecular Imaging. 2005;4(1):7–17. doi: 10.1162/15353500200504151. [DOI] [PubMed] [Google Scholar]
- 157.Walczak P, Kedziorek DA, Gilad AA, Lin S, Bulte JW. Instant MR labeling of stem cells using magnetoelectroporation. Magn Reson Med. 2005 Oct;54(4):769–74. doi: 10.1002/mrm.20701. [DOI] [PubMed] [Google Scholar]
- 158.Terreno E, Geninatti Crich S, Belfiore S, Biancone L, Cabella C, Esposito G, et al. Effect of the intracellular localization of a Gd-based imaging probe on the relaxation enhancement of water protons. Magn Reson Med. 2006 Mar;55(3):491–7. doi: 10.1002/mrm.20793. [DOI] [PubMed] [Google Scholar]
- 159.Gilad AA, Walczak P, McMahon MT, Na HB, Lee JH, An K, et al. MR tracking of transplanted cells with “positive contrast” using manganese oxide nanoparticles. Magn Reson Med. 2008 Jul;60(1):1–7. doi: 10.1002/mrm.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Erik M, Shapiro APK. Convertible manganese contrast for molecular and cellular MRI. Magnetic Resonance in Medicine. 2008;60(2):265–9. doi: 10.1002/mrm.21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Christopher H, Sotak KSAPK. Manganese cell labeling of murine hepatocytes using manganese(III)-transferrin. Contrast Media & Molecular Imaging. 2008;3(3):95–105. doi: 10.1002/cmmi.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang HT, Ding J, Chow GM, Dong ZL. Engineering Inorganic Hybrid Nanoparticles: Tuning Combination Fashions of Gold, Platinum, and Iron Oxide. Langmuir. 2008 Oct 17; doi: 10.1021/la802805w. [DOI] [PubMed] [Google Scholar]
- 163.Arbab AS, Yocum GT, Rad AM, Khakoo AY, Fellowes V, Read EJ, et al. Labeling of cells with ferumoxides-protamine sulfate complexes does not inhibit function or differentiation capacity of hematopoietic or mesenchymal stem cells. NMR Biomed. 2005;18(8):553–9. doi: 10.1002/nbm.991. [DOI] [PubMed] [Google Scholar]
- 164.Arbab AS, Yocum GT, Kalish H, Jordan EJ, Anderson SA, Khakoo AY, et al. Ferumoxides-protamine sulfate labeling does not alter differentiation of mesenchymal stem cells. Blood. 2004;104(10):3412–3. [Google Scholar]
- 165.Guzman R, Uchida N, Bliss TM, He D, Christopherson KK, Stellwagen D, et al. Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc Natl Acad Sci U S A. 2007 Jun 12;104(24):10211–6. doi: 10.1073/pnas.0608519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pawelczyk E, Arbab AS, Pandit S, Hu E, Frank JA. Expression of transferrin receptor and ferritin following ferumoxides-protamine sulfate labeling of cells: implications for cellular magnetic resonance imaging. NMR Biomed. 2006 Aug;19(5):581–92. doi: 10.1002/nbm.1038. [DOI] [PubMed] [Google Scholar]
- 167.Hsiao JK, Tai MF, Chu HH, Chen ST, Li H, Lai DM, et al. Magnetic nanoparticle labeling of mesenchymal stem cells without transfection agent: cellular behavior and capability of detection with clinical 1.5 T magnetic resonance at the single cell level. Magn Reson Med. 2007 Oct;58(4):717–24. doi: 10.1002/mrm.21377. [DOI] [PubMed] [Google Scholar]
- 168.Mailander V, Lorenz MR, Holzapfel V, Musyanovych A, Fuchs K, Wiesneth M, et al. Carboxylated superparamagnetic iron oxide particles label cells intracellularly without transfection agents. Mol Imaging Biol. 2008 May–Jun;10(3):138–46. doi: 10.1007/s11307-007-0130-3. [DOI] [PubMed] [Google Scholar]
- 169.Farrell E, Wielopolski P, Pavljasevic P, van Tiel S, Jahr H, Verhaar J, et al. Effects of iron oxide incorporation for long term cell tracking on MSC differentiation in vitro and in vivo. Biochem Biophys Res Commun. 2008 May 16;369(4):1076–81. doi: 10.1016/j.bbrc.2008.02.159. [DOI] [PubMed] [Google Scholar]
- 170.Schulze E, Ferrucci JT, Jr, Poss K, Lapointe L, Bogdanova A, Weissleder R. Cellular uptake and trafficking of a prototypical magnetic iron oxide label in vitro. Invest Radiol. 1995 Oct;30(10):604–10. doi: 10.1097/00004424-199510000-00006. [DOI] [PubMed] [Google Scholar]
- 171.Link G, Pinson A, Hershko C. Iron loading of cultured cardiac myocytes modifies sarcolemmal structure and increases lysosomal fragility. J Lab Clin Med. 1993 Jan;121(1):127–34. [PubMed] [Google Scholar]
- 172.Yu Z, Persson HL, Eaton JW, Brunk UT. Intralysosomal iron: a major determinant of oxidant-induced cell death. Free Radic Biol Med. 2003 May 15;34(10):1243–52. doi: 10.1016/s0891-5849(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 173.Arbab AS, Wilson LB, Ashari P, Jordan EK, Lewis BK, Frank JA. A model of lysosomal metabolism of dextran coated superparamagnetic iron oxide (SPIO) nanoparticles: implications for cellular magnetic resonance imaging. NMR Biomed. 2005;18(6):383–9. doi: 10.1002/nbm.970. [DOI] [PubMed] [Google Scholar]
- 174.Corot C, Petry KG, Trivedi R, Saleh A, Jonkmanns C, Le Bas JF, et al. Macrophage imaging in central nervous system and in carotid atherosclerotic plaque using ultrasmall superparamagnetic iron oxide in magnetic resonance imaging. Invest Radiol. 2004 Oct;39(10):619–25. doi: 10.1097/01.rli.0000135980.08491.33. [DOI] [PubMed] [Google Scholar]
- 175.Saleh A, Schroeter M, Jonkmanns C, Hartung HP, Modder U, Jander S. In vivo MRI of brain inflammation in human ischaemic stroke. Brain. 2004 Jul;127(Pt 7):1670–7. doi: 10.1093/brain/awh191. [DOI] [PubMed] [Google Scholar]
- 176.Trivedi RA, JM UK-I, Graves MJ, Cross JJ, Horsley J, Goddard MJ, et al. In vivo detection of macrophages in human carotid atheroma: temporal dependence of ultrasmall superparamagnetic particles of iron oxide-enhanced MRI. Stroke. 2004 Jul;35(7):1631–5. doi: 10.1161/01.STR.0000131268.50418.b7. [DOI] [PubMed] [Google Scholar]
- 177.Siglienti I, Bendszus M, Kleinschnitz C, Stoll G. Cytokine profile of iron-laden macrophages: implications for cellular magnetic resonance imaging. J Neuroimmunol. 2006 Apr;173(1–2):166–73. doi: 10.1016/j.jneuroim.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 178.Stein M, Gordon S. Regulation of tumor necrosis factor (TNF) release by murine peritoneal macrophages: role of cell stimulation and specific phagocytic plasma membrane receptors. Eur J Immunol. 1991 Feb;21(2):431–7. doi: 10.1002/eji.1830210227. [DOI] [PubMed] [Google Scholar]
- 179.Muller K, Skepper JN, Posfai M, Trivedi R, Howarth S, Corot C, et al. Effect of ultrasmall superparamagnetic iron oxide nanoparticles (Ferumoxtran-10) on human monocyte-macrophages in vitro. Biomaterials. 2007 Mar;28(9):1629–42. doi: 10.1016/j.biomaterials.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 180.Metz S, Bonaterra G, Rudelius M, Settles M, Rummeny EJ, Daldrup-Link HE. Capacity of human monocytes to phagocytose approved iron oxide MR contrast agents in vitro. Eur Radiol. 2004 Oct;14(10):1851–8. doi: 10.1007/s00330-004-2405-2. [DOI] [PubMed] [Google Scholar]
- 181.Janic B, Iskander AS, Rad AM, Soltanian-Zadeh H, Arbab AS. Effects of ferumoxides-protamine sulfate labeling on immunomodulatory characteristics of macrophage-like THP-1 cells. PLoS ONE. 2008;3(6):e2499. doi: 10.1371/journal.pone.0002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rieck B, Schlaak S. Measurement in vivo of the survival rate in autologous adipocyte transplantation. Plast Reconstr Surg. 2003 Jun;111(7):2315–23. doi: 10.1097/01.PRS.0000060797.59958.55. [DOI] [PubMed] [Google Scholar]
- 183.Burns TC, Ortiz-Gonzalez XR, Gutierrez-Perez M, Keene CD, Sharda R, Demorest ZL, et al. Thymidine analogs are transferred from prelabeled donor to host cells in the central nervous system after transplantation: a word of caution. Stem Cells. 2006 Apr;24(4):1121–7. doi: 10.1634/stemcells.2005-0463. [DOI] [PubMed] [Google Scholar]
- 184.Ebert SN, Taylor DG, Nguyen HL, Kodack DP, Beyers RJ, Xu Y, et al. Noninvasive tracking of cardiac embryonic stem cells in vivo using magnetic resonance imaging techniques. Stem Cells. 2007 Nov;25(11):2936–44. doi: 10.1634/stemcells.2007-0216. [DOI] [PubMed] [Google Scholar]
- 185.Amsalem Y, Mardor Y, Feinberg MS, Landa N, Miller L, Daniels D, et al. Iron-oxide labeling and outcome of transplanted mesenchymal stem cells in the infarcted myocardium. Circulation. 2007 Sep 11;116(11 Suppl):I38–45. doi: 10.1161/CIRCULATIONAHA.106.680231. [DOI] [PubMed] [Google Scholar]
- 186.Terrovitis J, Stuber M, Youssef A, Preece S, Leppo M, Kizana E, et al. Magnetic resonance imaging overestimates ferumoxide-labeled stem cell survival after transplantation in the heart. Circulation. 2008 Mar 25;117(12):1555–62. doi: 10.1161/CIRCULATIONAHA.107.732073. [DOI] [PubMed] [Google Scholar]
- 187.Lepore AC, Walczak P, Rao MS, Fischer I, Bulte JW. MR imaging of lineage-restricted neural precursors following transplantation into the adult spinal cord. Exp Neurol. 2006 Sep;201(1):49–59. doi: 10.1016/j.expneurol.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 188.Brekke C, Williams SC, Price J, Thorsen F, Modo M. Cellular multiparametric MRI of neural stem cell therapy in a rat glioma model. Neuroimage. 2007 Sep 1;37(3):769–82. doi: 10.1016/j.neuroimage.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 189.Pawelczyk E, Arbab AS, Chaudhry A, Balakumaran A, Robey PG, Frank JA. In vitro model of bromodeoxyuridine or iron oxide nanoparticle uptake by activated macrophages from labeled stem cells: implications for cellular therapy. Stem Cells. 2008 May;26(5):1366–75. doi: 10.1634/stemcells.2007-0707. [DOI] [PubMed] [Google Scholar]
- 190.Turetschek K, Roberts TP, Floyd E, Preda A, Novikov V, Shames DM, et al. Tumor microvascular characterization using ultrasmall superparamagnetic iron oxide particles (USPIO) in an experimental breast cancer model. J Magn Reson Imaging. 2001 Jun;13(6):882–8. doi: 10.1002/jmri.1126. [DOI] [PubMed] [Google Scholar]
- 191.Turetschek K, Huber S, Floyd E, Helbich T, Roberts TP, Shames DM, et al. MR imaging characterization of microvessels in experimental breast tumors by using a particulate contrast agent with histopathologic correlation. Radiology. 2001 Feb;218(2):562–9. doi: 10.1148/radiology.218.2.r01fe37562. [DOI] [PubMed] [Google Scholar]
- 192.Jendelova P, Herynek V, DeCroos J, Glogarova K, Andersson B, Hajek M, et al. Imaging the fate of implanted bone marrow stromal cells labeled with superparamagnetic nanoparticles. Magn Reson Med. 2003;50(4):767–76. doi: 10.1002/mrm.10585. [DOI] [PubMed] [Google Scholar]
- 193.Bowen CV, Zhang X, Saab G, Gareau PJ, Rutt BK. Application of the static dephasing regime theory to superparamagnetic iron-oxide loaded cells. Magnetic Resonance in Medicine. 2002;48(1):52–61. doi: 10.1002/mrm.10192. [DOI] [PubMed] [Google Scholar]
- 194.Heyn C, Bowen CV, Rutt BK, Foster PJ. Detection threshold of single SPIO-labeled cells with FIESTA. Magn Reson Med. 2005 Feb;53(2):312–20. doi: 10.1002/mrm.20356. [DOI] [PubMed] [Google Scholar]
- 195.Lebel RM, Menon RS, Bowen CV. Relaxometry model of strong dipolar perturbers for balanced-SSFP: application to quantification of SPIO loaded cells. Magn Reson Med. 2006 Mar;55(3):583–91. doi: 10.1002/mrm.20799. [DOI] [PubMed] [Google Scholar]
- 196.Cunningham CH, Arai T, Yang PC, McConnell MV, Pauly JM, Conolly SM. Positive contrast magnetic resonance imaging of cells labeled with magnetic nanoparticles. Magn Reson Med. 2005 May;53(5):999–1005. doi: 10.1002/mrm.20477. [DOI] [PubMed] [Google Scholar]
- 197.Stuber M, Gilson WD, Schar M, Kedziorek DA, Hofmann LV, Shah S, et al. Positive contrast visualization of iron oxide-labeled stem cells using inversion-recovery with ON-resonant water suppression (IRON) Magn Reson Med. 2007 Nov;58(5):1072–7. doi: 10.1002/mrm.21399. [DOI] [PubMed] [Google Scholar]
- 198.Zurkiya O, Hu X. Off-resonance saturation as a means of generating contrast with superparamagnetic nanoparticles. Magn Reson Med. 2006 Oct;56(4):726–32. doi: 10.1002/mrm.21024. [DOI] [PubMed] [Google Scholar]
- 199.Seppenwoolde JH, Viergever MA, Bakker CJ. Passive tracking exploiting local signal conservation: the white marker phenomenon. Magn Reson Med. 2003 Oct;50(4):784–90. doi: 10.1002/mrm.10574. [DOI] [PubMed] [Google Scholar]
- 200.Mani V, Briley-Saebo KC, Itskovich VV, Samber DD, Fayad ZA. Gradient echo acquisition for superparamagnetic particles with positive contrast (GRASP): sequence characterization in membrane and glass superparamagnetic iron oxide phantoms at 1.5T and 3T. Magn Reson Med. 2006 Jan;55(1):126–35. doi: 10.1002/mrm.20739. [DOI] [PubMed] [Google Scholar]
- 201.Posse S. Direct imaging of magnetic field gradients by group spin-echo selection. Magn Reson Med. 1992 May;25(1):12–29. doi: 10.1002/mrm.1910250103. [DOI] [PubMed] [Google Scholar]
- 202.Reichenbach JR, Venkatesan R, Yablonskiy DA, Thompson MR, Lai S, Haacke EM. Theory and application of static field inhomogeneity effects in gradient-echo imaging. J Magn Reson Imaging. 1997 Mar–Apr;7(2):266–79. doi: 10.1002/jmri.1880070203. [DOI] [PubMed] [Google Scholar]
- 203.Bakker CJ, Seppenwoolde JH, Vincken KL. Dephased MRI. Magn Reson Med. 2006 Jan;55(1):92–7. doi: 10.1002/mrm.20733. [DOI] [PubMed] [Google Scholar]
- 204.Liu W, Dahnke H, Jordan EK, Schaeffter T, Frank JA. In vivo MRI using positive-contrast techniques in detection of cells labeled with superparamagnetic iron oxide nanoparticles. NMR Biomed. 2008 Mar;21(3):242–50. doi: 10.1002/nbm.1187. [DOI] [PubMed] [Google Scholar]
- 205.Verdijk P, Scheenen TW, Lesterhuis WJ, Gambarota G, Veltien AA, Walczak P, et al. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. Int J Cancer. 2007 Mar 1;120(5):978–84. doi: 10.1002/ijc.22385. [DOI] [PubMed] [Google Scholar]
- 206.Dahnke H, Schaeffter T. Limits of detection of SPIO at 3.0 T using T2 relaxometry. Magn Reson Med. 2005 May;53(5):1202–6. doi: 10.1002/mrm.20435. [DOI] [PubMed] [Google Scholar]
- 207.Hoehn M, Kustermann E, Blunk J, Wiedermann D, Trapp T, Wecker S, Focking M, Arnold H, Hescheler J, Fleischmann BK, Schwindt W, Buhrle C. Monitoring of implanted stem cell migration in vivo: a highly resolved in vivo magnetic resonance imaging investigation of experimental stroke in rat. Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16267–72. doi: 10.1073/pnas.242435499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dodd SJ, Williams M, Suhan JP, Williams DS, Koretsky AP, Ho C. Detection of single mammalian cells by high-resolution magnetic resonance imaging. Biophys J. 1999 Jan;76(1 Pt 1):103–9. doi: 10.1016/S0006-3495(99)77182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Foster-Gareau P, Heyn C, Alejski A, Rutt BK. Imaging single mammalian cells with a 1.5 T clinical MRI scanner. Magnetic Resonance in Medicine. 2003;49(5):968–71. doi: 10.1002/mrm.10417. [DOI] [PubMed] [Google Scholar]
- 210.Rad AM, Arbab AS, Iskander AS, Jiang Q, Soltanian-Zadeh H. Quantification of superparamagnetic iron oxide (SPIO)-labeled cells using MRI. J Magn Reson Imaging. 2007 Aug;26(2):366–74. doi: 10.1002/jmri.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bos C, Delmas Y, Desmouliere A, Solanilla A, Hauger O, Grosset C, et al. In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver.[see comment] Radiology. 2004;233(3):781–9. doi: 10.1148/radiol.2333031714. [DOI] [PubMed] [Google Scholar]
- 212.Politi LS, Bacigaluppi M, Brambilla E, Cadioli M, Falini A, Comi G, et al. Magnetic-resonance-based tracking and quantification of intravenously injected neural stem cell accumulation in the brains of mice with experimental multiple sclerosis. Stem Cells. 2007 Oct;25(10):2583–92. doi: 10.1634/stemcells.2007-0037. [DOI] [PubMed] [Google Scholar]
- 213.Young IR, Cox IJ, Bryant DJ, Bydder GM. The benefits of increasing spatial resolution as a means of reducing artifacts due to field inhomogeneities. Magn Reson Imaging. 1988 Sep–Oct;6(5):585–90. doi: 10.1016/0730-725x(88)90133-6. [DOI] [PubMed] [Google Scholar]
- 214.Frahm J, Merboldt KD, Hanicke W. Direct FLASH MR imaging of magnetic field inhomogeneities by gradient compensation. Magn Reson Med. 1988 Apr;6(4):474–80. doi: 10.1002/mrm.1910060412. [DOI] [PubMed] [Google Scholar]
- 215.Kuhlpeter R, Dahnke H, Matuszewski L, Persigehl T, von Wallbrunn A, Allkemper T, et al. R2 and R2* mapping for sensing cell-bound superparamagnetic nanoparticles: in vitro and murine in vivo testing. Radiology. 2007 Nov;245(2):449–57. doi: 10.1148/radiol.2451061345. [DOI] [PubMed] [Google Scholar]
- 216.Nakamura K, Ito Y, Kawano Y, Kurozumi K, Kobune M, Tsuda H, et al. Antitumor effect of genetically engineered mesenchymal stem cells in a rat glioma model. Gene Ther. 2004;11(14):1155–64. doi: 10.1038/sj.gt.3302276. [DOI] [PubMed] [Google Scholar]
- 217.Brown AB, Yang W, Schmidt NO, Carroll R, Leishear KK, Rainov NG, et al. Intravascular delivery of neural stem cell lines to target intracranial and extracranial tumors of neural and non-neural origin. Hum Gene Ther. 2003;14(18):1777–85. doi: 10.1089/104303403322611782. [DOI] [PubMed] [Google Scholar]
- 218.Aboody KS, Brown A, Rainov NG, Bower KA, Liu S, Yang W, et al. Neural stem cells display extensive tropism for pathology in adult brain: evidence from intracranial gliomas. Proc Natl Acad Sci U S A. 2000;97(23):12846–51. doi: 10.1073/pnas.97.23.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ehtesham M, Kabos P, Kabosova A, Neuman T, Black KL, Yu JS. The use of interleukin 12-secreting neural stem cells for the treatment of intracranial glioma. Cancer Research. 2002;62(20):5657–63. [PubMed] [Google Scholar]
- 220.Schichor C, Birnbaum T, Etminan N, Schnell O, Grau S, Miebach S, et al. Vascular endothelial growth factor A contributes to glioma-induced migration of human marrow stromal cells (hMSC) Exp Neurol. 2006 199;2:301–10. doi: 10.1016/j.expneurol.2005.11.027. Epub 2006 Mar 29. [DOI] [PubMed] [Google Scholar]
- 221.Nakamizo A, Marini F, Amano T, Khan A, Studeny M, Gumin J, et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65(8):3307–18. doi: 10.1158/0008-5472.CAN-04-1874. [DOI] [PubMed] [Google Scholar]
- 222.Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, Honmou O, et al. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96(3):149–56. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ferrari N, Glod J, Lee J, Kobiler D, Fine HA. Bone marrow-derived, endothelial progenitor-like cells as angiogenesis-selective gene-targeting vectors. Gene Therapy. 2003;10(8):647–56. doi: 10.1038/sj.gt.3301883. [DOI] [PubMed] [Google Scholar]
- 224.Brentjens RJ, Latouche JB, Santos E, Marti F, Gong MC, Lyddane C, et al. Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat Med. 2003 Mar;9(3):279–86. doi: 10.1038/nm827. [DOI] [PubMed] [Google Scholar]
- 225.Daldrup-Link HE, Meier R, Rudelius M, Piontek G, Piert M, Metz S, et al. In vivo tracking of genetically engineered, anti-HER2/neu directed natural killer cells to HER2/neu positive mammary tumors with magnetic resonance imaging. Eur Radiol. 2005 Jan;15(1):4–13. doi: 10.1007/s00330-004-2526-7. [DOI] [PubMed] [Google Scholar]
- 226.Yamanaka R, Abe T, Yajima N, Tsuchiya N, Homma J, Kobayashi T, et al. Vaccination of recurrent glioma patients with tumour lysate-pulsed dendritic cells elicits immune responses: results of a clinical phase I/II trial. Br J Cancer. 2003;89(7):1172–9. doi: 10.1038/sj.bjc.6601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yamanaka R, Yajima N, Abe T, Tsuchiya N, Homma J, Narita M, et al. Dendritic cell-based glioma immunotherapy (review) Int J Oncol. 2003;23(1):5–15. [PubMed] [Google Scholar]
- 228.Ni HT, Spellman SR, Jean WC, Hall WA, Low WC. Immunization with dendritic cells pulsed with tumor extract increases survival of mice bearing intracranial gliomas. J Neurooncol. 2001;51(1):1–9. doi: 10.1023/a:1006452726391. [DOI] [PubMed] [Google Scholar]
- 229.Yang L, Ng KY, Lillehei KO. Cell-mediated immunotherapy: a new approach to the treatment of malignant glioma. Cancer Control. 2003;10(2):138–47. doi: 10.1177/107327480301000205. [DOI] [PubMed] [Google Scholar]
- 230.Yu JS, Wheeler CJ, Zeltzer PM, Ying H, Finger DN, Lee PK, et al. Vaccination of malignant glioma patients with peptide-pulsed dendritic cells elicits systemic cytotoxicity and intracranial T-cell infiltration. Cancer Res. 2001;61(3):842–7. [PubMed] [Google Scholar]
- 231.Yu JS, Liu G, Ying H, Yong WH, Black KL, Wheeler CJ. Vaccination with tumor lysate-pulsed dendritic cells elicits antigen-specific, cytotoxic T-cells in patients with malignant glioma. Cancer Res. 2004;64(14):4973–9. doi: 10.1158/0008-5472.CAN-03-3505. [DOI] [PubMed] [Google Scholar]
- 232.Fields RC, Shimizu K, Mule JJ. Murine dendritic cells pulsed with whole tumor lysates mediate potent antitumor immune responses in vitro and in vivo. Proc Natl Acad Sci U S A. 1998;95(16):9482–7. doi: 10.1073/pnas.95.16.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Smithers M, O’Connell K, MacFadyen S, Chambers M, Greenwood K, Boyce A, et al. Clinical response after intradermal immature dendritic cell vaccination in metastatic melanoma is associated with immune response to particulate antigen. Cancer Immunol Immunother. 2003;52(1):41–52. doi: 10.1007/s00262-002-0318-y. Epub 2002 Nov 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dembic Z, Schenck K, Bogen B. Dendritic cells purified from myeloma are primed with tumor-specific antigen (idiotype) and activate CD4+ T cells. Proc Natl Acad Sci U S A. 2000;97(6):2697–702. doi: 10.1073/pnas.050579897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Merchant RE, Baldwin NG, Rice CD, Bear HD. Adoptive immunotherapy of malignant glioma using tumor-sensitized T lymphocytes. Neurol Res. 1997;19(2):145–52. doi: 10.1080/01616412.1997.11740788. [DOI] [PubMed] [Google Scholar]
- 236.Geyer SJ, Landay A. Immunogenetic and immunologic aspects of gliosarcoma growth in rats. Laboratory Investigation. 1983;49(4):436–44. [PubMed] [Google Scholar]
- 237.Geyer SJ, Gill TJ, 3rd, Kunz HW, Moody E. Immunogenetic aspects of transplantation in the rat brain. Transplantation. 1985;39(3):244–7. doi: 10.1097/00007890-198503000-00005. [DOI] [PubMed] [Google Scholar]
- 238.de Vries IJ, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, Boerman OC, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol. 2005;23(11):1407–13. doi: 10.1038/nbt1154. Epub 2005 Oct 30. [DOI] [PubMed] [Google Scholar]
- 239.Zhu J, Zhou L, XingWu F. Tracking neural stem cells in patients with brain trauma. N Engl J Med. 2006 Nov 30;355(22):2376–8. doi: 10.1056/NEJMc055304. [DOI] [PubMed] [Google Scholar]
- 240.Toso C, Vallee JP, Morel P, Ris F, Demuylder-Mischler S, Lepetit-Coiffe M, et al. Clinical magnetic resonance imaging of pancreatic islet grafts after iron nanoparticle labeling. Am J Transplant. 2008 Mar;8(3):701–6. doi: 10.1111/j.1600-6143.2007.02120.x. [DOI] [PubMed] [Google Scholar]
- 241.Callera F, de Melo CM. Magnetic resonance tracking of magnetically labeled autologous bone marrow CD34+ cells transplanted into the spinal cord via lumbar puncture technique in patients with chronic spinal cord injury: CD34+ cells’ migration into the injured site. Stem Cells Dev. 2007 Jun;16(3):461–6. doi: 10.1089/scd.2007.0083. [DOI] [PubMed] [Google Scholar]
- 242.Arbab AS, Liu W, Frank JA. Cellular magnetic resonance imaging: current status and future prospects. Expert Rev Med Devices. 2006 Jul;3(4):427–39. doi: 10.1586/17434440.3.4.427. [DOI] [PubMed] [Google Scholar]
- 243.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008 Apr 10;2(4):313–9. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Haacke EM, Xu Y, Cheng YC, Reichenbach JR. Susceptibility weighted imaging (SWI) Magn Reson Med. 2004;52:612–8. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]