Abstract
The conversion of normal cellular prion protein to disease-associated prion protein (PrPSc) is a fundamental component of prion disease pathogenesis. The molecular mechanisms contributing to prion conversion and the impact of PrPSc accumulation on cellular biology are not fully understood. To further define the molecular changes associated with PrPSc accumulation in cultured cells, the transcriptional profile of PrPSc-accumulating primary ovine microglia was compared to the profile of PrPSc-lacking microglia using the Affymetrix Bovine Genome Array. The experimental design included three biological replicates, each with three technical replicates, and samples that were collected at the point of near maximal PrPSc accumulation levels as measured by ELISA. The array analysis revealed only 19 upregulated genes and 30 downregulated genes in PrPSc-accumulating microglia. The results support the hypothesis that chronic PrPSc accumulation in cultured microglia results in a limited transcriptional response.
Keywords: ovine, microglia, prion, scrapie, microarray
Introduction
Transmissible spongiform encephalopathies (TSE, prion diseases) are fatal, transmissible, neurodegenerative diseases, including scrapie in sheep and goats, and Creutzfeldt-Jakob disease in humans. Many of the underlying molecular mechanisms associated with the posttranslational conformational change of prion protein and the resulting cellular response are either unknown or poorly understood [1]. Although there are no known natural rodent prion diseases, previous studies into the transcriptomics associated with prion diseases have primarily employed the use of rodent models with only two studies using a sheep model [2, 3]. Neither of these sheep model experiments used cell cultures, but rather they used whole tissues. This prevents the determination of the cell type(s) responding to PrPSc and an inability to determine if the response is directly or indirectly related to PrPSc. To the authors’ knowledge, there is only one report of a large-scale transcriptomic study based on a cell line from a natural TSE host (human SH-SY5Y neuroblastoma cell line) [4]. Sheep scrapie is a logical experimental model for the investigation of a natural prion disease in a natural host. Currently there are no studies that have investigated the response of primary natural TSE-host cells to PrPSc accumulation.
Importantly, the results of previous rodent-based cell culture transcriptomic studies have conflicted. Not only are different transcripts often differentially regulated in these studies, but the overall transcriptional response varies from a robust response [4, 5] to a limited response [6, 7]. In light of these conflicting results and because PrPSc is a misfolded host protein, and not a foreign protein [1], the following study tests the hypothesis that microglia have a limited response to PrPSc accumulation. Additionally, this study extends the prion transcriptomic studies into sheep, the host of the prototypical prion disease, and focuses on the activation of microglia, which are likely contributors to prion-induced neuropathology [8].
Materials and Methods
Inoculation of primary microglia with PrPSc
Primary mixed glial cultures were obtained from a previously confirmed homozygous VRQ/VRQ [9] ovine fetal brain and cultured using a previously described technique [10]. VRQ inoculum derived from mechanical lysates of inoculated (Rov9Sc) and uninoculated (Rov9C) Rov9 cells [11] was used, as previously described [10].
Primary microglia were passed into 6-well plates, grown to approximately 60% confluency, and treated in triplicate. Briefly, microglia were rinsed with PBS, overlaid with 200 μl of a 1/20 dilution of either the Rov9Sc lysate (Inoc A, B, and C) or the Rov9C lysate (Mock A, B, and C) in OPTI-MEM I Reduced Serum Medium (Invitrogen, Carlsbad, CA), incubated for six hours, and then 200 μl of maintenance medium [10] were added to each well. After two days of incubation, 0.5 ml of maintenance medium was added, and microglia were incubated for four days at which time they were expanded into 25-cm2 tissue culture flasks. Microglia were fed every three to four days as necessary and serially passed 1/5.
As previously described [10], at selected passages following trypsinization, 4/5 of the microglial cell suspension from a 25-cm2 tissue culture flask was analyzed for PrPSc by commercial ELISA (HerdChek™ Scrapie Antigen Test Kit ELISA, IDEXX, Westbrook, ME) following the manufacturer's instructions.
RNA Collection, preparation, and microarray hybridization
Microarray experiments were performed in biological triplicates, each with its own set of technical triplicates. Near the maximal PrPSc levels, each biological replicate was passaged into three 75-cm2 flasks for technical triplicates of RNA collection (Inoc A1-A3 and so on, and Mock A1-A3 and so on). At approximately 90–100% confluency, each technical replicate was independently trypsinized, shredded (QIAshredder, Qiagen, Valencia, CA), and RNA was purified using RNeasy mini spin columns (Qiagen). Total RNA quantity and quality were determined by spectrophotometry and agarose gel electrophoresis, respectively. Samples for InocB and MockB were collected and analyzed by microarray at passage 8 post-PrPSc inoculation, and samples for InocA, InocC, MockA, and MockC were collected and analyzed at passage 6 post-inoculation.
Biotinylated cRNA targets were generated from total RNA using the One-Cycle Target Labeling kit (Affymetrix, Santa Clara, CA), fragmented, and then hybridized to the Affymetrix GeneChip Bovine Genome Array (representing approximately 23,000 transcripts). Arrays were processed on an Affymetrix GeneChip Fluidics Station 400 and scanned on a Gene/Array Scanner 2500A (Agilent, Santa Clara, CA). These steps were performed at the Laboratory for Biotechnology and Bioanalysis 1 at Washington State University (Pullman, WA). Complete microarray data is available online, Gene Expression Omnibus Accession Number GSE12688.
Microarray data analysis
Eighteen (two treatment groups, three biological replicates per treatment group, and three technical replicates per biological replicate) raw Affymetrix “.CEL” files were analyzed by the R programming language-based [12] Bioconductor [13] package “affy” [14]. The affyQCReport package (C. Parman, C. Halling, and R. Gentleman. affyQCReport: QC Report Generation for affyBatch objects. R package version 1.14.0) was used to assess quality control parameters.
The number of transcripts detected in this study was compared to previous reports that used ovine or bovine mRNA on the Affymetrix GeneChip Bovine array. Probe-level robust linear model fitting was accomplished via the affyPLM package (Ben Bolstad [2007]. affyPLM: Methods for fitting probe-level models. R package version 1.12.0. [http://bmbolstad.com]). Only those transcripts that were present in all three biological replicates were considered “detected”.
For determining differentially expressed transcripts, microarray data were preprocessed using the Micro Array Suite 5.0 (MAS5) [15] algorithm. Probe sets consistently present in at least one treatment group, as determined by the cutoff established by the MAS5 algorithm, and demonstrated at least a 0.5 log2 change in the interquartile range were analyzed further. Hierarchical clustering using the hclust function (Euclidean metrics, complete linkage) demonstrated a batch effect, which was corrected using the empirical Bayes-based ComBat algorithm [16]. Values for the technical replicates were averaged to attain a single value for each biological replicate. Differential transcriptional levels were determined by fitting the LIMMA linear model [17] to each probe set and correcting for multiple comparisons by controlling the false discover rate via the Benjamini and Hochberg method [18] with α < 0.05.
The results of this study using the Bovine Genome Array were compared to previous mouse and human model-based prion studies. The mouse and human homologues of the differentially expressed transcripts were determined via the annotations provided by Affymetrix NetAffx Analysis Center (accessed July 23, 2008 http://www.affymetrix.com/analysis/index.affx]). Probe sets representing the differentially expressed transcripts were also annotated to GO using the GO annotations available on NetAffx (accessed July 23, 2008).
Quantitative RT-PCR
Quantitative RT-PCR was performed on several genes to verify microarray results. RNA samples were treated with DNase (DNA-free kit, Ambion, Austin, TX) followed by Dnase Inactivation Reagent (Ambion) and centrifugation at 10,000 × g for 1.5 min. Samples were pooled for each set of technical replicates within a biological replicate and each reaction was conducted in triplicate. One microgram of RNA was reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis Supermix for qRT-PCR (Invitrogen). Quantitative real-time PCR was performed in an iCycler iQ (Bio-Rad, Hercules, CA). The 20 μl reaction mix contained 1× SYBR GreenER qPCR SuperMix for iCycler (Invitrogen), 200 nM of each specific primer, 8 μl of 1/100 diluted cDNA (1 μg), and water. Reaction conditions were 50°C for 2 min, 95°C for 8.5 min, 40 cycles of denaturation at 95°C for 15 sec and annealing at 59°C for 1 min followed immediately by a melt curve. GAPDH primers for sheep have been previously reported [19]; all other gene specific primers (Table 1) were designed by using PrimerQuest (Integrated DNA Technologies [http://www.idtdna.com/Scitools/Applications/Primerquest/]). Negative controls for quantitative RT-PCR included RNA processed without reverse transcriptase, and no-template controls for qRT-PCR. Relative transcript levels were calculated using the ΔΔCT method with normalization to GAPDH [20].
Table 1.
qRT-PCR primer information
| Gene Symbol |
Forward primer sequence | Reverse Primer Sequence | Amplicon size (bp) |
Reference sequence |
|---|---|---|---|---|
| PF4 | AATGCGTGTGCCTGAAGACCACTT | TTCCCGTCTTCAGCGTAGCTATCA | 159 | P30035a |
| MFGE8 | TCTCTGGCACCCAGTCCTCAA | AAAGGTCACCCACGCTCTGTACTT | 107 | NA |
| GALNT13 | TAGACAACATGGGCCGCAAGGAAA | ATCCAAGCACAAGTCATCGGTTCG | 125 | NA |
| RCAN1 | ATTCACACCTGTTGTGCACTTGGG | ATTAATGGGAAAGGCACTGCACCC | 184 | AY205232 |
| CCL2 | AGAAGAGTCACCACCAGCAAGTGT | TTGAGATGGTTTATGGCGTCCTGG | 119 | [39] |
| FAM14A | AAGCAACCAAAGGGACCACATCAG | AGATTCCTGTTCTGGTGAAGCCCA | 175 | BN000255 |
| MGC155285 | GTTTGGGCTCAGCAATTCCACTGT | CTGCTCCTTCGGATTAAAGGTTCAC | 132 | NA |
| TPM2 | ACCGAGCTATGAAGGATGAGGA | CTCTCCTTCCAGGATCACCAGTTT | 131 | NA |
| NR4A1 | CACACACACCGCCATGCTGTAAAT | AAGGAGGGCAGGAAGAGAGCAAGTA | 138 | NA |
| JUN | AACAGGTGGCCCAGCTTAAACAGA | TTGCAACTGCTGCGTTAGCATGAG | 80 | NA |
A protein sequence reference is supplied for PF4 as there is no nucleotide sequence is available for ovine PF4. The reference protein sequence was manually aligned with the bovine sequence to determine areas of amino acid similarity, and inferred nucleotide similarity. NA, not applicable and microarray probe sequences used to design primers.
Results
Primary sheep microglia were inoculated with PrPSc to determine their transcriptional response to PrPSc. Accumulation of PrPSc was monitored over time with a commercially available ELISA for PrPSc that was previously shown to correlate with PrPSc immunoblots [10].
Evaluation of Bovine Genome Array and verification of microglial cultures
Before identifying genes differentially regulated following PrPSc exposure, it was determined if the ovine mRNA used in this study yielded detectable transcripts when hybridized to the Affymetrix Bovine Genome Array. For each treatment group, over 13,000 (55%) transcripts were detected in all three of the biological replicates. This was consistent with previous reports of using ovine mRNA [21] and bovine mRNA [22] on the Affymetrix Bovine Genome Array.
While the cell isolation procedure used in this study has been previously described in sheep and shown to result in cultures of microglia [10], the transcript levels of five genes commonly used to identify microglia were analyzed to further characterize the cells. Toll-like receptor 4, CD68, and cathepsin K were detected in all technical replicates for each of the six biological groups. CD14 and CD163L1 were detected in the majority of technical replicates for each of the biological groups. The possibility of significant contamination of the cultures with astrocytes and endothelial cells was further excluded by the lack of transcripts for glial fibrillary acidic protein and von Willebrand factor, respectively. Based on these transcript features, the primary cell cultures were classified as uniformly sheep microglia and it was demonstrated that the Bovine Genome Array provides useful data from ovine samples.
Identification of differentially regulated genes associated with PrPSc
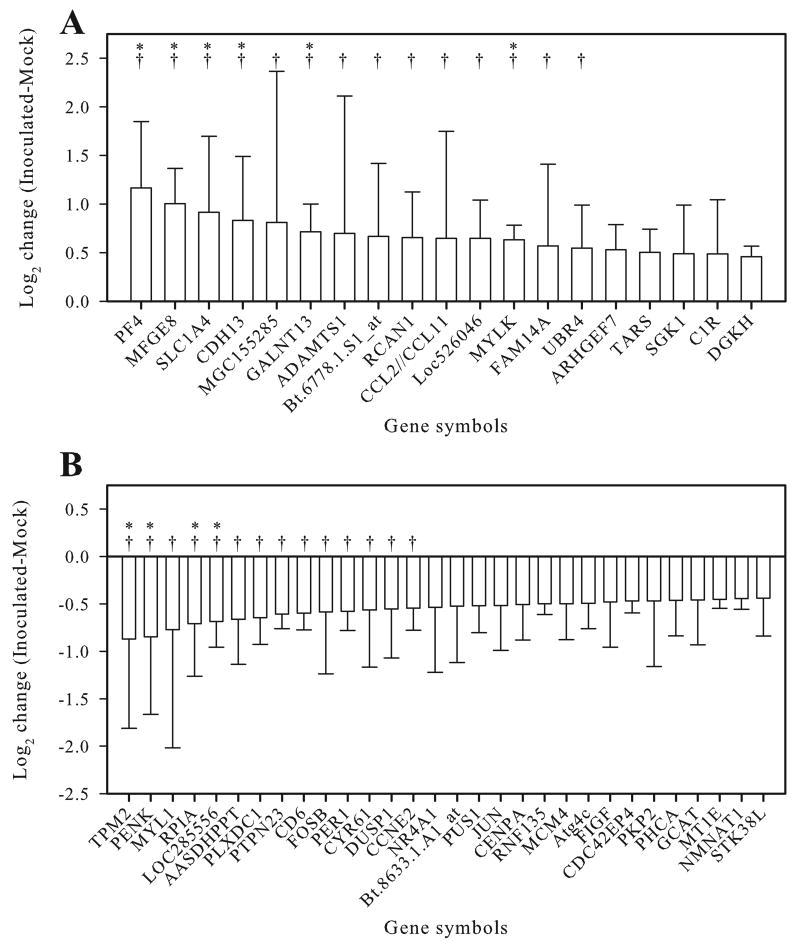
To determine which transcripts were significantly upregulated or downregulated in PrPSc-accumulating microglia, the array data were subjected to statistical analysis with threshold values for significance defined as a Benjamini and Hochberg (BH) corrected P < 0.05. Using this criterion, nineteen genes were significantly upregulated in PrPSc accumulating microglia (Fig. 1A, Supplementary Table 1). Of these 19 genes, six had BH corrected P < 0.01. The maximum fold upregulation was observed for platelet factor 4 (PF4, CXCL4), which was upregulated 2.2-fold. Only one other transcript, milk fat globule-EGF factor 8 protein (MFGE8, lactadherin), was detected with at least a twofold increase. Thirty transcripts were considered significantly (BH corrected P < 0.05) downregulated in the PrPSc treated group (Fig. 1B, Supplementary Table 2). None of the transcripts in the PrPSc treated group were decreased more than twofold.
Fig. 1.
Transcripts upregulated (A) and downregulated (B) in PrPSc-accumulating primary sheep microglia as compared to mock-treated microglia. Error bars represent one standard deviation based on the three pairwise comparisons between biological replicates; * = BH corrected P < 0.01, † = fold change greater than 1.5.
Functional categorization of differentially regulated genes
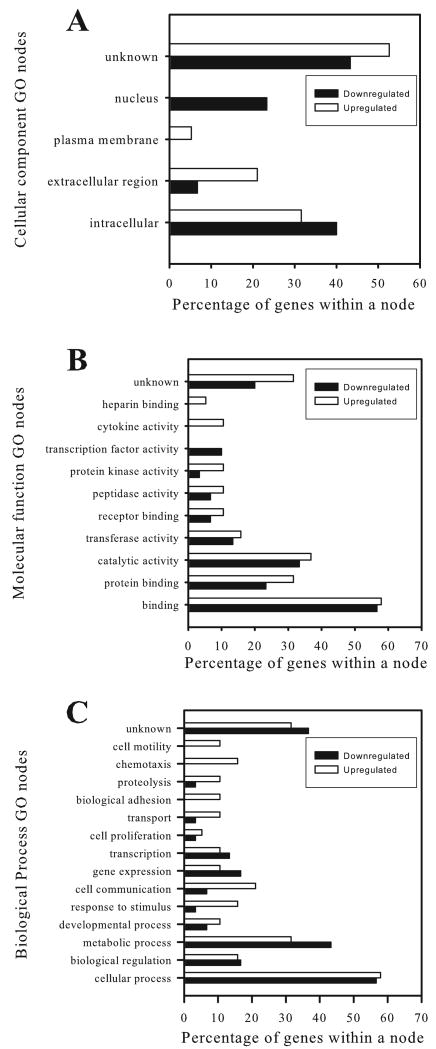
To functionally categorize the differentially regulated transcripts and identify patterns in the cellular response to PrPSc accumulation, the gene ontology (GO) categories were clustered to select GO parent categories (nodes) and then graphed as the percentage of probe sets within a node (Fig. 2). Although there was a slight trend towards microglial activation (cytokine activity, chemotaxis, cell communication, proteolysis in the upregulated transcripts), no definitive pattern of microglial response is evident.
Fig. 2.
Functional categorization of differentially expressed transcripts using gene ontology (GO) annotations. Differentially expressed transcripts were annotated to GO categories for cellular component (A), molecular function (B), and biological process (C) using NetAffx. Transcripts without functional annotations are listed as unknown.
Verification of microarray results by RT-PCR
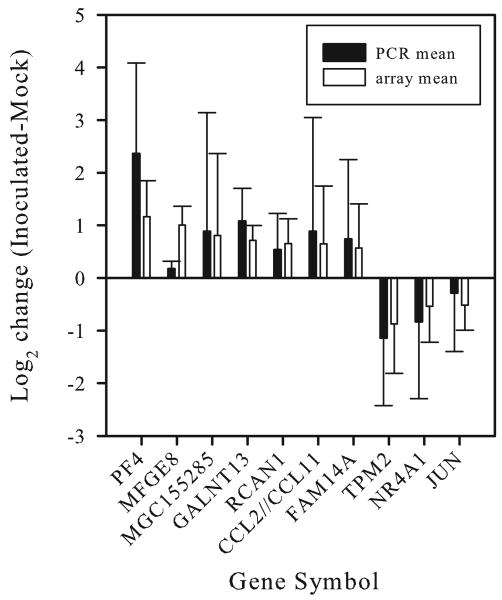
To verify the microarray results, quantitative RT-PCR was used to assay seven of the upregulated transcripts and three of the downregulated transcripts. Qualitative differences (up- or downregulation) were confirmed in all cases (Fig. 3), although for MFGE8 the qPCR results demonstrated a minimal increase in transcript levels.
Fig. 3.
Verification of microarray analysis by quantitative RT-PCR (qRT-PCR). qRT-PCR results for each gene were normalized to GAPDH, and the log2 change in transcript levels between Inoc cells and Mock cells was calculated using the ΔΔCT method (black bars). The microarray data (unfilled bars) from Figure 1 are included for comparison. Error bars represent one standard deviation of the biological replicates for each treatment group.
Discussion
Prion diseases manifest as chronic neurodegenerative diseases with limited, localized inflammation characterized by activation of astrocytes and microglia [23]. The complete pathogenesis leading to this neurodegeneration and limited inflammation is poorly understood. Previous studies investigating the transcriptional response of murine neuronal cells have yielded conflicting results [4-7], and it can not be assumed that the ovine microglial response would be similar to the murine neuronal response. Thus, to further understand the microglial responses to PrPSc accumulation in a natural host we investigated the transcriptional response of primary ovine microglia to determine if microglia have a limited response to PrPSc accumulation.
The transcriptional response by the primary microglia was limited with only a total of 49 of approximately 13,000 transcripts being differentially regulated with relatively small magnitudes of change (largest absolute value fold change = 2.2) and no distinct pattern of activation. This is markedly different than what is found when microglia or macrophages are activated in culture either by lipopolysaccharide (LPS) [24] or when infected by viruses [25] or bacteria [26], in which 200–600 genes are upregulated with 10-fold to 100-fold changes in transcript levels. In light of the conflicting results from previous studies that investigated the transcriptional response of neuronal cell cultures to PrPSc [4-7], the limited transcriptional response using sheep microglial cultures support the more recent findings [6, 7] and indicate that PrPSc fails to induce a significant transcriptional response in neuronal cells [6, 7] and microglia (current study).
The minimal transcriptional response to PrPSc in cultured neurons and microglia contrasts with the robust transcriptional response from whole brain tissue in which hundreds of transcripts have been shown to be differentially regulated [27-31]. This discrepancy is likely multifactorial. Contributing factors to this discrepancy include, but are not limited to, lack of multiple cell types and associated cell to cell signaling (e.g. neurons signaling microglia), lack of normal cellular functions in culture conditions, and time of disease progression (often months to years in organismal studies). It is speculated that the in vivo transcriptional response likely represents the response to neurodegeneration as much, if not more than, the response specifically to PrPSc.
While the overall transcriptomic response was limited, there were several specific genes that warrant further study to determine their relevance to prion diseases based on previous prion-based studies, or studies relating to the molecular functions of the transcripts. Three transcripts differentially regulated in the current study (CCL2, SGK1, and AASDHPPT) were differentially regulated in a manner similar to previous reports using rodent-adapted scrapie (CCL2: [27], SGK1: [29, 30] and AASDHPPT: [4]). To identify additional transcripts of interest, each differentially regulated gene was searched via Pubmed (July 30th, 2008) and The Database for Annotation, Visualization and Integrated Discovery (DAVID; July 29th, 2008) for associations with Alzheimer's disease (another amyloid plaque neurological disease). It was found that CCL2 [32, 33]) and MFGE8 [34] were also upregulated in studies of Alzheimer's disease. Interestingly, the MFGE8 gene encodes the lactadherin protein, whose homologue in yeast (SED1) increases heterologous protein secretion [35]. MFGE8 is also present on exosomes of dendritic cells [36] and in the PrPC enriched exosomes of Mov cells [37]. PrPSc is also found in exosomes released from various cell types, and it is thought that these exosomes are at least one possible mechanism of PrPSc transfer [38]. Additionally, MFGE8 functions in the phagocytosis of Alzheimer's disease-associated amyloid β-peptide [34]. The colocalization of MFGE8 with PrPC in exosomes, its ability to stimulate protein expression from yeast, and the data indicating a specific role for MFGE8 in the phagocytosis of amyloid β-peptide suggest a possible role in the pathogenesis of prion disease; however, further experiments are required to test any hypotheses concerning the role of MFGE8 in PrPSc accumulation.
Taken together, these data show that like two previous reports using neuronal cell cultures [6, 7], but unlike two other earlier reports [4, 5], the transcriptional response of primary sheep microglia to PrPSc is limited both in number of genes differentially expressed and the magnitude of change. This limited response is in stark contrast to microglia and monocyte cultures exposed to LPS [24], viruses [25], and bacteria [26]. It is proposed that this limited response is consistent with the unique pathogenesis and limited inflammation of prion diseases. Additionally, based on previous information concerning CCL2 and MFGE8, further investigation into the specific role(s) these genes play in the pathogenesis PrPSc accumulation is warranted.
Supplementary Material
Acknowledgments
This work was supported by NIH grant K08 AI064729, USDA ARS-CRIS 5348-32000-026-00D and USDA ARS-SCA 5348-32000-026-08S.
We thank Dr. Matt Settles for assistance with microarray data analysis, and Mr. Derek Pouchnik for performing the microarray processing. We also thank Dr. Didier Vilette and Dr. Byron Caughey for use of the Rov9 cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weissmann C. Birth of a prion: spontaneous generation revisited. Cell. 2005;122:165–168. doi: 10.1016/j.cell.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Austbo L, Kampmann A, Muller-Ladner U, Neumann E, Olsaker I, Skretting G. Identification of differentially expressed genes in ileal Peyer's patch of scrapie-infected sheep using RNA arbitrarily primed PCR. BMC Vet Res. 2008;4:12. doi: 10.1186/1746-6148-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosseddu GM, Andreoletti O, Maestrale C, Robert B, Ligios C, Piumi F, Agrimi U, Vaiman D. Gene expression profiling on sheep brain reveals differential transcripts in scrapie-affected/not-affected animals. Brain Res. 2007;1142:217–222. doi: 10.1016/j.brainres.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Martinez T, Pascual A. Identification of genes differentially expressed in SH-SY5Y neuroblastoma cells exposed to the prion peptide 106-126. Eur J Neurosci. 2007;26:51–59. doi: 10.1111/j.1460-9568.2007.05646.x. [DOI] [PubMed] [Google Scholar]
- 5.Greenwood AD, Horsch M, Stengel A, Vorberg I, Lutzny G, Maas E, Schadler S, Erfle V, Beckers J, Schatzl H, Leib-Mosch C. Cell line dependent RNA expression profiles of prion-infected mouse neuronal cells. J Mol Biol. 2005;349:487–500. doi: 10.1016/j.jmb.2005.03.076. [DOI] [PubMed] [Google Scholar]
- 6.Julius C, Hutter G, Wagner U, Seeger H, Kana V, Kranich J, Klohn P, Weissmann C, Miele G, Aguzzi A. Transcriptional stability of cultured cells upon prion infection. J Mol Biol. 2008;375:1222–1233. doi: 10.1016/j.jmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Fasano C, Campana V, Griffiths B, Kelly G, Schiavo G, Zurzolo C. Gene expression profile of quinacrine-cured prion-infected mouse neuronal cells. J Neurochem. 2008;105:239–250. doi: 10.1111/j.1471-4159.2007.05140.x. [DOI] [PubMed] [Google Scholar]
- 8.Williams AE, Lawson LJ, Perry VH, Fraser H. Characterization of the microglial response in murine scrapie, Neuropathol. Appl Neurobiol. 1994;20:47–55. doi: 10.1111/j.1365-2990.1994.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 9.Alverson J, O'Rourke KI, Baszler TV. PrPSc accumulation in fetal cotyledons of scrapie-resistant lambs is influenced by fetus location in the uterus. J Gen Virol. 2006;87:1035–1041. doi: 10.1099/vir.0.81418-0. [DOI] [PubMed] [Google Scholar]
- 10.Stanton JB, Knowles DP, O'Rourke KI, Herrmann-Hoesing LM, Mathison BA, Baszler TV. Small-ruminant lentivirus enhances PrPSc accumulation in cultured sheep microglial cells. J Virol. 2008;82:9839–9847. doi: 10.1128/JVI.01137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilette D, Andreoletti O, Archer F, Madelaine MF, Vilotte JL, Lehmann S, Laude H. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci U S A. 2001;98:4055–4059. doi: 10.1073/pnas.061337998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Development Core Team. R: A Language and Environment for Statistical Computing, ed. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- 13.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 15.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 16.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 17.Smyth G. Bioinformatics and computational biology solutions using r and bioconductor, ed. Springer; New York: 2005. [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 19.Budhia S, Haring LF, McConnell I, Blacklaws BA. Quantitation of ovine cytokine mRNA by real-time RT-PCR. J Immunol Methods. 2006;309:160–172. doi: 10.1016/j.jim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Applied Biosystems. Applied Biosystems. Foster City, CA: 2001. [Google Scholar]
- 21.Fleming-Waddell JN, Wilson LM, Olbricht GR, Vuocolo T, Byrne K, Craig BA, Tellam RL, Cockett NE, Bidwell CA. Analysis of gene expression during the onset of muscle hypertrophy in callipyge lambs. Anim Genet. 2007;38:28–36. doi: 10.1111/j.1365-2052.2006.01562.x. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt JA, de Avila JM, McLean DJ. Analysis of gene expression in bovine testis tissue prior to ectopic testis tissue xenografting and during the grafting period. Biol Reprod. 2007;76:1071–1080. doi: 10.1095/biolreprod.106.058222. [DOI] [PubMed] [Google Scholar]
- 23.Eikelenboom P, Bate C, Van Gool WA, Hoozemans JJ, Rozemuller JM, Veerhuis R, Williams A. Neuroinflammation in Alzheimer's disease and prion disease. Glia 40. 2002:232–239. doi: 10.1002/glia.10146. [DOI] [PubMed] [Google Scholar]
- 24.Lund S, Christensen KV, Hedtjarn M, Mortensen AL, Hagberg H, Falsig J, Hasseldam H, Schrattenholz A, Porzgen P, Leist M. The dynamics of the LPS triggered inflammatory response of murine microglia under different culture and in vivo conditions. J Neuroimmunol. 2006;180:71–87. doi: 10.1016/j.jneuroim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol. 2008;181:698–711. doi: 10.4049/jimmunol.181.1.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmann O, von Kockritz-Blickwede M, Holtje C, Chhatwal GS, Geffers R, Medina E. Transcriptome analysis of murine macrophages in response to infection with Streptococcus pyogenes reveals an unusual activation program. Infect Immun. 2007;75:4148–4157. doi: 10.1128/IAI.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, Dierkes N, Westner IM, Kretzschmar HA. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J Virol. 2004;78:11051–11060. doi: 10.1128/JVI.78.20.11051-11060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawiris GP, Becker KG, Elliott EJ, Moulden R, Rohwer RG. Molecular analysis of bovine spongiform encephalopathy infection by cDNA arrays. J Gen Virol. 2007;88:1356–1362. doi: 10.1099/vir.0.82387-0. [DOI] [PubMed] [Google Scholar]
- 29.Booth S, Bowman C, Baumgartner R, Sorensen G, Robertson C, Coulthart M, Phillipson C, Somorjai RL. Identification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infection. J Gen Virol. 2004;85:3459–3471. doi: 10.1099/vir.0.80110-0. [DOI] [PubMed] [Google Scholar]
- 30.Skinner PJ, Abbassi H, Chesebro B, Race RE, Reilly C, Haase AT. Gene expression alterations in brains of mice infected with three strains of scrapie. BMC Genomics. 2006;7:114. doi: 10.1186/1471-2164-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang W, Hummel M, Mitteregger G, Pace C, Windl O, Mansmann U, Kretzschmar HA. Transcriptome analysis reveals altered cholesterol metabolism during the neurodegeneration in mouse scrapie model. J Neurochem. 2007;102:834–847. doi: 10.1111/j.1471-4159.2007.04566.x. [DOI] [PubMed] [Google Scholar]
- 32.Ishizuka K, Kimura T, Igata-yi R, Katsuragi S, Takamatsu J, Miyakawa T. Identification of monocyte chemoattractant protein-1 in senile plaques and reactive microglia of Alzheimer's disease. Psychiatry Clin Neurosci. 1997;51:135–138. doi: 10.1111/j.1440-1819.1997.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 33.Smits HA, Rijsmus A, van Loon JH, Wat JW, Verhoef J, Boven LA, Nottet HS. Amyloid-beta-induced chemokine production in primary human macrophages and astrocytes. J Neuroimmunol. 2002;127:160–168. doi: 10.1016/s0165-5728(02)00112-1. [DOI] [PubMed] [Google Scholar]
- 34.Boddaert J, Kinugawa K, Lambert JC, Boukhtouche F, Zoll J, Merval R, Blanc-Brude O, Mann D, Berr C, Vilar J, Garabedian B, Journiac N, Charue D, Silvestre JS, Duyckaerts C, Amouyel P, Mariani J, Tedgui A, Mallat Z. Evidence of a role for lactadherin in Alzheimer's disease. Am J Pathol. 2007;170:921–929. doi: 10.2353/ajpath.2007.060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wentz AE, Shusta EV. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl Environ Microbiol. 2007;73:1189–1198. doi: 10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 37.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 39.Dunphy J, Horvath A, Barcham G, Balic A, Bischof R, Meeusen E. Isolation, characterisation and expression of mRNAs encoding the ovine CC chemokines, monocyte chemoattractant protein (MCP)-1alpha and -2. Vet Immunol Immunopathol. 2001;82:153–164. doi: 10.1016/s0165-2427(01)00356-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.