Abstract
The human genome is enriched in interspersed segmental duplications that sensitize approximately 10% of our genome to recurrent microdeletions and microduplications as a result of unequal crossing over. We review the recent discovery of recurrent rearrangements within these genomic hotspots and their association with both syndromic and non-syndromic diseases. Studies of common complex genetic disease show that a subset of these recurrent events plays an important role in autism, schizophrenia and epilepsy. The genomic hotspot model may provide a powerful approach for understanding the role of rare variants in common disease.
Introduction
Development of cytogenetic techniques, including high resolution karyotyping and fluorescence in situ hybridization (FISH), in the early 1980s resulted in the identification of microdeletions responsible for Prader-Willi (15q11–q13 deletions) [1] and Smith-Magenis (17p11.2 deletions) [2] syndromes. The term genomic disorder was originally introduced to describe conditions resulting from non-allelic homologous recombination (NAHR) or unequal crossing over between segmental duplications (a.k.a. low copy repeats) [3]. Over the next decade, continued efforts to fine-map recurrent deletions implicated NAHR for recurrent rearrangements in Charcot-Marie-Tooth disease [4], hereditary neuropathy with liability to pressure palsies [5], and Prader-Willi [6], Angelman [7], Smith-Magenis [8], velocardiofacial [9], Williams-Beurens [10] and Sotos [11] syndromes as well as spinal muscular atrophy [12] and juvenile nephronophthisis type I [13] (Figure 1) to name a few. Molecular diagnosis became possible but relied on (1) suspecting a specific disorder based on clinical features, and (2) using a targeted FISH assay for the chromosomal region to confirm the suspected diagnosis - a “phenotype first” approach.
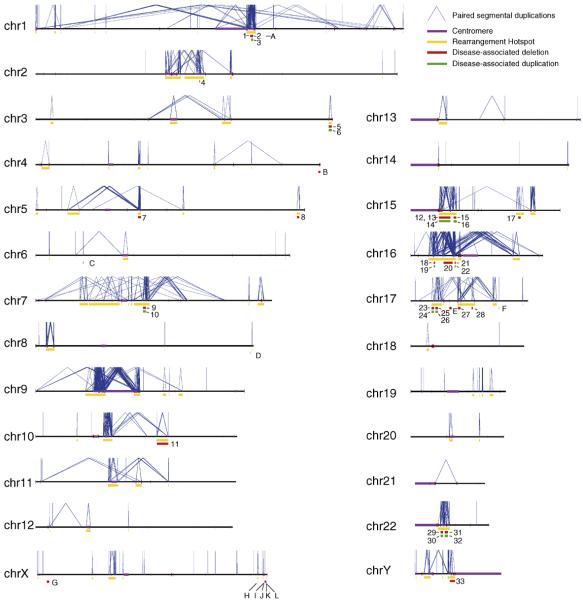
Figure 1.
Rearrangement hotspots and their associated disease phenotypes. Each chromosome is depicted as a horizontal line with intrachromosomal segmental duplications connected by blue lines. Gold bars represent “rearrangement hotspots” defined as unique regions (50 kb–10 Mb) flanked by intrachromosomal segmental duplications >10 kb with >95% sequence identity. Disease-associated deletions and duplications are represented by red or green bars, respectively. Numbers represent rearrangements within hotspot regions as defined above: (1) TAR syndrome, (2) 1q21.1 deletion and (3) reciprocal duplication, (4) Juvenile Nephronophthisis, (5) 3q29 deletion and (6) reciprocal duplication, (7) Spinal Muscular Atrophy, (8) Sotos syndrome, (9) Williams syndrome and (10) reciprocal duplication, (11) 10q22–q23 microdeletion, (12) Prader-Willi syndrome, (13) Angelman syndrome, (14) duplication 15q11, (15) 15q13.3 microdeletion and (16) reciprocal duplication, (17) 15q24 microdeletion, (18) 16p13.11 deletion and (19) reciprocal duplication, (20) 16p11.2–p12.2 deletion and (21) 16p11.2 deletion and (22) reciprocal duplication, (23) HNPP, (24) CMT1A, (25) Smith-Magenis syndrome, (26) Potocki-Lupski syndrome, (27) renal cysts and diabetes (RCAD) syndrome, (28) 17q21.31 microdeletion syndrome, (29) velocardiofacial/DiGeorge/deletion 22q11 syndrome and (30) reciprocal duplication, (31) distal 22q11.2 deletion and (32) reciprocal duplication, and (33) azoospermia. Letters represent NAHR-mediated deletions that occur outside of hotspots (as defined above): (A) Gaucher disease, (B) Fascioscapulohumeral dystrophy, (C) congenital adrenal hyperplasia, (D) glucocorticoid-remediable aldosteronism, (E) Neurfibromatosis type I microdeletion syndrome, (F) pituitary dwarfism, (G) X-linked ichthyosis, (H) Hunter syndrome, (I) red-green color-blindness, (J) Emery-Dreifuss muscular dystrophy, (K) incontinentia pigmenti and (L) hemophilia A.
Advances in technology - most notably the introduction of array comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) microarrays - now allow rapid evaluation of many targeted loci or the entire genome for submicroscopic deletions and duplications. A significant advantage of these approaches is that a suspected diagnosis is not necessary before performing the diagnostic test. The application of both targeted and whole-genome technologies to large series of patients with mental retardation or developmental delay [14–19], autism [20–25], congenital anomalies [26–29] and schizophrenia [30–32] has had several important consequences. First, the rate of discovery of novel disorders has increased dramatically. Since 2005, eighteen new genomic disorders involving twelve regions of the genome have been described, more than doubling the number of disorders described in the previous 20 years (Table 1). Perhaps more importantly, whole-genome approaches have led to a remarkable shift from a “phenotype first” to a “genotype first” definition of genomic disorders. Whereas previously, disorders were described using clinical features, new disorders are described by their genomic rearrangement and clinical features are compared among patients after a common rearrangement is identified. As the diversity of phenotypes evaluated for pathogenic copy number changes expands, so does the phenotypic diversity associated with at least a subset of recurrent rearrangements - in fact, for some of the rearrangements described below, the “phenotype first” approach would have been nearly impossible.
Table 1.
Genomic hotspot rearrangements and their associated phenotypes
| Locus | Del or dup | Coordinates (Build 36) and size of critical region | Associated phenotypes | Possible candidate genes | Size and % sequence identity of flanking repeats | References |
|---|---|---|---|---|---|---|
| 1q21.1 | del | chr1: 144.10–144.60 Mb 500 kb | TAR syndrome: hypomegakaryocytic thrombocytopenia, upper extremity abnormalities ranging from bilateral absent radii to phocomelia; normal intellect | PIAS3, Lix1L | 19 kb >95.0% | [27] |
| 1q21.1 | del | chr1: 145.0–146.35 1.35 Mb | Deletion: variable phenotypes: two groups report mild to severe MR, microcephaly, occasional congenital heart disease; two studies find enrichment of the deletion in schizophrenia | GJA5, GJA8, HYDIN2 | 281 kb >99.9% | [30,31,51–53] |
| dup | Duplication: macrocephaly, mild to moderate delays, autistic features; unlike the deletion, has not been seen in schizophrenia | |||||
| 3q29 | del | Chr3: 197.4–198.9 1.5 Mb | Deletion: mild to moderate MR, microcephaly, mild dysmorphic features | PAK2, DLG3 | 21 kb >97.1% | [46–48] |
| dup | Duplication: mild to moderate MR | |||||
| 10q22–q23 | del | Chr10: 81.12–89.07 Mb 7.95 Mb | Two families reported: deletion carriers have cognitive and behavioral abnormalities of varying severity including: learning disabilities, speech and language delay, mild developmental delays | NRG3, GRID1, BMPR1, ASNCG, GLUD1 | 107 kb >98% | [41] |
| 15q13.3 | del | Chr15: 28.7–30.2 Mb 1.5 Mb | Deletion: variable phenotypes - mild to severe MR, mild dysmorphism, digital abnormalities, autism; schizophrenia; IGE | CHRNA7 | 218 kb >99.4% | [18,30,31,54–57] |
| dup | Duplication: few patients reported, mild to moderate delays; unlike deletion of the same region, has not been reported in schizophrenia or IGE | |||||
| 15q24 | del | Chr15: 72.2–73.8 Mb 1.8 Mb | Mild to moderate MR, high anterior hairline, downslanting PF, long philtrum, digital abnormalities, genital abnormalities, loose connective tissue | MAN2C1, CYP11A1, STRA6 | 51 kb >94.0% | [38,39] |
| 16p13.11 | del | chr16:15.4–16.4 Mb | Deletion: MR, autism, brain abnormalities | NDE1, NTAN | 138 kb >99.0% | [49,50] |
| dup | 1 Mb | Duplication: autism, MR; decreased penetrance | ||||
| 16p11.2 | del | chr16: 29.50–30.10 Mb 600 Kb | Deletion: detected in 0.5–1% of individuals with autism; also seen in 0.1% of individuals with psychiatric or language disorders, 0.01% of controls | MAPK3, MAZ, DOC2A, SEZ6L2, HIRIP3 | 146 kb >99.4% | [21,25] |
| dup | Duplication: autism, psychiatric or language disorders (0.04%); also seen in 0.03% of population controls | |||||
| 16p11.2–p12.2 | del | chr16: 22.0–28.0 Mb 6 Mb | Severe developmental delay; hypotonia; flat facies, downslanting palpebral fissures, posteriorly rotated ears | Many genes | 146 kb >99.4% | [40] |
| 17q12 | del | Chr17: 31.8–33.3 Mb 1.5 Mb | Deletion: Renal abnormalities ranging from severe multicystic dysplastic kidneys to occasional renal cysts; renal cysts and diabetes (RCAD) syndrome | HNF1B | 76 kb >99% | [28] |
| 17q21.31 | del | chr17: 41.0–41.7 Mb 700 kb | Mild to severe global developmental delay, childhood hypotonia, long face, tubular or pear-shaped nose, bulbous nasal tip, friendly/amiable behavior | MAPT, CRHR1 | 38 kb >98% | [15,18,19,34] |
| Distal 22q11.2 | del | chr22:19.8–22.0 Mb 2.2 Mb | Deletion: prematurity, growth delay, learning problems and/or developmental delay, various skeletal abnormalities | MAPK1 | 10 kb >95.9% | [44,45] |
| dup | Duplication: mild to moderate MR, mild dysmorphic features | |||||
The underlying genomic architecture in each of the genomic disorders identified to date is similar: a stretch of unique sequence (50 kb–10 Mb) flanked by large (>10 kb), highly homologous (>95%) segmental duplications that provide the substrate for NAHR. In 2002, we used these criteria to identify rearrangement “hotspots” - regions predicted to be susceptible to recurrent rearrangement based on the flanking genomic architecture [33] - and developed a targeted array CGH assay to evaluate copy number variation in both affected and unaffected individuals. An updated map of predicted hotspots and associated disorders is shown in Figure 1; there are now 21 discrete regions of the genome that undergo recurrent rearrangement, resulting in 33 diseases, and at least ten additional diseases are the result of NAHR in regions of the genome that are flanked by duplications but do not meet our strict definition of a hotspot.
Mental Retardation Syndromes
The majority of the genomic disorders identified before 2006 were characterized by developmental delay, learning disability and/or mental retardation (MR). Interestingly, the genetic basis for MR is still unknown in well over 50% of clinical cases. Therefore, many studies have been aimed at identifying submicroscopic copy number changes in this population [14–19], and it is now estimated that large microdeletions and microduplications underlie >15% of MR. We note that many potential pathogenic copy number changes are non-recurrent (i.e. private mutations seen only once) and likely occur by a mechanism other than NAHR since segmental duplications have not been found at the junctions. Although significant in the aggregate, the pathogenicity for any one specific event can be difficult to prove. Here, we focus on those genomic disorders mediated by segmental duplications where the pathogenic significance is unambiguous. Sixteen of the eighteen new genomic disorders identified since 2005 are associated with MR (Table 1). Several of these appear to be highly penetrant with recognizable syndromic features.
In 2006, three groups simultaneously reported recurrent microdeletions of chromosome 17q21.31 detected by array CGH [15,18,19], kicking off a flurry of discovery of novel genomic disorders. The 17q21.31 microdeletion, with an estimated prevalence of 1 in 16,000, fits the definition of a classic genomic disorder: the microdeletion has breakpoints in flanking segmental duplications, is always de novo in affected individuals and has never been seen in controls, and patients harboring 17q21.31 microdeletions have very similar phenotypes (Table 1) [15,18,19,34]. Notably, within the same genomic region is an inversion of ~900 kb observed in approximately 20% of individuals of European ancestry [35]. Further emphasizing the importance of regional genomic architecture, the inversion has been found in every parent who transmits a de novo deletion to an affected child and appears to be a prerequisite to facilitate microdeletion [34,36]. The reciprocal duplication has also been reported in one patient with severe psychomotor delay and craniofacial dysmorphism [37]; whether individuals with the reciprocal duplication have syndromic features will require the identification of additional patients.
Reports of recurrent microdeletions in individuals with developmental delay or mental retardation continued steadily throughout 2007 and 2008. Microdeletions of 15q24, although rare, are also highly penetrant. To date, five individuals with overlapping deletions [38,39] and one patient with autism and a larger but overlapping deletion [22] have been reported. As with 17q21.31, all deletions appear to be de novo, and affected individuals have similar facial features in addition to developmental delays (Table 1). Another rare but recognizable syndrome involves deletions of chromosome 16p11.2–p12.2. Ballif and colleagues [40] reported four individuals (from 8789 analyzed) with severe developmental delays and similar facial features; each had a large deletion sharing the same distal breakpoint at 16p12.2, ranging from 7.1 to 8.7 Mb in size. Deletions of a large hotspot region on chromosome 10q22–q23 are also rare but recurrent. Two families with inherited deletions and one individual with an overlapping deletion have been reported; deletion carriers have varying degrees of cognitive and behavioral abnormalities [41].
The long arm of chromosome 22 is rich with segmental duplications, some of which are responsible for recurrent rearrangements seen in velocardiofacial syndrome, reciprocal 22q11 duplications and cat-eye syndrome [9,42,43]. More recently, recurrent deletions distal to the velocardiofacial syndrome region were reported [44]; most affected individuals had developmental and growth delays and were born prematurely (Table 1). Reciprocal duplications have also been reported and tend to result in milder, more variable phenotypes [45]. Because of the number and density of segmental duplications on 22q, there are several possible rearrangements due to NAHR; many appear to be associated with disease, but collecting information on individuals with the same events is critical to determine features associated with each. Two additional regions for which reciprocal deletions and duplications have been recently reported include 3q29 [46–48] and 16p13.11 [49,50]. Deletions of 3q29 are associated with mild to moderate MR, microcephaly, mild dysmorphic features and possibly autism; duplications may also be associated with MR but with decreased penetrance. Deletions of 16p13.11 are highly (though not fully) penetrant and have been seen in individuals with autism, mental retardation, dysmorphic features and brain abnormalities. Individuals harboring duplications tended to have mental retardation, autism and/or behavioral problems, but the duplication is also seen rarely in controls suggesting decreased penetrance and/or variable expressivity (Table 1).
Non-MR Genomic Disorders
Although neurocognitive and neurobehavioral diseases appear to be enriched for genomic disorders, this may simply be a result of ascertainment bias. Recent investigations of other diseases suggest that recurrent genomic rearrangements also underlie some disorders that do not include cognitive deficits as a primary phenotype. Array CGH studies of individuals with thrombocytopenia-absent radius (TAR) syndrome found that 30/30 affected probands shared a ~500-kb deletion on chromosome 1q21.1 [27]. The deletion is not sufficient to cause disease, as it is inherited from an unaffected parent in at least half of cases. It is thought that one or more as-of-yet unidentified genetic modifiers must play a role. A second disorder described in 2007 is associated primarily with pediatric renal abnormalities and renal cysts and diabetes (RCAD) syndrome [28]. We identified a 1.5-Mb microdeletion of 17q12 encompassing the HNF1B gene in a fetus with severe multicystic dysplastic kidneys. This led us to screen individuals with pediatric renal abnormalities or RCAD syndrome, and we found the identical microdeletion in a subset of patients. The microdeletion appears to be highly penetrant and a frequent cause of early cystic renal disease.
Genomic Disorders Defying Syndromic Classification
One of the most intriguing developments over the past two years has been the discovery of at least three new recurrent microdeletions that are enriched in multiple neuropsychiatric diseases but elude syndromic classification. Although each microdeletion was first identified in a series of individuals with similar phenotypes, the application of whole-genome copy number variation analysis to a wider range of neurocognitive disorders has revealed unprecedented phenotypic diversity.
16p11.2 rearrangements
An exciting development in the autism field was the discovery by multiple groups of a recurrent microdeletion of 16p11.2 found in 0.5–1% of affected individuals [21–23,25]. This is one of the most common cytogenetic findings, second only to the 15q11.2 microduplication, for a disorder that has been difficult to tackle from a genetics perspective. However, although significantly enriched in patients with autism, both deletions and duplications are also found in individuals with a psychiatric or language disorder (0.1% and 0.04%, respectively) and in the general population (0.01% and 0.03%, respectively) [25], suggesting extensive variability in expressivity. It is now clear that the deletion is not specific for autism as it is enriched in individuals diagnosed with mental retardation, autism as well as schizophrenia.
Rearrangements of chromosome 1q21.1
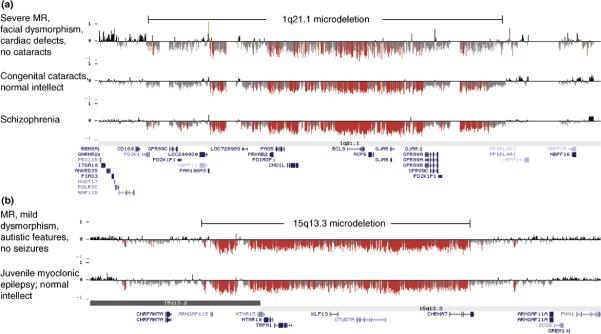
Rearrangements of a 1.35-Mb region on 1q21.1, just distal to the deletion found in TAR syndrome, have also been associated with a wide range of phenotypes, including mental retardation and developmental delay [14,18,51,52], schizophrenia [30,31] and congenital heart disease [52,53]. Based on our targeted array of hotspot regions, we identified a de novo deletion in a single patient with developmental delay and mental retardation [18]. Later, we reported a significant enrichment of both deletions and duplications of the same region in a larger series of patients with developmental delay, mental retardation and/or congenital anomalies [52], a finding replicated by Brunetti-Pierri and colleagues [51]. In two large studies of patients with schizophrenia, deletions of 1q21.1 were found in 0.26% of affected individuals compared to 0.02% of population controls [30,31]. At least three of the deletion carriers also had mild cognitive impairment and one had epilepsy. Detailed analysis of deletion breakpoints revealed that individuals with very different phenotypes appear to carry the exact same deletion (Figure 2).
Figure 2.
Identical deletions are associated with diverse phenotypic outcomes. Oligonucleotide array CGH results are depicted for (A) chromosome 1q21.1 (hg18, chr1: 143,000,000–145,500,000) and (B) chromosome 15q13.3 (hg18, chr15: 28,000,000–31,000,000). X-axis, genomic location; y-axis, log2 ratio of fluorescence intensity; red and green bars represent log2 ratio less than or greater than 1.5 standard deviations from the mean, respectively. Clinical features for the individuals shown are listed to the left of the array CGH results for that individual. Segmental duplication blocks are depicted by orange/yellow/gray bars at the top (90–94.9%, 95–99% and >99% sequence identity respectively); genes are shown below each figure.
Rearrangements of chromosome 15q13.3
Another microdeletion, first described by Sharp and colleagues in a series of patients with mental retardation, mild dysmorphic features and seizures [18,54], may have even greater phenotype diversity than rearrangements of 1q21.1. Three additional studies have confirmed that the deletion is relatively common in individuals with mild to moderate mental retardation and is also found in a subset of individuals with autism [55–57]. In contrast to the first series of patients reported, very few of the patients in these subsequent studies suffered from seizures. As with 1q21.1 deletions, 15q13.3 deletions are also enriched in individuals with schizophrenia: combined, two studies found 15q13.3 deletions in 0.2% of affected individuals compared to 0.01% of unaffected individuals [30,31]. Five of nine deletion carriers in one of the studies [31] also had mild cognitive impairment and one had epilepsy. Finally, yet another large study found enrichment of the same deletion in patients with idiopathic generalized epilepsy (IGE), the most common form of epilepsy [58]. In fact, 15q13.3 microdeletions are more common in IGE (1% of affected individuals) than in mental retardation, autism or schizophrenia. Again, detailed analysis of breakpoints reveals identical deletions despite highly variable phenotypes (Figure 2). The reciprocal duplication has also been reported in individuals with developmental delays and autistic features [55,57] and rarely in controls, but not in schizophrenia or epilepsy.
Genomic Hotspot Model of Common and Rare Disease
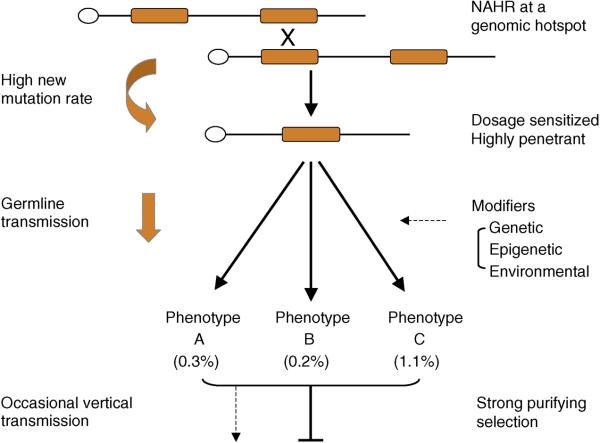
A slight majority of the rearrangements that have been shown to be disease-causing are mediated by segmental duplications. This is simply a consequence of the fact that duplicated sequences promote recurrent rearrangements (Figure 3) requiring far fewer patients and controls to be tested in order to prove pathogenicity when compared to large copy number variants (CNVs) not flanked by segmental duplications. The wide range of phenotypes associated with rearrangements of 16p11.2, 1q21.1 and 15q13.3 points to a common disease mechanism for a wide range of neurocognitive deficits, including autism, mental retardation, epilepsy, schizophrenia and other psychiatric disorders. While individually, each of these lesions may contribute only 0.1% to 1% of the total genetic basis of a specific disease, the fact that they influence so many diverse diseases (autism, epilepsy, mental retardation, schizophrenia, etc.) means the overall disease burden is significant, warranting intense scrutiny of these genomic intervals. We propose that these large microdeletions and microduplications are primarily responsible for disease, but the actual specificity of disease is determined by other perhaps more common modifiers - genetic, epigenetic and environmental. Depending on the severity of the outcome, mildly affected individuals may transmit these alleles to the next generation (explaining both the inherited and de novo aspects), but due to their high penetrance there is a strong purifying selection operating against the persistence of these alleles in the population. As a result, a high frequency of new mutations or evolutionarily young mutations, as opposed to ancient inherited mutations, is the primary basis for both common and rare diseases associated with neurodevelopmental (and perhaps other) human diseases.
Figure 3.
Model for recurrent rearrangements in common disease. Orange blocks represent segmental duplications flanking a unique stretch of sequence. Mis-alignment at meiosis results in non-allelic homologous recombination (NAHR) resulting in deletion (shown) or duplication (not shown). See text for additional details.
Future Directions and Conclusions
As we forge ahead in this “genotype first” era of rapid CNV discovery, we should anticipate the need to screen large disease cohorts (10,000–50,000 affected individuals) in order to assess the pathogenicity of other rare CNVs, especially those not flanked by segmental duplications. Some of these numbers may be achieved by leveraging CNV datasets from seemingly disparate disease cohorts (i.e. autism, mental retardation, schizophrenia and epilepsy). Until such large supracollaborations are established, targeting even smaller hotspots systematically for high-throughput CNV detection may provide a cost-effective way of identifying other pathogenic CNVs. In our work, for example, we identified 107 rearrangement hotspot regions in the human genome, 31% of which (33/107) are now associated with a variety of diseases. Advances in oligonucleotide microarray technology now allow a much larger number of smaller hotspot regions to be assessed for disease association. While genotyping first by array CGH is important, the ability to access extensive phenotypic information for individuals carrying an individual lesion is critical. Simply lumping individuals into categories of disease or controls with no ability to go back to patients (a design common to some genome-wide association studies) is inadequate. As evidenced by the 15q13.3 microdeletion and its role in epilepsy, detailed clinical information from families provides important clues into other related diseases. High-quality phenotype-genotype correlation is a reiterative process requiring the three-way participation of patient (family), clinician and researcher. This is something that has been recognized by human geneticists for decades but is worth reiterating as we contemplate the role of duplication hotspots and genomic disorders in common disease.
References
- 1.Butler MG, Meaney FJ, Palmer CG. Clinical and cytogenetic survey of 39 individuals with Prader-Labhart-Willi syndrome. Am J Med Genet. 1986;23:793–809. doi: 10.1002/ajmg.1320230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith AC, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, Reiss J, Lahr M, Allen L, Magenis E. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet. 1986;24:393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- 3.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]; **Key paper defining “genomic disorders” as conditions that result from DNA rearrangements due to regional genomic architecture.
- 4.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]; *CMT1A (duplication of 17p11) and HNPP (deletion of 17p11; ref. 5) were the first two genomic disorders described to result from reciprocal rearrangements mediated by segmental duplications.
- 5.Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]; *CMT1A (duplication of 17p11; ref. 4) and HNPP (deletion of 17p11) were the first two genomic disorders described to result from reciprocal rearrangements mediated by segmental duplications.
- 6.Carrozzo R, Rossi E, Christian SL, Kittikamron K, Livieri C, Corrias A, Pucci L, Fois A, Simi P, Bosio L, et al. Inter- and intrachromosomal rearrangements are both involved in the origin of 15q11–q13 deletions in Prader-Willi syndrome. Am J Hum Genet. 1997;61:228–231. doi: 10.1086/513907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]; *This paper provided the first direct evidence for non-allelic homologous recombination as the mechanism for deletions in Smith-Magenis syndrome.
- 9.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 10.Perez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet. 1996;59:781–792. [PMC free article] [PubMed] [Google Scholar]
- 11.Kurotaki N, Harada N, Shimokawa O, Miyake N, Kawame H, Uetake K, Makita Y, Kondoh T, Ogata T, Hasegawa T, et al. Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum Mutat. 2003;22:378–387. doi: 10.1002/humu.10270. [DOI] [PubMed] [Google Scholar]
- 12.Melki J, Lefebvre S, Burglen L, Burlet P, Clermont O, Millasseau P, Reboullet S, Benichou B, Zeviani M, Le Paslier D, et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994;264:1474–1477. doi: 10.1126/science.7910982. [DOI] [PubMed] [Google Scholar]
- 13.Saunier S, Calado J, Benessy F, Silbermann F, Heilig R, Weissenbach J, Antignac C. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet. 2000;66:778–789. doi: 10.1086/302819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]; *One of the first genome-wide studies of copy-number variation among patients which showed that 10% of mental retardation was due to large CNVs which were infrequent in a normal control group.
- 15.Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 16.Sagoo G, Butterworth A, Sanderson S, Shaw-Smith C, Higgins J, Burton H. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med. 2009;11:139–146. doi: 10.1097/GIM.0b013e318194ee8f. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]; *Authors use the duplication architecture to predict hotspot regions in patients with idiopathic mental retardation and developmental delay; five de novo events were identified including the 17q21.31, 15q13.3, 15q24, 1q21.1 and 17q12 deletion/duplication; all five have since been shown to be recurrent and pathogenic.
- 19.Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 20.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr., Dobyns WB, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 22.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Paper shows that 10% of sporadic autism may be due to de novo large copy-number variants.
- 24.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 26.Erdogan F, Larsen LA, Zhang L, Tumer Z, Tommerup N, Chen W, Jacobsen JR, Schubert M, Jurkatis J, Tzschach A, et al. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J Med Genet. 2008;45:704–709. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- 27.Klopocki E, Schulze H, Strauss G, Ott CE, Hall J, Trotier F, Fleischhauer S, Greenhalgh L, Newbury-Ecob RA, Neumann LM, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Evidence of recurrent rearrangement resulting in a non-neurologic, non-mental retardation syndrome.
- 28.Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R, Pinkel D, Cooper GM, Ventura M, Ropers HH, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, Hauser NS, Zinn AR, Garg V. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr Res. 2008;64:358–363. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 30.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DH, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein P, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]; *Provides evidence of an enrichment of large CNVs that disrupt genes in cases of schizophrenia.
- 33.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]; **Describes the segmental duplication architecture of the human genome and predicts rearrangement “hotspots” defined by this architecture.
- 34.Koolen DA, Sharp AJ, Hurst JA, Firth HV, Knight SJ, Goldenberg A, Saugier-Veber P, Pfundt R, Vissers LE, Destree A, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 36.Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchhoff M, Bisgaard AM, Duno M, Hansen FJ, Schwartz M. A 17q21.31 microduplication, reciprocal to the newly described 17q21.31 microdeletion, in a girl with severe psychomotor developmental delay and dysmorphic craniofacial features. Eur J Med Genet. 2007;50:256–263. doi: 10.1016/j.ejmg.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Klopocki E, Graul-Neumann LM, Grieben U, Tonnies H, Ropers HH, Horn D, Mundlos S, Ullmann R. A further case of the recurrent 15q24 microdeletion syndrome, detected by array CGH. Eur J Pediatr. 2008;167:903–908. doi: 10.1007/s00431-007-0616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, Striano P, Coppola A, Regan R, Price SM, Knoers NV, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 40.Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 41.Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, Bill BR, Easterday MC, Staaf J, Oseth L, Czapansky-Beilman D, et al. Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet. 2007;80:938–947. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, Lenzi T, Keegan CE, Sutton VR, Belmont J, et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- 46.Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, FitzPatrick DR, Maher E, Martin H, Parnau J, et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lisi EC, Hamosh A, Doheny KF, Squibb E, Jackson B, Galczynski R, Thomas GH, Batista DA. 3q29 interstitial microduplication: a new syndrome in a three-generation family. Am J Med Genet A. 2008;146A:601–609. doi: 10.1002/ajmg.a.32190. [DOI] [PubMed] [Google Scholar]
- 49.Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, et al. Recurrent reciprocal deletions and duplications of 16p13.11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 51.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Detailed assessment of phenotypic details of patients within 1q21.1 deletion shows extreme variability in expressivity including developmental delay, congenital heart defects, cataracts and/or schizophrenia..
- 53.Christiansen J, Dyck JD, Elyas BG, Lilley M, Bamforth JS, Hicks M, Sprysak KA, Tomaszewski R, Haase SM, Vicen-Wyhony LM, et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res. 2004;94:1429–1435. doi: 10.1161/01.RES.0000130528.72330.5c. [DOI] [PubMed] [Google Scholar]
- 54.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pagnamenta AT, Wing K, Akha ES, Knight SJ, Bolte S, Schmotzer G, Duketis E, Poustka F, Klauck SM, Poustka A, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.228. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Bon BWM, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, Innis JW, de Ravel TJL, Mercer CL, Fichera M, et al. Further delineation of the 15q13 microdeletion an duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009 doi: 10.1136/jmg.2008.063412. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]