Abstract
Over the past three decades new fungal diseases have emerged that now constitute a major threat, especially for patients with chronic diseases and/or underlying immune defi ciencies. Despite the epidemiologic data, the emergence of stable drug-resistant or hyper-virulent fungal strains in human disease has not been demonstrated as seen in emerging viral and bacterial infections. Fungi are eukaryotic microbes that capitalize on a sophisticated built-in ability to generate phenotypic variability. This successful strategy allows them to undergo rapid adaptation in response to environmental challenges, such as individual body locations that may exhibit drastic differences in temperature and pH. Rapid microevolution can also confer drug resistance and protect them from the host’s immune response. This review explores phenotypic switching in pathogenic fungi, including Candida spp and Cryptococcus spp, and how phenotypic switching contributes to the pathogenesis of fungal diseases.
Introduction
High-frequency phenotypic switching has been observed in prokaryotic and eukaryotic microbes, including pathogenic fungi. Phenotypic switching enables micro-organisms to undergo rapid microevolution and to adapt to a constantly changing microenvironment; it also facilitates pathoadaptation in the host. For fungi, switching was first described more than 20 years ago in Candida albicans [1,2]. In more recent years, phenotypic switching also has been demonstrated in other fungi, including Candida glabrata and Cryptococcus neoformans [3–8].
Phenotypic switching in fungi is an in vitro reversible phenomenon that is defined as the spontaneous emergence of colonies with altered colony morphology at rates higher than somatic mutation rates [9]. (Phenotypic switching in molds may occur, but diagnosis would be difficult because plating of many single colonies is technically not feasible.) Phenotypic switching in fungi can occur at any time during colony growth, which results in sectored colonies. The switch often is associated with a diverse array of changes on the cellular level, resulting in complex phenotypes. What distinguishes phenotypic switching from other phenotypic changes (eg, the yeast-hyphal transition or capsule induction) is that switching only occurs in a small percentage of the population; it also is reversible but the colony morphologies are stably passed on for many generations after the initial switching event. Fungi have evolved multiple regulatory pathways that recognize a range of environmental changes, including pH, temperature, and the presence of serum. These environmental signals can promote yeast-hyphal transition or capsule induction, which are coordinated morphological changes of the entire fungal population. At the same time, phenotype variability may occur randomly at a low frequency, and altered phenotypes that have biological advantages in certain settings can then be selected. This allows these fungi to rapidly adapt to different host microenvironments with both heritable and nonheritable mechanisms, which helps to promote tissue invasion and evasion of the host immune system. Thus, phenotypic switching represents one of several different mechanisms by which “phenotype heterogeneity” can be achieved in a fungal population.
Phenotypic Switching in the Human Host
Of the estimated 1.5 million fungal species, fewer than 200 species cause disease in the human host because many fungi cannot grow above 37°C. Thus, the host represents a microenvironment of particular interest for fungal pathogens and commensals. Most Candida spp are commensals and naturally colonize certain niches in the human host. Colonization of the host occurs without damage to the host, but under certain circumstances when the host is weakened, the commensal can dominate the microbiome and become a pathogen that causes disease or death. The environmental fungus C. neoformans is ubiquitous in the environment and thus is more of an “accidental” pathogen, although its main virulence factor, the polysaccharide capsule, is upregulated in the host. Both exposure and usually an immunocompromised host are required for this fungus to become an opportunistic human pathogen. It is important to acknowledge that the boundaries of commensal, opportunistic, and primary pathogens are not well defined and in part depend on our ability to recover the fungus from the host.
Regardless of the particular classification of the fungus, phenotypic switching within the colonized or infected host most certainly can affect the host-pathogen relationship. Phenotypic switching can either augment damage to the host (and thus change a commensal into a pathogen) or augment baseline virulence of an opportunistic pathogen, which may facilitate persistence. Classic examples of how phenotypic switching can alter the host-pathogen relationship are provided by 1) Trypanosoma cruzi, in which phenotypic switching generates antigenically different variants that can escape recognition by specific antibody [10], and 2) Neisseria meningitidis, in which phenotypic phase variation is pivotal in determining whether the microorganism is a commensal or an invasive pathogen [11].
Most fungal infections are chronic, and even though switching may occur at a low frequency, there is ample time for switch variants to emerge and evade the immune response. Studies addressing the phenotypic diversity of an isogenic population can be difficult to undertake because it is hard to examine a pathogen population in vivo. This review presents findings on phenotypic switching of human fungal pathogens. Extensive work has been done with the human commensal and opportunistic pathogen C. albicans and with the human pathogen C. neoformans.
Phenotypic Switching in Candida albicans
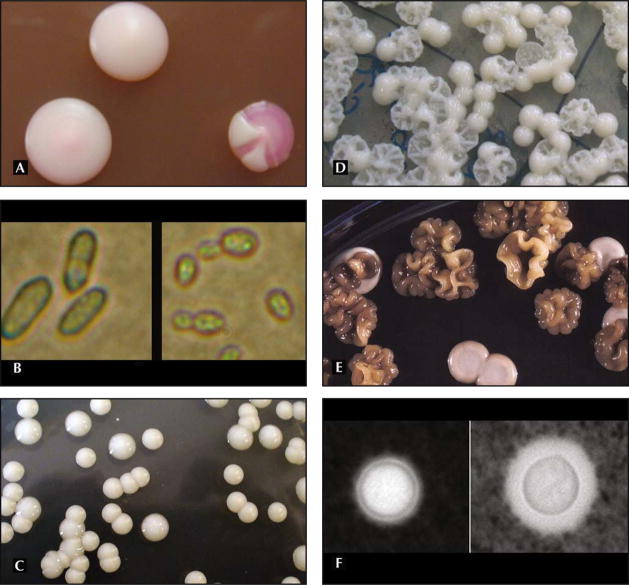
Candida albicans is a commensal that colonizes the human host in various niches. In 1985, it was first demonstrated that common strains of C. albicans switched reversibly at high frequency between a number of variant phenotypes [1,12]. Candida strains exhibited different variant phenotypes in their switching repertoires, but switching in all strains showed common characteristics [9]. Two switching C. albicans strains have been intensely investigated (Table 1). The first Candida model switching strain, the “3153A switching system,” is quite heterogeneous and includes many variant phenotypes. Cells switch between seven general phenotypes, including “original smooth,” “star,” “irregular wrinkle,” “ring,” “mottled,” “fuzzy,” and “revertant smooth” [1,9,13]. In recent years, most research has been performed on the white-opaque switching strain because the phenotypes are numerous and variable. The W0-1 switching strain switches between a white colony phenotype and an opaque colony phenotype. Switching in the white-to-opaque direction occurs at a frequency of 104 to 105 per cell generation, and switching from the opaque to white phase occurs at the slightly higher frequency of 5 × 104 per cell generation. However, appearances of white/opaque-sectored colonies are common in switching. This strain also has a clearly distinguishable phenotype at a microscopic level (Fig. 1). The white colony phenotype is composed of round budding cells, and the opaque colony phenotype contains large, elongated, asymmetrical budding cells [2,14,15].
Table 1.
Characteristics of switching Candida and Trichosporon strains
| Species | Switching strain | Parent | Variants | Switching frequency |
|---|---|---|---|---|
| Candida albicans | 3153A | Smooth | Star, ring, irregular wrinkled, stopple, hat, and fuzzy | 1.4 × 10−4 (all variants) |
| WO1 | White (W) | Opaque (O) | 4.6 × 10−4 to 4.4 × 10−3 (W→O) | |
| 2.1 × 10−2 to 1.3 × 10−1 (O→W) | ||||
| Strain 1001 | Smooth (S) | Rough (R) | < 10−4 (S→R) | |
| < 9 × 10−4 (R→S) | ||||
| 3.5 × 10−3 (sectored colony) | ||||
| NUM 961 | Smooth (S) | Semi-rough (SRT-1) | 3 × 10−5 (S→SRT-1) | |
| Candida glabrata | 35B11 | Dark brown (DB) | White (Wh) | 2 × 10−3 (DB→Wh) |
| Light brown (LB) | 9 × 10−3 (DB→LB) | |||
| Very dark brown (vDB) | 8 × 10−3 (DB→vDB) | |||
| Irregular wrinkled (IWr) | 1 × 10−4 (DB→IWr) | |||
| Candida lusitaniae | JHH Clu-3B | Light brown (LB) | White (W) | 1 × 10−4 (LB→W) |
| Light brown (LB) | 6.6 × 10−2 (LB→DB) | |||
| Dark brown (DB) | 1.4 × 10−2 (DB→LB) | |||
| 2 × 10−4 (W→LB) | ||||
| Trichosporon asahii | White farinose (WF) | White, radial, fi ssure (WRF) | 10−2 to 10−4 (all variants) | |
| White pustular (WP) | White farinose (WF) | |||
| Yellowish white (Y) | White (W); yellowish white (Y); smooth (S); radial fi ssure (RF) |
Figure 1.
Colony morphology of switch variants in Candida species and Cryptococcus neoformans. Candida spp: (A) white-opaque switch variants and sectored colonies; and (B) microscopic phenotype differences of the white and opaque variant. Cryptococcus neoformans: (C) RC-2 smooth and mucoid colonies; (D) SB4 SM and wrinkled colonies; (E) differences in melanization of SM and WR switch variants of 24067A; and (F) capsule differences of SM and MC switch variants.
Phenotypic switching in Candida albicans is important for mating
The ability of a C. albicans strain to undergo white-opaque switching is regulated by the mating-type–like (MTL) locus. Opaque-phase a- and α-type Candida cells mate 106 times more efficiently than white-phase a and α cells. In contrast, white-phase cells appear to be more robust and thus survive better within a mammalian host. Approximately 3% and 7% of clinical C. albicans isolates can undergo white-opaque switching. Switching Candida strains that are found in nature are homozygous a/a or α/α strains. Candida strains that are heterozygous (a/α) at the MTL locus cannot undergo white-opaque switching [16,17]. Opaque cells have been isolated from inside mammalian hosts, and elegant studies in mice have confirmed that mating can actually occur in the host [18]. Certain signals from the host can impact the expression of MTL genes, and other signals may also exist that stabilize the opaque-phase in specific locations within the host. Taken together, white-to-opaque switching represents a critical step in the mating process; the opaque cells are the mating-competent form of C. albicans, whereas the white cells are better suited to survive within the mammalian host environment but can generate mating-competent cells when needed.
Effects of phenotypic switching in Candida albicans on host-pathogen interaction
Switching involves the transcriptional regulation of a number of phase-specific genes, which are regulated by phase-specific trans-acting factors [19–21]. Several of these genes have been implicated in virulence [19,22–24]. Kvaal et al. [25] extensively studied the role of white-opaque switching with respect to virulence, demonstrating that opaque-phase cells were more virulent in newborn mouse cutaneous models of infection and that white-phase cells were more virulent in an intravenous systemic infection model [25]. Opaque-phase cells better colonize the skin and mate on the skin of newborn mice (surface temperature 31.5°C) in a highly efficient manner; up to 40% of opaque a and α cells can fuse during a 24-hour period [26]. Furthermore, switching can affect other virulence traits, including the bud-hyphal transition [27], the yeast’s sensitivity to neutrophils and oxidants [28], antigenicity [27], adhesion [29], secretion of proteinase [22,30], drug susceptibility, and phagocytosis by macrophages [31]. All of these altered traits could potentially affect the survival within the mammalian host.
Molecular mechanisms and regulation of Candida albicans switching
In the white-opaque phenotypic switching strain, more than 373 genes are differentially expressed in the switch variants. Analysis determined that 221 genes were expressed at a higher level in opaque cells and 152 were more highly expressed in white cells [32]. These genes represent diverse functions, including metabolism, adhesion, cell surface composition, stress response, signaling, mating type, and virulence. Approximately one third of the differences among cell types are related to metabolic pathways. Opaque cells expressed a transcriptional profile consistent with oxidative metabolism, and white cells expressed a transcription profile that was consistent with a fermentative metabolism. Metabolic specialization of switch phenotypes may enhance selection in relation to the nutrients available at different anatomical sites, especially because the transcriptional bias was obtained regardless of carbon source. Zordan et al. [33] demonstrated that an interlocking feedback loop network is responsible for stably maintaining the epigenetic state of switch variants through many cell divisions. MTLa1, MTLα2, WOR1, CZF1, WOR2, and EFG1 were found to constitute a genetic circuit that regulates the white-opaque switch. This circuit, formed from a network of interlocking positive feedback loops, appears to be distinct for C. albicans and is not present in closely related fungi. It was proposed that because EFG1 and CZF1, for example, have other key functions in the Candida life cycle, their involvement in white-opaque switching could be a recent adaptation that occurred in the warm-blooded host [33,34••].
Candida glabrata
Candida glabrata is the second most prevalent Candida spp that colonizes humans [35]. Candida glabrata is more closely related to Saccharomyces cerevisiae than to C. albicans, and it employs mechanisms different from those of C. albicans to generate phenotypic plasticity during pathogenesis. Candida glabrata switches between a number of phenotypes distinguished best by graded colony coloration on agar containing 1 mM CuSO4. This phenomenon is referred to as core switching [36], and the core phenotypes are white (Wh), light brown (LB), dark brown (DB), and very dark brown (vDB). A core phenotype can spontaneously and reversibly switch to an irregular wrinkle (IWr) phenotype, a transition referred to as irregular wrinkle switching. The graded brown coloration of the core phenotypes is thought to be the result of graded levels of conversions of Cu2+ to Cu1+ and the associated reduction of SO42− to S1−. It is important to note that CuSO4 does not induce switching per se. Switch-colony variants can be best identified on CuSO4 supplemented agar, which stains Wh, LB, DB, and vDB colonies in an intensity order that is reversed from the pink-graded colors on phloxine B-containing agar. The coloration of the IWr phenotype is white on CuSO4-containing agar, regardless of the core phenotype from which the IWr strain arose. However, IWr exhibits a propensity for switching back to the core phenotype from which it arose, suggesting that although it expresses characteristics of Wh, it retains, or “remembers,” its original core phenotype. A microscopic analysis revealed that during the first 3 days of colony development, cells in the centers of Wh, LB, DB, and vDB colonies expressed almost exclusively the budding yeast phenotype; on day 3, pseudohyphae and cells extending tubes accumulated, so that by day 7 the proportions of these two cellular phenotypes reached 40% to 50% and 10% to 20%, respectively. In contrast, IWr colonies were composed almost exclusively of pseudohyphae in the first 6 days and then accumulated both budding yeast cells and tubes. Core switching, pseudohyphae formation, and tube formation occurred in the majority of 62 tested clinical C. glabrata strains, suggesting that core switching is a general characteristic of most C. glabrata strains. Data from transcriptional profiling showed that the majority of genes upregulated in DB (78%) were involved in copper assimilation, sulfur assimilation, and stress responses; these genes also were differentially regulated in the various phenotypes. DB is the most commonly expressed phenotype at sites of host colonization. With respect to differences in host-pathogen interaction among the various phenotypes, little is known to date and future studies will have to delineate how upregulation of these genes in the context of phenotypic switching may play a role in colonization and virulence.
Candida lusitaniae
Candida lusitaniae is an emerging human pathogen that, unlike other fungal pathogens, either has intrinsic resistance or frequently develops resistance to the commonly used antifungal agent amphotericin B. It is infrequently found as a human commensal (skin, respiratory, gastrointestinal, and genitourinary tracts) and less than 1% of yeasts recovered from hospitalized patients are C. lusitaniae [37] The mechanism(s) of this resistance to amphotericin B is not well understood [38]. Studies have shown that C. lusitaniae undergoes phenotypic switching when plated on yeast peptone dextrose (YPD) agar with 1 mM CuSO4. Three colony colors are observed: LB to DB to W, which are similar to C. glabrata. The study showed that phenotypic switching affected amphotericin B resistance. High resistance (minimum inhibitory concentration [MIC] of 256 μg/mL) was associated with W, whereas LB and DB colonies exhibited lower MICs of 2 to 8 μg/mL and 2 to 16 μg/mL, respectively. Filamentation (pseudo-hyphae) was associated with DB colonies. All phenotypes occurred spontaneously with greater frequency (10−2 to 10−4) than spontaneous mutations, and all phenotypes were reversible, fulfilling the two phenotypic-switching criteria. High amphotericin B MICs were always associated with W colonies but not all W colonies exhibited high MICs. Data suggest that phenotypic switching in C. lusitaniae could represent a key strategy that confers a selective advantage during chronic infection in a host that is treated with amphotericin B.
Trichosporon asahii
Infections with Trichosporon spp have been recognized with increasing frequency over the past two decades [39,40]. These non–Candida yeasts have a broad spectrum of clinical manifestations, from self-limiting cutaneous infections to life-threatening invasive disease in the immunocompromised host. A recent study investigated phenotypic switching in 61 clinical isolates of T. asahii, producing four different morphological phenotypes on Sabouraud dextrose agar (SDA). These are white farinose (69%), white pustular (18%), yellowish white (10%), and white cerebriform (3%). Strains of the three major types (white farinose, white pustular, and yellowish white) produced two to five colony types when cultured on SDA at 37°C. The frequency of switching among colony types was 102 to 104, similar to C. albicans and C. neoformans. The colonies switch to the smooth type—perhaps irreversibly—at frequencies of 102 to 103. The enzymatic activity of beta-N-acetyl-hexosaminidase in the smooth type was significantly greater than that of the parent type in all strains. This could potentially confer enhanced virulence, but more data would be required to determine this.
Phenotypic Switching in Various Cryptococcus Species
Cryptococcus neoformans var neoformans and Cryptococcus neoformans var grubii are encapsulated pathogenic yeasts that can be found in soil worldwide and can cause chronic meningoencephalitis, pneumonia, and disseminated disease syndromes predominantly in immunocompromised patients. Cryptococcus neoformans var gattii causes a similar disease syndrome and used to be found mainly in subtropical climates like Australia, but more recently, out-breaks have been described in the northwestern part of the United States [41]. Phenotypic switching has been reported in C. neoformans var grubii, which are serotype A strains (SB4 and J32) [4], in C. neoformans var neoformans, which are serotype D strains (24067A, RC-2) [6], and in the sibling species C. neoformans var gattii [42]. In these strains, colonies with altered morphology arise spontaneously at a frequency of about 1 in 10−4 to 10−5, and reversion to the parent smooth colony type occurs at a comparable frequency (approximately 1 in 10−3 to 10−5) (Table 2). The smooth colonies (S and SM) of SB4, 24067, and NP-1 are round with a smooth dome surface and smooth edges. The mucoid (M and MC) colonies are round with smooth edges and a shiny and mucoid-appearing colony surface. They represent the parent colony morphology in the C. neoformans var gattii strain (NP-1). Wrinkled (WR), serrated (C), and pseudohyphal (PH) colonies exhibit an irregular dome surface with or without serrated margins. Both smooth and mucoid colonies represent standard colony types common to many C. neoformans strains, whereas the WR, C, S, and PH colony types of SB4 and 24067A are rarely observed in clinical isolates. Phenotypes and switching frequencies are most stable in the RC-2 switching strains; therefore, most of the in vivo work has been done with that strain.
Table 2.
Characteristics of switching Cryptococcus strains
| Switching strain | Serotype | Phenotype | Switching frequencies | Virulence (in murine models) | Infl ammatory response | GXM SRG |
|---|---|---|---|---|---|---|
| SB-4 | A | Smooth (S) | 2.2 × 10−3 to 9 × 10−6 | WR > S > C | C > S > WR in rats* | ND |
| Serrated (C) | ||||||
| Wrinkled (WR) | ||||||
| J32 | A | Smooth (NM) | 3.4 × 10−3 to 4.1 × 10−5 | ND | ND | ND |
| Mucoid (MC) | ||||||
| 24067A | D | Smooth (SM) | 2.2 × 10−3 to 6 × 10−6 | WR persists; SM and PH are cleared in rats | WR > PH > SM in rats† | M1, M2, M5, M6 |
| Pseudohyphal (PH) | M1, M5 | |||||
| Wrinkled (WR #0) | M1, M5, M6 | |||||
| RC-2 | D | Smooth (SM) | 10−4 to 0.5 × 10−4 | MC > SM in mice | MC > SM in mice‡ | ND |
| Mucoid (MC) | ||||||
| NP1 | B | Smooth (NP1-SM) | MC to SM ≫ SM to MC Range (10−3 to 10−5) | MC > SM (mice) | MC > SM in mice | M3, M1, M6 |
| Mucoid (NP1-MC) | M3, M1, M6 |
Minimal infl ammation in wrinkled (WR)-infected rats in contrast to intense caseous necrosis in serrated (C)-infected rats.
No infl ammation in smooth (SM)-infected rats in contrast to extensive granuloma formation in wrinkled (WR)-infected rats.
Organized granuloma formation in smooth (SM)-infected mice in contrast to extensive and destructive infl ammatory response in mucoid (MC)-infected mice.
GXM—glucuronoxylomannan; ND—not done; SRG—structural reporter group.
Phenotypic switch variants of Cryptococcus exhibit distinct phenotypes with altered polysaccharide capsule
All switching strains in C. neoformans exhibit changes in the polysaccharide capsule, which is important because it may affect phagocytosis and rapid destruction by macrophages [43,44]. For RC-2, the MC capsule is larger, and excessive production of viscous exo-polysaccharide results in a shiny surface (Fig. 1). Careful analysis of newly generated MC switch variants demonstrates that other phenotypic characteristics are also changed. The MC switch variant also more slowly and exhibits increased sensitivity to lysing enzymes when compared with the SM parent. In a similar fashion, the SB4, 24067A, and NP1 variants exhibit changes in polysaccharide capsule that affect virulence. Glucuronoxylomannan (GXM) is the predominant capsular polysaccharide and is composed of (1→3)-linked linear α-D-mannopyranan with β-D-xylopyranosyl (Xylp) and β-D-glucupyranosyluronic acid (GlcpA) residues added to the mannose at various positions. Six structural reporter groups (SRGs) are defined based on the amount of 2-0–linked, 4-0–linked Xylp residues, and 2-0–linked GlcpA residues [45]. In C. neoformans strains SB4 and 24067A, phenotypic switching resulted in significant changes of the biochemical composition of GXM [5]. The GXMs of the C colony type of SB4 are composed of mixtures of SRGs (M2 and M3 for the C colony), whereas SB4-SM exhibits predominately SRG M2. In a similar fashion, the PH and WR colonies of 24067A exhibit a mix of SRGs (M1 and M5), whereas the SM parents are predominately M1 and M2. The addition of a Xylp group at the 4-0 position in M3 and M5 most likely requires a different enzyme than linkage to the 2-0 position. M3 SRGs are traditionally thought to be present only in the GXM of C. neoformans var gattii isolates (serotype B and C) and not in GXM of C. neoformans var neoformans isolates (serotype A and D). In RC-2, phenotypic switching alters the biophysical and biochemical properties of GXM [46]. Viscosity data in solutions of different ionic strength suggest that the spacing of GlcpA along the mannose backbone differs between the GXM of the SM and that of the MC strain. Because NMR measures an average repeat unit, no differences between SM and MC of RC-2 were detected.
Phenotypic switch variants exhibit difference in virulence in animal infection models
In all the “switching” C. neoformans var neoformans and C. neoformans var gattii strains studied so far, phenotypic switching results in variants that exhibit altered virulence in murine infection models (Table 2). For RC2, the MC variant is consistently more virulent than the SM parent in murine intratracheal infection models, intravenous infection models, and intracisternal infection models. Similar effects are seen for 24067A, in which the WR variant is more virulent, and for SB4, in which the serrated and wrinkled variants are more virulent. For C. neoformans var gattii strain NP1, MC was also found to be more virulent in the pulmonary model, but only the switched strain NP1-SM was able to elicit central nervous system infection. This suggests that switching in this case was required for crossing of the blood-brain barrier. Studies also indicate that RC-2 MC (but not the RC-2 SM variant) can promote intracranial pressure in a rat model of cryptococcal meningitis. In humans, elevated intracranial pressure is the leading cause of high morbidity and mortality in patients with chronic cryptococcosis; these studies suggest phenotypic switching may contribute to the development of intracranial pressure [47]. Antifungal interventions do not enhance phenotypic switching per se, and antifungal susceptibility is not affected by phenotypic switching. However, in mice infected with a switching strain (RC2-SM), both antifungal drug therapy and administration of anticapsular monoclonal antibody promoted the emergence of RC2-MC variants in chronic murine infection models [48]. These findings are important because they further our understanding on how phenotypic switching may affect the outcome of infection in chronically infected hosts who are treated. Phenotypic switching may contribute to the frequent treatment failures observed in immunocompromised hosts because treatment affects selection in the host.
Molecular mechanism of phenotypic switching
The molecular mechanisms mediating phenotypic switching in C. neoformans are currently not understood. Although karyotype instability was observed in strain 24067A and SB4 [4,49] similar to the switching C. albicans strain 3153A, it could not consistently be correlated with phenotypic variability and was irreversible. Investigations involving differential display of RC2-SM and RC2-MC messager RNA (RC-2 strain) demonstrated that phenotypic switching was associated with downregulation of genes in the MC switch variant relative to the SM switch variant. The functions of most of these genes are still unknown; however, some may represent immunogenic epitopes. Future studies are directed at determining the specific function of the individual genes.
Conclusions
Phenotypic switching has been described for several pathogenic fungi, and it enables these pathogens to adapt to changing environments. Such environments include different hosts that will employ sophisticated immunological tools to rid themselves of the pathogen and variable environmental niches where microbes can be subjected to many types of selection pressures. This process allows the fungus to be resilient while maintaining the ability to generate diversity. Data from C. albicans emphasize the relevance of phenotypic switching for key biological functions such as mating, in which phenotypic switching can essentially protect a vulnerable phenotype of a species and thereby maintain its ability to mate. Data from chronic infection models of C. neoformans demonstrate how phenotypic switching in a fungus during experimental infection could potentially alter outcome and promote persistence.
Acknowledgments
This work was supported by R0-1 AI 59681 for Dr. Fries, and Fahmi Hassan by the AIDS International Training and Research Program (NID D43-TW01403) of the Albert Einstein College of Medicine.
Footnotes
Disclosures
No potential conflicts of interest relevant to this article have been reported.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 2.Slutsky B, Staebell M, Anderson J, et al. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachke SA, Srikantha T, Tsai LK, et al. Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect Immun. 2000;68:884–895. doi: 10.1128/iai.68.2.884-895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman DL, Fries BC, Franzot SP, et al. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci U S A. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fries BC, Goldman DL, Cherniak R, et al. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun. 1999;67:6076–6083. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fries BC, Taborda CP, Serfass E, Casadevall A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J Clin Invest. 2001;108:1639–1648. doi: 10.1172/JCI13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kugler S, Schurtz Sebghati T, Groppe Eissenberg L, Goldman WE. Phenotypic variation and intracellular parasitism by Histoplasma Capsulatum. Proc Natl Acad Sci U S A. 2000;97:8794–8798. doi: 10.1073/pnas.97.16.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha H, Pain A, Johnstone K. Analysis of the role of recA in phenotypic switching of Pseudomonas tolaasii. J Bacteriol. 2000;182:6532–6535. doi: 10.1128/jb.182.22.6532-6535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soll DR. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myler P, Allison J, Agabian N, Stuart K. Antigenic variation in African trypanosomes by gene replacement or activation of alternate telomeres. Cell. 1984;39:203–211. doi: 10.1016/0092-8674(84)90206-x. [DOI] [PubMed] [Google Scholar]
- 11.Swartley L, Marfin A, Edupuganti S, et al. Capsule switching of Neisseria meningitidis. PNAS. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomes R, Gil C, Nombela C. Genetic analysis of Candida albicans morphological mutants. J Gen Microbiol. 1985;131:2107–2113. doi: 10.1099/00221287-131-8-2107. [DOI] [PubMed] [Google Scholar]
- 13.Ramsey H, Morrow B, Soll DR. An increase in switching frequency correlates with an increase in recombination of the ribosomal chromosomes of Candida albicans strain 3153A. Microbiology. 1994;140(Pt 7):1525–1531. doi: 10.1099/13500872-140-7-1525. [DOI] [PubMed] [Google Scholar]
- 14.Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legrand M, Lephart P, Forche A, et al. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol Microbiol. 2004;52:1451–1462. doi: 10.1111/j.1365-2958.2004.04068.x. [DOI] [PubMed] [Google Scholar]
- 17.Lockhart SR, Pujol C, Daniels KJ, et al. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 19.Srikantha T, Chandrasekhar A, Soll DR. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol Cell Biol. 1995;15:1797–1805. doi: 10.1128/mcb.15.3.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockhart SR, Nguyen M, Srikantha T, Soll DR. A MADS box protein consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J Bacteriol. 1998;180:6607–6616. doi: 10.1128/jb.180.24.6607-6616.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow B, Srikantha T, Soll DR. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hube B, Monod M, Schofield DA, et al. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 24.Balan I, Alarco AM, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvaal C, Lachke SA, Srikantha T, et al. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson J, Mihalik R, Soll DR. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolotila MP, Diamond RD. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy MJ, Rogers AL, Hanselmen LR, et al. Variation in adhesion and cell surface hydrophobicity in Candida albicans white and opaque phenotypes. Mycopathologia. 1988;102:149–156. doi: 10.1007/BF00437397. [DOI] [PubMed] [Google Scholar]
- 30.Vargas K, Messer SA, Pfaller M, et al. Elevated phenotypic switching and drug resistance of Candida albicans from human immunodeficiency virus-positive individuals prior to first thrush episode. J Clin Microbiol. 2000;38:3595–3607. doi: 10.1128/jcm.38.10.3595-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohse MB, Johnson AD. Differential Phagocytosis of White versus Opaque Candida albicans by Drosophila and Mouse Phagocytes. PLoS ONE. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan CY, Newport G, Murillo LA, et al. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Zordan RE, Miller MG, Galgoczy DJ, et al. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. This paper delineates the molecular mechanism of the transcriptional feedback loop that controls white-opaque switching. The authors demonstrate that the master regulator gene WOR1 occupies a central position. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fidel PL, Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachke SA, Joly S, Daniels K, Soll DR. Phenotypic switching and filamentation in Candida glabrata. Microbiology. 2002;148(Pt 9):2661–2674. doi: 10.1099/00221287-148-9-2661. [DOI] [PubMed] [Google Scholar]
- 37.Merz WG. Candida lusitaniae: frequency of recovery, colonization, infection, and amphotericin B resistance. J Clin Microbiol. 1984;20:1194–1195. doi: 10.1128/jcm.20.6.1194-1195.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young LY, Hull CM, Heitman J. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother. 2003;47:2717–2724. doi: 10.1128/AAC.47.9.2717-2724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichikawa T, Sugita T, Wang L, et al. Phenotypic switching and beta-N-acetylhexosaminidase activity of the pathogenic yeast Trichosporon asahii. Microbiol Immunol. 2004;48:237–242. doi: 10.1111/j.1348-0421.2004.tb03519.x. [DOI] [PubMed] [Google Scholar]
- 40.Ebright JR, Fairfax MR, Vazquez JA. Trichosporon asahii, a non-Candida yeast that caused fatal septic shock in a patient without cancer or neutropenia. Clin Infect Dis. 2001;33:E28–30. doi: 10.1086/322640. [DOI] [PubMed] [Google Scholar]
- 41.Upton A, Fraser JA, Kidd SE, et al. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J Clin Microbiol. 2007;45:3086–3088. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain N, Li L, McFadden DC, et al. Phenotypic switching in a Cryptococcus neoformans variety gattii strain is associated with changes in virulence and promotes dissemination to the central nervous system. Infect Immun. 2006;74:896–903. doi: 10.1128/IAI.74.2.896-903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levitz SM, Nong SH, Seetoo KF, et al. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherniak R, Valafar H, Morris L, Valafar F. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronxylomannans with a computer-simulated artificial neural network. Clinical and Diagnostic Laboratory Immunology. 1998;5:146–159. doi: 10.1128/cdli.5.2.146-159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell. 2007;6:1464–1473. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fries BC, Lee SC, Kennan R, et al. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect Immun. 2005;73:1779–1787. doi: 10.1128/IAI.73.3.1779-1787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fries BC, Cook E, Wang X, Casadevall A. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob Agents Chemother. 2005;49:350–357. doi: 10.1128/AAC.49.1.350-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franzot SP, Mukherjee J, Cherniak R, et al. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]