Abstract
Heat shock proteins protect cells from various conditions of stress. Hsp70, the most ubiquitous and highly conserved Hsp, helps proteins adopt native conformation or regain function after misfolding. Various co-chaperones specify Hsp70 function and broaden its substrate range. We discuss Hsp70 structure and function, regulation by co-factors and influence on propagation of yeast prions.
Keywords: Hsp70, chaperone, co-chaperone, prion
Introduction
The three-dimensional structure of protein is determined by amino acid sequence but the crowded cytoplasmic environment presents obstacles to successful protein folding [1]. Although many proteins fold on their own, others require assistance of accessory proteins called chaperones to reach their native state. Chaperones are a functionally related group of proteins that assist protein folding under both physiological and stressful conditions [2].
An important subgroup of chaperones are the heat shock proteins (Hsps), which were originally identified as proteins whose expression is induced by heat stress. When cells are exposed to stress such as high temperature, starvation, inflammation, water deprivation, or nitrogen deficiency, any of which can cause proteins to become partially unfolded, Hsps, primarily of the Hsp70 family, bind partially exposed hydrophobic surfaces, to prevent non-productive interactions that would lead to aggregation, and to promote protein refolding. Like many other chaperones, the ability to bind somewhat non-specifically to hydrophobic peptides allows Hsp70 to act on a broad range of substrates. As the predominant group of Hsps in all cell types from bacteria and plants to humans, Hsp70s have very high structural homology and have conserved functional properties across species. For example, Drosophila Hsp70 can complement mammalian Hsp70 in protection from heat stress [3].
In addition to their protective role under stress conditions, Hsp70s are involved in essential cellular processes where proteins are partially or completely unfolded such as translation, translocation across membranes, presentation of substrates for degradation, assembly and disassembly of macromolecular complexes or aggregates, and in gene induction and apoptosis [4–7]. Isoforms of Hsp70 (Hsc70) are therefore expressed constitutively at moderate to low levels in all organisms. In eukaryotes, Hsp70 resides in or is associated with all subcellular compartments. It is also well established that Hsp70s play important roles in the propagation of yeast prions [8–10].
Hsp40s are obligatory co-chaperone partners of Hsp70 that regulate Hsp70 activities and functional specificity [11]. Some organisms also encode Hsp100 family members that function in cooperation with Hsp70 and Hsp40 to resolubilize aggregated proteins [12–14]. The present review summarizes our current knowledge about various aspects of Hsp70 proteins, its regulation by different co-factors and role in prion propagation.
Eukaryotes contain multiple Hsp70 isoforms
Humans encode at least eight Hsp70 homologs, two of which are organelle specific whereas the remaining six reside in the cytosol and nucleus. Similarly, S. cerevisiae contains two organelle specific and six cytosolic Hsp70s. Yeast cytosolic Hsp70s are subdivided into two classes Ssa (Ssa1, Ssa2, Ssa3, and Ssa4) and Ssb (Ssb1 and Ssb2). Ssa and Ssb proteins are approximately 60% identical to each other and possess some overlapping functions, although they are not functionally identical [15]. Within the Ssa or Ssb subfamily, sequence similarity is near 100% and therefore proteins within each subfamily were considered to have the same functions but vary in expression so Hsp70 abundance could be adjusted to match the need for chaperone activity. However, functional differences among Ssa proteins are being uncovered [16–18], (Sharma and Masison unpublished). The constitutive Ssa1 and Ssa2 are 98% identical whereas identity between Ssa3 and Ssa4 is around 88%. Any one of the Ssas can support cell growth but the Ssbs cannot compensate for essential Ssa function, which exemplifies the overlapping and distinct functions of cytosolic Hsp70s. Likewise, many prokaryotic organisms also contain several Hsp70s. For example, E.coli K12 encodes three Hsp70s; DnaK, HscA and HscC, and three Hsp70 proteins have been identified in the unicellular green alga Chlamydomonas reinhardtii.
Mechanism of action
Hsp70-dependent folding of polypeptides occurs through repeated cycles of binding and release of substrate at the expenditure of ATP. Much of our knowledge about Hsp70 action is based upon studies on E. coli Hsp70 DnaK. However, since bacterial and mammalian Hsp70s are 50% identical and have essentially superimposable structures and similar enzymatic functions, Hsp70 chaperones across species are believed to work through similar mechanisms. This notion is supported by biochemical and structural studies of Hsp70s from many different organisms. Hsp70s consist of three structural domains: a 44kDa amino-terminal ATPase domain (NBD) followed by an 18kD substrate binding domain (SBD) and a 10kD C-terminal domain (CTD), which forms a lid-like structure over the substrate-binding pocket that helps trap substrate in the SBD [19–21].
Hsp70 function requires coordinated action of all three domains (Figure 1). Substrate binding occurs in a hydrophobic pocket in the SBD with an affinity and kinetics dependent upon the nucleotide state of the NBD. The ATP-bound form of Hsp70 has low substrate binding affinity and a fast exchange rate while the ADP-bound form exhibits higher affinity and slower exchange rates. The major difference between the ATP and ADP-bound states is the location of the CTD, which is positioned over the substrate binding pocket when ADP is bound and thus reduces release of bound substrate [22]. Structural changes in the NBD upon ATP hydrolysis induce the conformational changes in the SBD and CTD that lead to this substrate trapping, pointing to a strong communication between the domains. Correspondingly, substrate binding to the SBD alters SBD structure in a way that transmits a signal to the NBD that stimulates ATP hydrolysis and thus increases its affinity of binding to Hsp70. Subsequently, nucleotide exchange factors promote release of ADP, facilitating rebinding of ATP and thereby restoring Hsp70 to the low affinity state allowing substrate release. Released substrate is then provided the opportunity to assume the native state or else undergoes a kinetic partitioning between rebinding to Hsp70, binding to other chaperones and aggregation.
Figure 1.

Model representing ATPase cycle of Hsp70. Binding of ATP to the NBD of Hsp70 opens the lid over the substrate-binding domain and allows substrate to bind. ATP hydrolysis, catalyzed by various co-chaperones leads to the closing of lid and trapping substrate in the substrate binding pocket. Further action of nucleotide exchange factors exchanges ADP with ATP that releases the bound substrate and a new cycle begins.
The intrinsic ability of Hsp70 to hydrolyze ATP and release ADP is low, which severely limits the rate of the Hsp70 reaction cycle and provides key regulatory points for fine tuning of Hsp70 activity by various co-factors. A 2–6 fold activation is achieved by substrate binding to the SBD. Further stimulation is achieved by transient interaction of Hsp70s with various co-factors such as the Hsp40s and tetratricopeptide repeat (TPR) co-chaperones [23, 24], and simultaneous interaction with Hsp40 and substrate stimulate ATPase synergistically. In turn, NEFs can enhance ATPase activity by accelerating return to the ATP-bound state [25, 26]. In addition to influencing the Hsp70 reaction cycle, these various cofactors provide functional variation and specificity among different Hsp70 proteins [27].
Regulatory roles of Hsp70 co-factors
Nucleotide Exchange Factors
Hsp70s bind weakly to substrate when in the ATP-bound state and more strongly when in the ADP state, so ADP-ATP exchange is an important step in regulating Hsp70's reaction cycle. The slow intrinsic release of ADP by Hsp70 is enhanced by nucleotide exchange factors (NEFs). By shifting the equilibrium toward the ATP-bound state, NEFs trigger substrate release and restart the chaperone cycle. Well known NEFs are GrpE in E. coli, Fes1 and Sse1 in S. cerevisiae [28], Bag-1 in higher eukaryotes, and the human Fes1 ortholog HspBP1 [29].
GrpE and Bag-1 are the first protein candidates identified as nucleotide exchange factors for E. coli DnaK and mammalian Hsc70, respectively[30, 31]. GrpE acts in coordination with DnaJ to stimulate ATPase activity of DnaK by 5000-fold and control the ATP dependent substrate release of DnaK [31–34]. Interaction of GrpE with DnaK reduces its affinity for ADP by 200-fold and ADP similarly reduces DnaK affinity for GrpE by 200-fold [33]. Although GrpE and Bag-1 facilitate release of ADP by a similar mechanism, they vary in both specificity and nucleotide exchange ability. For example, although the ATPase domains of inducible Hsp70 and constitutive Hsc70 are nearly identical, GrpE and Bag-1 accelerate exchange exclusively of their respective Hsp70 and Hsc70 binding partners, and GrpE activates release of both ATP and ADP while Bag-1 catalyzes only ADP release [35]. Additionally, although GrpE augments DnaK-DnaJ mediated protein folding and assembly, Bag-1 can exert either positive or negative effects on Hsp70 [35, 36]. It appears that nucleotide exchange function of Bag-1 is not simply a general regulator of Hsp70 during productive folding of protein substrates, but rather it assists Hsp70 in substrate unloading on certain chaperone pathways.
Like most organisms, S. cerevisiae has multiple NEFs such as Snl1, Sls1, Sil1, Lhs1, Fes1, and Sse1 [37–40]. The significance of possessing multiple NEFs is not completely understood but different NEFs probably have specificity for interaction with different Hsp70s. For example, although Lhs1 enhances Hsp70 function of the endoplasmic reticulum (ER) resident Hsp70 Kar2 [40], Sse1 stimulates yeast and mammalian cytosolic Hsp70s [41, 42]. Some NEFs also have partially overlapping functions. Nucleotide exchange activity of Lhs1 and Sil1 is partially redundant but Lhs1 is more potent than Sil1 with Kar2 [40]. Sse1 and Fes1 also exhibit functional redundancy in that overexpression of Fes1 confers viability to the otherwise lethal sse1Δ sse2Δ double deletion, but Sse1 is more potent with Ssa Hsp70s than Fes1 [43].
Hsp40 co-chaperones
One important family of co-chaperones that plays a major role in regulating Hsp70 interaction with client proteins is Hsp40. Hsp40s are crucial partners of all Hsp70s that not only regulate Hsp70 activity but also broaden the scope of their functions [44]. Like Hsp70s, Hsp40s are ubiquitous and represented by multiple isoforms. Humans, S. cerevisiae and E. coli contain 41, 22 and 6 Hsp40 homologs respectively. The best characterized Hsp40 is E. coli DnaJ, and the Hsp40 co-chaperone family, or “J proteins”, are defined by a conserved stretch of about 70 residues (the J domain) with similarity to the initial 73 amino acids of E. coli DnaJ [11, 45, 46].
Hsp40 binds hydrophobic peptides with specificity that overlaps that of Hsp70 and can prevent protein aggregation, defining it as a chaperone itself [47, 48]. As a co-chaperone Hsp40 interacts with Hsp70's NBD and CTD as well as with Hsp70 client proteins and thereby facilitates transfer of substrate to the SBD of Hsp70 [49–51]. Hsp40 binding to Hsp70 stimulates ATP hydrolysis up to 1000-fold and coordinates ATP hydrolysis with substrate binding [31, 52]. The J-domain is the minimal region required for Hsp70 interaction and sequence variation within this domain provides specificity to Hsp70–Hsp40 interactions [53–55]. Though, some Hsp70s interact with multiple J proteins, others show specificity to a single J protein [45, 56]. For example, yeast Hsp70 Ssz1 and not Ssa1/2, interacts with J-protein Zou1 [57], and Ssa1/2 interact with Hsp40s Sis1, Ydj1, Swa2, Hlj1 and Djp1 [58–60]. Selectivity of Hsp70–Hsp40 interactions enables Hsp70 to perform a variety of functions and localize to specific places in the cell. For example, while Ssa1/2 have general protein folding functions in the cytosol, Sis1 assists Ssa1/2 on ribosomes in translation [59], Ydj1 recruits Ssa1/2 to protein folding on ER membranes [61] and Djp1 stimulates Ssa1/2 to assist in peroxisome biogenesis [58]. Interaction of Hsp40 with various substrates as well as its ability to stimulate the Hsp70 reaction cycle allows Hsp70 to trap a wider range of substrates than it would be capable of binding on its own [62].
Structural analysis of Hsp70 function
Initial understanding of Hsp70 structure was deduced from crystal structures of the NBD and SBD obtained separately [19, 20]. Recent structures of intact bovine Hsc70 without the C-terminal lid [63], and NMR structure of modified two-domain Thermos thermophilus DnaK [64] and two-domain modified E. coli DnaK [65] have been solved, which provides deeper insight into communication between the NBD and SBD of different Hsp70s, broadening our understanding about Hsp70 action with its substrate.
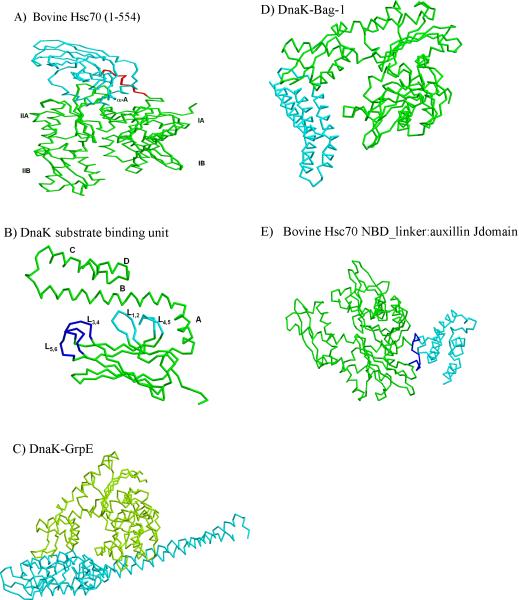
The ATPase domain of Hsp70 consists of four subdomains arranged in two lobes, I and II. Subdomain IA and IB strongly interact each other and make up lobe I (Figure 2A). Similarly IIA and IIB make the other lobe. The two lobes are connected at their bases through subdomains IA and IIA, which have similar secondary structure and topology. Binding of ATP occurs at the bottom of a cleft between subdomains IB and IIB [63, 66]. Superposition of bovine Hsc70 NBDs with and without ATP shows that IB and IIB shift toward each other when bound to ATP [63].
Figure 2.
Structural features of Hsp70 with and without various co-factors. A) Backbone representation of bovine Hsc70 with NBD (green), SBD(cyan) and linker (red) (PDB code 1YUW). B) Backbone representation of substrate binding domain and part of C-terminal domain (389–591) of DnaK (PDB code 1DKY). Inner and outer loops of substrate binding domain are shown in cyan and blue repectively. C) Structure of nucleotide exchange factor GrpE (cyan) bound to ATPase domain (green) of DnaK (PDB code 1DKG). D) Structural representation of the complex of Hsc70 ATPase domain (green) in complex with Bag domain fragment (Cyan) of NEF Bag-1 (PDB code 1HX1). E) Strucutral presentation of auxillin J domain (cyan) bound to bovine Hsc70 NBD (green) containing interdomain linker (blue) (PDB code 2QWO).
The DnaK SBD forms a β-sandwich of two antiparallel β-sheets comprised of four β-strands each, the first sheet consisting of β3, β6, β7, β8, and the second of β5, β4, β1, β2, with four upward protruding loops [20]. The substrate binding cavity, formed by the β-sheet 1 and 2 and loops L1,2 and L3,4, is primarily hydrophobic (Figure 2B). However, the surface potential surrounding the cavity is mainly negative and therefore positive charges in substrate also contribute to affinity with Hsp70. Peptides having an interior hydrophobic core flanked with basic residues possess highest affinity binding to Hsp70 DnaK and the optimal substrate length for ATPase stimulation is at least eight residues [67].
Comparison of substrate-bound and isolated SBDs shows an overall similar fold, but in the apo form the side chains in loops L1,2 and L3,4 that contact either substrate or CTD lid protrude to partially occupy the space that is taken by the substrate in the peptide-bound form of DnaK [68]. Predominant differences were observed for strand β3, where it does not participate in β-sheet formation and occludes the substrate binding cleft in the apo form. For the CTD, only helix α-B is necessary for the primary peptide-SBD interactions. Furthermore, interaction between SBD and CTD domains is not necessary for several functions of DnaK such as interdomain communication. For example, DnaK(1–507), which lacks the C-terminal lid, binds peptide with affinity only five fold lower than wild type DnaK and can support bacterophage λ propagation in vivo [68]. Also, addition of ATP to DnaK(1–507) lowers peptide binding affinity by a magnitude similar to wild type DnaK, and peptide binding to DnaK(1–507) stimulates ATPase activity. These studies indicate that allosteric interface is functional in both directions even in truncated DnaK, arguing that the SBD itself controls a significant portion of ATP induced allosteric signal.
The CTD lid is linked to the β-sandwich SBD through a network of hydrogen bonds and ionic interactions between the lid and residues in loops L4,5 and L1,2 of the SBD. The isolated CTD of DnaK consists of 5 α-helices (αA-αE) and a flexible subdomain of approximately 30 amino acids (Figure 2B). Ahough the α-helical lid is not neccesary for the interdomain communication between NBD and SBD, it is important for the kinetics of peptide binding. Deletion of this region of DnaK increases ATP-induced peptide release by approximately 100 fold [68]. Contribution of the lid domain to overall Hsp70 chaperone activity has been studied in detail by sequentially deleting the disordered subdomain and the individual lid helices [69]. It was observed that αB and αD play significant roles in peptide binding to SBD and the rate of ATP-induced high to low-affinity conformational changes, respectively. The residues in αE helix and disordered domain modulate the kinetics of ATP binding as their deletion in DnaK shifted the biphasic nature of ATP binding to monophasic, presumably due to a decrease in conformational heterogeneity upon deletion of the disordered tail. In eukaryotes the EEVD motif present at the extreme C-terminus mediates Hsp70 interaction with Hsp40 and TPR-containing co-chaperones [21, 70–72].
The exposed 10 amino acid linker of bovine Hsc70 that connects NBD to SBD lies at the opposite end of the structure from the substrate channel and lid [63] (Figure 2A). Communication between the NBD and SBD is mediated by a 12 amino acid SBD helix (helix A) that rests in a grove between lobes IA and IIA of NBD. Helix B forms part of the substrate binding domain and may transmit conformational changes from the NBD to the substrate binding pocket. These structural data indicate that conformational changes induced by ATP binding are a necessary part of Hsp70 chaperonin function. The significance of surface involved in the interaction was shown by mutational analysis of interdomain residues and the linker, which indicates communication occurs through the interdomain surface and involves the adjacent NBD-SBD linker [63]. The work supports an allosteric mechanism in which the linker occupies and disrupts the interdomain surface upon binding of ATP to the NBD.
The bacterial NEF GrpE interacts primarily at the cleft between subdomain IB and IIB, and the loop inserted within β-strand 3 of DnaK is required for this binding [73] (Figure 2C). Although eukaryotic Bag-1 is structurally unrelated to GrpE, it binds to the same subdomains of mammalian Hsc70 [73, 74]. However, although Hsc70 residues that make contact with Bag-1 are conserved in all eukayotic cytosolic Hsp70s, they are highly diverged from DnaK (Figure 2D). Binding of these NEFs induces an outward 14° rotation of subdomain IIB. Subdomain IIB in the ADP-bound form contributes three side chains to the binding of adenine and ribose sugar of nucleotide and these contacts are disrupted by the NEF-induced conformational changes. Free Hsp70 in the absence of ATP can maintain this open conformation, indicating that NEFs are not required for Hsp70 to remain “open”.
Structural studies of Hsp40–Hsp70 complexes are hindered by the transience and ATP dependence of the interaction [75]. Much current knowledge of Hsp40–Hsp70 interaction is from studies involving mutagenesis of Hsp40 or Hsp70 together with structural information of fragments of different Hsp40s, with or without Hsp70s, and biochemical analysis of effects of the mutations on functional interaction [76–78]. A screen to restore functional association of DnaJ mutant D35N with Hsp70 uncovered R167 as an important DnaK NBD residue for interaction with DnaJ [76]. DnaK R167H is unable to interact with wild type DnaJ, and DnaJ D35 requires interaction with R167 for proper ATP hydrolysis and conversion to a high affininty substrate-binding conformation of DnaK.
In yeast, the peptide binding fragment of type II Hsp40 Sis1 interacts with the extreme C-terminal octapeptide G634PTVEEVD641 of Ssa1 [72, 79]. The interaction involves both charge-charge as well as hydrophobic interactions. The presence of the glycine rich region preceding the polypeptide binding region of Sis1 is thought to be important to provide flexibility to transfer non-native polypeptides from Hsp40 to Hsp70. Conservation of residues important for Sis1-Ssa1 interaction suggests the mechanism of interaction is similar for other type II Hsp40s and their Hsp70 partners.
The J-domain of neuronal specific co-chaperone auxilin binds to the cleft between lobes IA and IIA of mammalian Hsc70 near the NBD-SBD linker region [80] (Figure 2E). Various residues in the linker are important for J domain stimulation of ATPase activity, indicating that it is required not only for binding to J proteins but also to mediate the J signal to nucleotide in the NBD. Indeed, in the J-domain bound form the Hsc70 linker was directed toward the hydrophobic patch on NBD surface and interacts with residues Y371 and I181.
Hsp70 functions
Because of its ability to bind hydrophobic stretches commonly present in polypeptides, Hsp70 performs a variety of functions in many different cellular processes. E. coli DnaK has also been shown to possess cis-trans isomerase activity for non-prolyl bonds [81]. Hsp70 chaperones are also implicated in a range of protein misfolding diseases and have received particular attention in the area of prion formation and propagation. The following sections discuss several examples of these many important Hsp70 functions.
Protein folding and prevention of aggregation
Hsp70 is important for helping proteins adopt and maintain native conformations from the time they are sythesized until they are eventually degraded. During translation, an elongating chain is susceptible to aggregation because patches of hydrophobic residues remain exposed until all the amino acids required for forming native contacts emerge from the ribosome. In prokaryotes, Hsp70, in coordination with its co-factors, interacts with about 20–30% of proteins as they are synthesized to help them attain their native conformations. The need for Hsp70 is even greater for eukaryotes where more proteins are bigger than those of prokaryotes. Hsp70 binds transiently to these structurally immature polypeptides and prevents them from aggregating or adopting non-functional conformations. In addition, even upon folding, natively folded proteins still remain prone to aggregation as they have a relatively small margin of stability that can be affected by minor physicochemical intracellular conditions. Therefore, Hsp70s and other chaperones continue their essential role of preventing aggregation beyond translation.
Different Hsp70 isoforms can act during different stages of a protein's life. For example, although S. cerevisiae cytosolic Hsp70s Ssa and Ssb are highly identical, Ssb facilitates elongation of nascent polypeptide chains by interacting with polypeptides at early stages of synthesis while Ssa facilitates translocation by interacting with proteins at later stages after the polypeptide chain adopts some aspect of secondary structure [15, 82]. Therefore, by interacting sequentially with the same substrate, Hsp70s of different subfamilies perform different functions in the same pathway. Moreover, as discussed below, we found that nearly identical members within the same Ssa subfamily have distinct functions with regard to yeast prion propagation.
Signal transduction and apoptosis
Hsp70 and its co-factors interact with various components of signaling pathways to facilitate conformational changes required for their activation. Chaperones play a key role in either presenting signalling proteins to or protecting them from the degradative machinery. Through this balancing of folding and degradation, chaperones provide a means for extensive regulation of signaling pathways. For example, Hsp70 in coordination with Hsp90 associates with certain kinases and maintains steroid hormone receptors in a ligand-binding conformation. Similarly, the constitutive Hsc73 interacts with retinoblastoma (pRB), which is a major regulatory molecule at the cell cylce starting point, and protects it from degradation. When conjugated to a peptide, Hsp70 can also act as an antigen carrier and induce cytokine production. For example, p24-Hsp70 fusion protein elicits both humoral and cellular responses against p24 in mice. Similarly tumor derived Hsp70 is able to produce Th1 cytokines such as IFN-γ [83].
Another Hsp70-chaperoning substrate is p53, a transcription factor that regulates the cell cycle and functions as tumor suppressor. Missense mutations in p53 that inactivate its tumor suppressor function increase propensity for cancer. In combination with Hsp90 or CHIP, Hsp70 regulates p53 stability in a positive or negative way, respectively. Hsp70 in complex with CHIP promotes degradation of unfolded p53 mutants, In a complex with Hsp90 it has little or no effect on degradation of wild type or mutant p53 that adopt a folded conformation [84]. Therefore, by contributing to stabilization of some p53 mutants, Hsp70, in combination with Hsp90 or CHIP, can influence tumorogenicity.
There also have been a number of studies suggesting an anti-apoptotic role of Hsp70. Hsp70 may affect apoptosis through its interaction with co-chaperone Bag-1, which is known to interact with antiapoptotic protein bcl-2. Elevated Hsp70 protects cells from cytotoxicity induced by apoptosis inducing agents; TNF, monocytes, radiation and chemotherapeutic agents [85, 86]. Hsp70 binds directly to apoptotic protease activating factor-1 (apaf-1), and prevents the formation of functional apoptosome [87]. Hsp70 inhibition is, therefore, a promising therapeutic approach for cancer treatment. Hsp70 inhibitors strongly increase the sensitivity of cancer cells to chemotherapy in vitro. For example triptolide induces apoptosis in pancreatic cancer cells by decreasing Hsp70 mRNA and protein levels [88].
Role of Hsp70 in protein degradation
Hsp70s help co-ordinate the ubiquitin-proteosome pathway to promote protein degradation, although the role of Hsp70 in transfer of substrates to the proteosome is specific to substrates such as actin, α-crystallin and glyceraldehyde-3-phosphate dehydrogenase [89]. Other substrates such as BSA and lysozyme do not require Hsp70 for degradation. The degradation acitivities of Hsp70 are primarily mediated by its co-factors such as CHIP, BAG domain containing proteins and Hsj1 [90, 91].
CHIP, which was originally identified in a screen of human TPR containing proteins, acts as a ubiquitin ligase and ubiquitinates Hsp70 captured proteins and subsequently targets them to 19S subunit of the proteosome [84]. CHIP contains an Hsp70-binding domain and a U-box required for ubiquitin ligase activity [92]. CHIP interaction inhibits Hsp70's ability to refold proteins by interfering with the ATPase cycle [93]. Tau protein, whose aggregation in neuronal cells leads to neurological disorders, is targeted by Hsp70-CHIP complex for degradation [91]. Increased expression of Hsp70 leads to a decrease in Tau levels and a selective reduction of detergent insoluble Tau. CHIP also promotes ubiquitination and degradation of immature ER-localized cycstic fibrosis transmembrane membrane conductance regulator, CFTR [94].
Bag-1 contains both a BAG domain, required for Hsp70 interaction, and a ubiquitin-like domain, and thereby provides a link for Hsp70 association to the proteolytic complex [90]. However, there is no experimental evidence showing a direct role of Bag-1 in transfer of substrate from Hsp70 to the proteosome. Instead it is believed that Bag-1 accepts substrate proteins from Hsc/Hsp70 and transfers them to CHIP, which promotes their ubiquitination. In contrast to its role in ubiquitination, Bag-1 also has been shown to inhibit proteosomal degradation of Tau protein, possibly due to enhancement of refolding activity of Hsc70 [95].
BiP, an Hsp70 of the ER lumen, facilitates ER-associated degradation. One well known substrate associated with BiP in this degradation pathway is BACE457, a pancreatic isoform of human β-secretase [96]. BACE457, when misfolded, associates with BiP before being retrotranslocated to the cytosol for proteosome mediated degradation. A possible role of BiP in BACE457 retrotranslocation to is to keep it in a degradation competent state.
It should be noted that, although homologous, Hsp70s show substrate specificity in protein degradation. For example, wild type yeast strains show normal CFTR degradation at 26° and 40°, but in ssa1-45 temperature sensitive mutant cells only 60% of CFTR was degraded irrespective of temperature. Mutants of Kar2, the yeast BiP, have no effect on CFTR degradation indicating that yeast Ssa Hsp70, and not BiP, mediates CFTR degradation [97].
The role of yeast Ssb and Ssa proteins in cytosolic protein degradation has also been investigated. Overproduction of Ssb1, but not Ssa1, suppresses the temperature sensitive growth of proteosome y7 and y13 mutants [98]. In addition, extra Ssb1 significantly improved the turnover of short lived proteins in y7 mutant cells, indicating that Ssb1 promotes proteosomal protein degradation.
Since Hsps interact transiently with a wide variety (10–20%) of cellular proteins, their capacity to promote protein degradation is carefully controlled. The critical balance between protein folding and degradation activity of Hsp70 is regulated mainly by Hsp70 co-factors. This control is achieved primarily by competitive binding at the same sites on Hsp70 of co-chaperones that stimulate protein folding with those that promote degradation. For example, Hsp70 interacting protein Hip, which promotes folding capacity of Hsp70, competes with Bag-1 for binding to the Hsp70 ATPase domain [99]. Similarly, Hop, an Hsp70–Hsp90 organizing protein that facilitates transfer of client substrates from Hsp70 to Hsp90, competes with CHIP for binding at the Hsp70 CTD [49, 100]. Under normal conditions the intracellular concentration of Hip and Hop is around 5–10 times more than Bag-1 and CHIP, so the folding pathway of Hsp70 should be favored. Therefore changes in abundance of different cofactors triggered by changes in environmental conditions can in turn affect the balance of regulation. Regulation can occur at even higher levels as illustrated by the Hsp70 NEF HspBP1, which by forming a ternary complex with Hsp70 and CHIP can interfere with ubiquitin ligase activity of CHIP [101].
Role of Hsp70s in formation and propagation of yeast prions
Prions are infectious proteins that cause a number of mammalian neurodegenerative diseases such as bovine spongiform encephalopathy, Creutzfeldt-Jacob disease in humans, scrapie in sheep and chronic wasting disease in deer and elk. Prion diseases also share important cell biological features with Alzheimer's and other diseases that involve accumulation of amyloid forms of pathogenic protein. At present prion diseases are untreatable and ultimately fatal.
Prion formation occurs when the soluble form of a protein abnormally refolds into an alternative structure comprised primarily of β-sheet. Solid-state NMR studies show that yeast prion proteins form a parallel in-register β-sheet amyloid core [102, 103]. This misfolded structure self-perpetuates by acting as a nucleation center to convert soluble protein into the same altered form [104] (Figure 3). Fibers grow by recruiting and converting the soluble form of the protein. In yeast, iteraction of these growing fibers with cytosolic chaperones, in particular the Hsp104/70/40 machinery, leads to their fragmenting into smaller fibers, increasing the number of nuclei, or seeds, onto which more of the soluble protein is further depleted. An increase in number of seeds also increases the efficiency of transmission of fibers to daughter cells during cell division.
The discovery that proteins in yeast can behave as prions has greatly improved our understanding of prion biology [105]. Work by us and others clearly shows that Hsp70 strongly influences propagation of the two most studied prions in Saccharomyces cerevisiae [PSI+] and [URE3], which are infectious amyloid forms of Sup35 and Ure2 proteins, respectively [8, 18, 106, 107]. Sup35 is a translation-termination factor (eRF3) whose conversion to the prion state reduces its activity, thus increasing read-through of nonsense codons [108]. Ure2 is a regulator of nitrogen catabolite repression [109]. The presence of [URE3] similarly reduces activity of Ure2, causing derepression of genes involved in nitrogen utilization.
In order to be maintained in a yeast population, these prions depend on normal functions of chaperones, and altered function or abundance of Hsp70 considerably interferes with prion propagation [8–10]. Hsp70 forms a part of a larger chaperone network, consisting of Hsp104, Hsp40s and TPR-containing proteins, that directly impact prion propagation and its effects on prions might be direct or through alterations of its cooperativity with other chaperones [110]. For example, an in vitro study of the effect of Ssa1 on Sup35 fibrillization shows that Ssa1 in coordination with Ydj1 blocks Sup35 polymerization [111]. This inhibitory effect was seen only in the presence of Ydj1, demonstrating a strong requirement of Ydj1 for this Ssa1 affect on Sup35 fibrillization.
A specific role for Hsp70 in prion propagation is pointed to by various studies. We isolated a dominant mutant of Ssa1, Ssa1–21, containing an L483W substitution in the SBD, that impairs [PSI+] mitotic stability and weakens allosuppression caused by [PSI+] [8, 107]. The prion-weakening effect of Ssa1–21 is even more pronounced when Ssa1–21 is the only source of cytosolic Ssa1 Hsp70. We further found that Ssa1–21 weakens [URE3], and that the analogous Ssa2–21 mutation weakens both [PSI+] and [URE3], pointing to a general role of Ssa Hsp70s in prion propagation (Sharma and Masison unpublished). Ssa1–21 has no effect on thermotolerance or resolubilization of heat-aggregated luciferase in vivo, indicating it functions well in these processes and that its effects on prions are independent of these Hsp104 activities. Yeast expressing Ssa1–21 contain several fold fewer inheritable prion seeds and larger than normal aggregates of prion polymers, suggesting that Ssa1–21 causes the polymers to more readily self-associate into large aggregates [8, 18, 106, 107]. Thus, Ssa1 might normally act to prevent such higher-order aggregation of individual polymers or to recover prion seeds from such aggregates.
Although yeast cytosolic Ssa and Ssb subfamilies are highly homologous, these Hsp70s vary with regard to their effect on prion formation and propagation. Overexpression of Ssa, but not Ssb, increases de novo formation of [PSI+] [112]. In contrast, Ssb proteins inhibit [PSI+] propagation. Moreover, while overexpression of Ssa1 reduces the curing effect of excess Hsp104 by inhibiting the shift of soluble sup35 to the insoluble prion form [113], Ssb family proteins enhanced the curing effect of Hsp104. It was recently found that Hsp70 physically interacts with Sup35 and influences its ability to form prions [114]. Similar amounts of Ssb were associated with Sup35 in both [psi−] and [PSI+] cells while the amount of Ssa associated with Sup35 was less in [psi−] cells. In contrast, another study found that similar amounts of Ssa and Ssb co-purify with Sup35 that is immunoprecipitated from [psi−] yeast cell lysates [112]. The discrepancy might be due to use in the latter study of elevated expression of Sup35, which could cause partial aggreagation of Sup35 that produces as Ssa substrate even in [psi−] cells. On the whole the evidence suggests the Ssas have preference for binding the prion form of Sup35 while Ssbs interact with both prion and properly folded forms. Understanding how these differences relate to the opposite effects Ssa and Ssb have on prions will require further experiments.
The way Hsp70s affect yeast prions differs not only among members of different subfamilies but also within those of the same subfamily. We showed that overexpression of Ssa1, but not Ssa2 antagonized [URE3], while neither affected [PSI+] [17]. Additionally, deleting Ssa1 weakens [PSI+] but not [URE3] while deleting Ssa2 weakens [URE3] but not [PSI+]. Effects of Ssa3p and Ssa4p, though somewhat different, paralleled those of Ssa1p and Ssa2p, respectively, suggesting that Ssa3 has Ssa1-like activity while Ssa4 functions more like Ssa2 (Sharma and Masison unpublished). In addition to uncovering differences in how different Hsp70 isoforms affect yeast prions, this work also suggests that certain subtle differences in constitutive Hsp70 function that might be important for optimal fitness under ideal growth conditions are also important in inducible isoforms under stress. The different effects of Ssa1 and Ssa2 on prion propagation could reflect differences in affinity of Ssa1 and Ssa2 for the different prion proteins as substrates, or that they interact differently with Hsp40s or other co-factors that in turn determine the specificity. Because Ssa1 and Ssa2 SBDs differ by only a single methyl group outside their binding pocket, we suspect the latter explanation to be more likely. Alternatively, Ssa1 and Ssa2 might differentially regulate the activity or expression of other stress response factors that in turn affect the prions.
Further support for a role of Ssa Hsp70 in [PSI+] propagation was obtained when a collection of Ssa1 mutants were identified in a screen for [PSI+]-impairing effects [9, 10]. Yeast strains expressing these mutant proteins as the only Ssa grow normally, indicating that these mutants perform well in all functions essential for cell growth. These results imply that Ssa Hsp70 interacts with the highly-ordered prion aggregates differently than with other cellular aggregates or substrates. All of the mutations that impaired [PSI+] were located in the NBD, showing that regulation of substrate interactions is more important than specificity of substrate binding per se. Similar conclusions were derived from another study where almost all of many additional Ssa1 and Ssa2 mutations identified that impaired [PSI+] propagation were in the NBD [115].
We and others also showed that co-factors that interact with Hsp70 and influence its reaction cycle can strongly influence prion propagation. Conditions expected to promote conversion to, or stabilize, Hsp70's closed ADP-bound form, such as overexpression of Hsp40s or TPR co-chaperones, or depletion of NEFs Fes1 or Sse1, impair [PSI+] propagation [10, 47, 60]. It is proposed that prolonged binding of Hsp70 to abnormally folded Sup35 decreases its conversion back into a prion state. Alternatively, excessive binding of Ssa Hsp70 to Sup35 aggregates might inhibit the Hsp104 machinery in generating newer seeds. Therefore, proper Hsp70 activity, alone or in cooperation with other chaperones, seems necessary to prevent individual polymers from aggregating or to regenerate prion seeds from aggregated polymers. By any scenario, the considerable amount of available data establish Hsp70 as a key element influencing prion propagation in yeast.
Summary
Protein misfolding is a major cause of a number of human diseases. By maintaining cellular proteins in a folding competent state Hsp70 proteins, in coordination with other chaperones, play important roles in cellular viability and therefore much current research is aimed at understanding the molecular basis of Hsp70 action. A vast amount of structural, biochemical and genetic studies carried out on Hsp70s have significantly broadened our understanding about Hsp70 and related proteins. However various aspects of Hsp70 still remain to be investigated. For example, although it is well established that various co-factors regulate Hsp70 action, the mechanistic details about the interaction of co-factors with Hsp70 is still limited. As such, a primary area of current research is the role of co-factors in regulating Hsp70 action. Structural information of intact Hsp70 is still lacking and several studies are underway to obtain structural information of the complex of full length Hsp70 and co-factors. Deeper understanding of Hsp70 action will provide us avenues to develop new drugs for treatment of various diseases. Some of the compounds, such as 15-DSG (15-deoxyspergualin) and NSC-630668-R/1, which stimulate ATPase activity of Hsc70 and inhibit Hsp40 stimulated ATPase activity of yeast Hsp70 respectively, are currently under study. Understanding the details of chaperone action has provided us mechanism to manipulate several biological pathways that can be used in future as a therapeutic tool for the treatment of various diseases. Yeast prions have proven to be a uniquely powerful genetic system for identifying and characterizing alterations that affect protein chaperones in a way that considerably impairs ability of amyloid to propagate in vivo without affecting their normal cellular functions. Such mutations, and the insight they provide into chaperone function and cooperation, would be missed by most other systems that identify mutations on the basis of phenotypic effects on growth.
Abbreviations
- Hsp
heat shcok protein
- NBD
N-terminal domain
- SBD
substrate-binding domain
- CTD
C-terminal domain
- TPR
tetratricopeptide repeat
- NEF
nucleotide exchange factor
- ER
endoplasmic reticulum
References
- [1].Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem. 2006;387:485. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- [2].Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [3].Pelham H, Lewis M, Lindquist S. Expression of a Drosophila heat shock protein in mammalian cells: transient association with nucleoli after heat shock. Philos Trans R Soc Lond B Biol Sci. 1984;307:301. doi: 10.1098/rstb.1984.0131. [DOI] [PubMed] [Google Scholar]
- [4].Nollen EA, Morimoto RI. Chaperoning signaling pathways: molecular chaperones as stress-sensing `heat shock' proteins. J Cell Sci. 2002;115:2809. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- [5].Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- [6].De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA. 2006;103:6166. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci USA. 2008;105:2975. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- [12].Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- [13].Gurley WB. HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell. 2000;12:457. doi: 10.1105/tpc.12.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol Chem. 2005;386:739. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- [15].Craig E, Ziegelhoffer T, Nelson J, Laloraya S, Halladay J. Complex multigene family of functionally distinct Hsp70s of yeast. Cold Spring Harb Symp Quant Biol. 1995;60:441. doi: 10.1101/sqb.1995.060.01.049. [DOI] [PubMed] [Google Scholar]
- [16].Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roberts BT, Moriyama H, Wickner RB. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- [19].Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- [20].Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- [22].Han W, Christen P. Interdomain communication in the molecular chaperone DnaK. Biochem J. 2003;369:627. doi: 10.1042/BJ20020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- [24].Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278:25970. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- [25].Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531:339. doi: 10.1016/s0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- [26].Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. Embo J. 2006;25:2519. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- [28].Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22:4677. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- [30].Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. Embo J. 1989;8:1601. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995;249:126. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- [33].Packschies L, Theyssen H, Buchberger A, Bukau B, Goody RS, Reinstein J. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry. 1997;36:3417. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- [34].Brehmer D, Gassler C, Rist W, Mayer MP, Bukau B. Influence of GrpE on DnaK-substrate interactions. J Biol Chem. 2004;279:27957. doi: 10.1074/jbc.M403558200. [DOI] [PubMed] [Google Scholar]
- [35].Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- [36].Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275:24728. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- [37].Kabani M, Beckerich JM, Gaillardin C. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol. 2000;20:6923. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. Embo J. 2000;19:6440. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sondermann H, Ho AK, Listenberger LL, Siegers K, Moarefi I, Wente SR, Hartl FU, Young JC. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J Biol Chem. 2002;277:33220. doi: 10.1074/jbc.M204624200. [DOI] [PubMed] [Google Scholar]
- [40].Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- [41].Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- [42].Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. Embo J. 2006;25:2510. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- [45].Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lian HY, Zhang H, Zhang ZR, Loovers HM, Jones GW, Rowling PJ, Itzhaki LS, Zhou JM, Perrett S. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem. 2007;282:11931. doi: 10.1074/jbc.M606856200. [DOI] [PubMed] [Google Scholar]
- [48].Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Demand J, Luders J, Hohfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Russell R, Wali Karzai A, Mehl AF, McMacken R. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry. 1999;38:4165. doi: 10.1021/bi9824036. [DOI] [PubMed] [Google Scholar]
- [51].Landry SJ. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry. 2003;42:4926. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- [52].Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Karzai AW, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- [54].Hennessy F, Cheetham ME, Dirr HW, Blatch GL. Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones. 2000;5:347. doi: 10.1379/1466-1268(2000)005<0347:aotloc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci. 2005;14:1697. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA. 2007;104:7163. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6:157. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- [58].Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I. The cytosolic DnaJ-like protein djp1p is involved specifically in peroxisomal protein import. J Cell Biol. 1998;142:421. doi: 10.1083/jcb.142.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Horton LE, James P, Craig EA, Hensold JO. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem. 2001;276:14426. doi: 10.1074/jbc.M100266200. [DOI] [PubMed] [Google Scholar]
- [60].Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem. 2002;277:23702. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- [61].Brodsky JL, Lawrence JG, Caplan AJ. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- [62].Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- [63].Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Revington M, Zhang Y, Yip GN, Kurochkin AV, Zuiderweg ER. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J Mol Biol. 2005;349:163. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- [65].Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].O'Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis. J Biol Chem. 1996;271:15874. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- [67].Jordan R, McMacken R. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J Biol Chem. 1995;270:4563. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- [68].Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, Gierasch LM, Zuiderweg ER. Structural insights into substrate binding by the molecular chaperone DnaK. Nat Struct Biol. 2000;7:298. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- [69].Slepenkov SV, Patchen B, Peterson KM, Witt SN. Importance of the D and E helices of the molecular chaperone DnaK for ATP binding and substrate release. Biochemistry. 2003;42:5867. doi: 10.1021/bi034126v. [DOI] [PubMed] [Google Scholar]
- [70].Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. Embo J. 1995;14:2281. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 × Hop × Hsp90 complexes. J Biol Chem. 2002;277:19265. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- [72].Li J, Wu Y, Qian X, Sha B. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem J. 2006;398:353. doi: 10.1042/BJ20060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- [74].Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- [75].Misselwitz B, Staeck O, Matlack KE, Rapoport TA. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem. 1999;274:20110. doi: 10.1074/jbc.274.29.20110. [DOI] [PubMed] [Google Scholar]
- [76].Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;95:15223. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Genevaux P, Schwager F, Georgopoulos C, Kelley WL. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics. 2002;162:1045. doi: 10.1093/genetics/162.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Awad W, Estrada I, Shen Y, Hendershot LM. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci USA. 2008;105:1164. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Qian X, Hou W, Zhengang L, Sha B. Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. Biochem J. 2002;361:27. doi: 10.1042/0264-6021:3610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Schiene-Fischer C, Habazettl J, Schmid FX, Fischer G. The hsp70 chaperone DnaK is a secondary amide peptide bond cis-trans isomerase. Nat Struct Biol. 2002;9:419. doi: 10.1038/nsb804. [DOI] [PubMed] [Google Scholar]
- [82].Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. Embo J. 1998;17:3981. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Suzue K, Young RA. Adjuvant-free hsp70 fusion protein system elicits humoral and cellular immune responses to HIV-1 p24. J Immunol. 1996;156:873. [PubMed] [Google Scholar]
- [84].Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008 doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- [85].Jaattela M, Wissing D. Heat-shock proteins protect cells from monocyte cytotoxicity: possible mechanism of self-protection. J Exp Med. 1993;177:231. doi: 10.1084/jem.177.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Simon MM, Reikerstorfer A, Schwarz A, Krone C, Luger TA, Jaattela M, Schwarz T. Heat shock protein 70 overexpression affects the response to ultraviolet light in murine fibroblasts. Evidence for increased cell viability and suppression of cytokine release. J Clin Invest. 1995;95:926. doi: 10.1172/JCI117800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol. 2000;2:476. doi: 10.1038/35019510. [DOI] [PubMed] [Google Scholar]
- [88].Phillips PA, Dudeja V, McCarroll JA, Borja-Cacho D, Dawra RK, Grizzle WE, Vickers SM, Saluja AK. Triptolide induces pancreatic cancer cell death via inhibition of heat shock protein 70. Cancer Res. 2007;67:9407. doi: 10.1158/0008-5472.CAN-07-1077. [DOI] [PubMed] [Google Scholar]
- [89].Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- [90].Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- [91].Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann WH, Browne SE, Hall A, Voellmy R, Tsuboi Y, Dawson TM, Wolozin B, Hardy J, Hutton M. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- [92].Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- [93].Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- [95].Elliott E, Tsvetkov P, Ginzburg I. BAG-1 associates with Hsc70. Tau complex and regulates the proteasomal degradation of Tau protein. J Biol Chem. 2007;282:37276. doi: 10.1074/jbc.M706379200. [DOI] [PubMed] [Google Scholar]
- [96].Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ohba M. Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett. 1997;409:307. doi: 10.1016/s0014-5793(97)00535-8. [DOI] [PubMed] [Google Scholar]
- [99].Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. Embo J. 1997;16:6209. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- [101].Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol Biol Cell. 2004;15:4003. doi: 10.1091/mbc.E04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Yeast prions act as genes composed of self-propagating protein amyloids. Adv Protein Chem. 2001;57:313. doi: 10.1016/s0065-3233(01)57026-6. [DOI] [PubMed] [Google Scholar]
- [105].Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- [106].Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Song Y, Wu YX, Jung G, Tutar Y, Eisenberg E, Greene LE, Masison DC. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell. 2005;4:289. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- [109].Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 2002;26:223. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1) J Biol Chem. 2005;280:34178. doi: 10.1074/jbc.M505420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. Embo J. 2006;25:822. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] Infection is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Loovers HM, Guinan E, Jones GW. Importance of the Hsp70 ATPase domain in yeast prion propagation. Genetics. 2007;175:621. doi: 10.1534/genetics.106.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]