Abstract
Dietary Restriction (DR) extends longevity in diverse species suggesting that there is a conserved mechanism for nutrient regulation and prosurvival responses1. We have discovered a role for the HECT E3 ubiquitin ligase WWP-1 as a positive regulator of lifespan in C. elegans in response to diet restriction. We find that overexpression of wwp-1 in worms extends lifespan up to 20% under conditions of ad libitum feeding. This extension is dependent upon the FoxA transcription factor pha-4, and independent of the FoxO transcription factor, daf-16. Reduction of wwp-1 completely suppresses the extended longevity of diet-restricted animals. However, loss of wwp-1 does not affect the long lifespan of animals with compromised mitochondrial function or reduced insulin/IGF-1 signaling. Overexpression of a mutant form of WWP-1 lacking catalytic activity suppresses the increased lifespan of diet-restricted animals, indicating that WWP-1 ubiquitin ligase activity is essential for longevity. Additionally, we find that the E2 ubiquitin conjugating enzyme, UBC-18, is essential and specific for DR induced longevity. UBC-18 interacts with WWP-1 and is required for the ubiquitin ligase activity of WWP-1 and the extended longevity of worms overexpressing wwp-1. Taken together, our results indicate that WWP-1 and UBC-18 function to ubiquitinate substrates that regulate DR induced longevity.
HECT (homologous to E6AP C-terminus) E3 ligases promote the ubiquitination of proteins that are essential in a variety of cellular events. The mammalian WWP1, WWP2 and Itch family of WW domain HECT ligases (WWP ligases) were initially identified in a search for novel proteins containing WW domains, which are modular protein interaction domains recognizing short proline motifs in their partners2. WWP ligases have an N-terminal C2 domain, a phospholipid membrane interaction motif, followed by four WW domains. To identify cellular pathways in which WWP E3 ligases are required, we have taken advantage of C. elegans as a model organism, which contains a single HECT WWP E3 ligase orthologue, wwp-1 (Y65B4BR.4). Disruption of wwp-1 using RNA interference (RNAi) yields a lethal phenotype late in embryogenesis characterized by abnormal embryogenesis despite normal cell proliferation3. The wwp-1(ok1102) mutant allele has a partially penetrant embryonic lethal phenotype4. Independent of the early developmental function of wwp-1, we found that loss of wwp-1 decreased stress resistance during adulthood (Supplementary Fig. 1 a-b, e), leading us to investigate a possible role in longevity. Loss of wwp-1 function by RNAi or mutation reduced lifespan at 25°C (Supplementary Fig. 2 a-b), but not at 20°C (Supplementary Fig. 3 a-b), consistent with a role for wwp-1 in stress resistance. To investigate if increased expression of wwp-1 extended longevity in N2 (WT) worms, we created stable transgenic lines which express an N-terminal GFP-WWP-1 fusion protein, under the control of the endogenous wwp-1 promoter in which expression of wwp-1 mRNA is increased by approximately 50% (Supplementary Fig. 4). Overexpressing wwp-1 transgenic lines (GFP∷WWP-1) lived up to 20% longer than controls expressing gfp under the same promoter (Fig. 1a), indicating that wwp-1 is a positive regulator of lifespan.
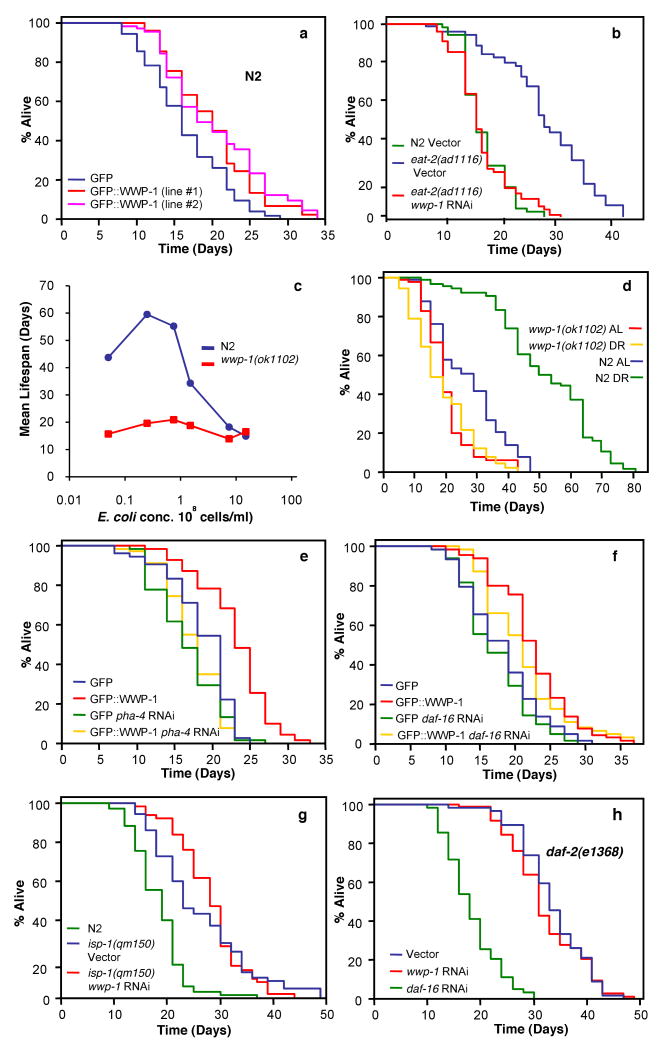
Figure 1. wwp-1 is required and specific for the extension of lifespan by dietary restriction (DR).
Lifespan values are given in Supplementary Tables 1 and 2. Two-way ANOVA analysis is presented in Supplementary Table 7. Supplementary Fig. 15a,c show data confirming specific knockdown of wwp-1 expression by RNAi. a, Two independent wwp-1 overexpressing strains (GFP∷WWP-1) can extend longevity compared to control worms expressing gfp. b, Lifespan analysis of eat-2(ad1116) mutant animals fed bacteria expressing wwp-1 dsRNA or control vector. c, Lifespans of N2 and wwp-1(ok1102) mutant worms grown in S basal buffer with different E. coli concentrations. d, Lifespan analysis of N2 and wwp-1(ok1102) mutant worms grown in DR or AL (ad libitum) E. coli concentrations. e,f, Lifespan analysis of wwp-1 overexpressing worms (GFP∷WWP-1) or a control line fed bacteria expressing pha-4 dsRNA (e), or daf-16 dsRNA (f). g,h, Lifespan analysis of isp-1(qm150) (g) and daf-2(e1368) (h) fed bacteria expressing wwp-1 dsRNA or control vector.
When diet is restricted lifespan is extended in diverse species suggesting that there is a conserved mechanism for nutrient regulation of aging. DR in worms can be reproduced genetically using eat-2(ad1116) mutant worms5,6. Reduced levels of wwp-1 completely suppressed the extended longevity of eat-2 mutant animals (Fig. 1b). Suppression of DR extended lifespan by wwp-1 depletion is unlikely to be due to increased food intake, since no difference in pharyngeal pumping rates with loss or knockdown of wwp-1 in N2 or eat-2(ad1116) worms was observed (Supplementary Table 4).
We tested whether loss of wwp-1 suppressed the extended longevity of animals subjected to DR by reduced food intake imposed by bacterial dilution in liquid culture. N2 animals exhibited a bell-shaped curve for lifespan in response to varying bacterial concentrations (Fig. 1c)7-9. The lifespan of N2 animals grown under DR conditions was more than double that of animals fed ad libitum (AL) (Fig. 1c-d). In contrast, the lifespan of wwp-1(ok1102) mutant worms across the entire food concentration range did not change noticeably (Fig. 1c-d), indicating that WWP-1 plays an essential role in regulating the response to nutrient intake and longevity. Similar results were seen using an additional method of DR, solid plate DR10 (Supplementary Fig. 5). To determine if DR could affect wwp-1 expression we used qPCR to quantify wwp-1 mRNA and found no difference in wwp-1 expression in animals grown at DR and AL conditions (Supplementary Fig. 6). Finally, expression of a GFP∷WWP-1 transgene partially rescued the suppression of DR longevity in wwp-1(ok1102) mutants (Supplementary Figs. 4 and 7). Since loss of wwp-1 prevented the extension of lifespan of animals grown using three different DR methods, we conclude that wwp-1 is essential for the increased longevity response to DR.
The Foxa transcription factor PHA-4 is required to specifically mediate DR induced longevity in C. elegans8. RNAi reduction of pha-4 suppressed the increased longevity of worms overexpressing wwp-1 (Fig. 1e), but not when these worms were fed bacteria expressing dsRNA against daf-16, the forkhead transcription factor required for the increased longevity due to reduced insulin/IGF1 signaling11,12 (Fig. 1f). Mutations in the iron sulfur component of complex III, isp-1, increase longevity by reducing mitochondrial function13-15. RNAi of wwp-1 did not suppress the extended lifespan of isp-1(qm150) mutant animals (Fig. 1g), and had only minor suppressive effects on lifespan extension of another mitochondrial mutant, clk-1(qm30), and in cyc-1 RNAi-treated worms (Supplementary Fig. 8). Partial loss of function mutations in the insulin/IGF-1 receptor homolog, DAF-2, increase lifespan in a daf-16 dependent, pha-4 independent manner8,16. RNAi depletion of wwp-1 had no effect on the long lifespan of daf-2 mutant animals (Fig. 1h and Supplementary Fig. 9a). Our results indicate that loss of wwp-1 does not make animals sick, but rather specifically regulates the response to DR that results in extended longevity.
Ubiquitination by HECT ligases requires the intermolecular transfer of ubiquitin from an associated E2 to the E3 ligase prior to transfer to a lysine in the target protein17. These transfers depend on the formation of a thioester bond between ubiquitin and a conserved cysteine in the HECT domain. Mutation of this cysteine renders HECT ligases catalytically inactive and the mutants act as dominant negatives in vivo18. We established an in vitro ubiquitination assay for WWP-1 ligase activity using C. elegans embryo extract as a source of substrates. In the presence of extract, bacterially-expressed GST-WWP-1 had very robust ligase activity, which was abolished by mutation of the catalytic cysteine (C762A) of WWP-1 (Fig. 2a). We then compared the longevity of eat-2(ad1116) transgenic animals that overexpress a GFP-WWP-1(C762A) fusion protein driven by the wwp-1 promoter to a control line expressing gfp under the same promoter. Two independent eat-2(ad1116) transgenic lines expressing the dominant negative wwp-1 had a significantly shorter lifespan, comparable to WT animals (Fig. 2b). Therefore, the ubiquitin ligase activity of WWP-1 is essential for DR-induced longevity.
Figure 2. WWP-1 ubiquitin ligase activity is essential for DR induced longevity.

a, Mutation of the conserved catalytic cysteine of WWP-1 abolishes ubiquitin ligase activity. In vitro ubiquitination assay of recombinant WT WWP-1 or mutant WWP-1 (C762A) using C. elegans embryo extract. b, eat-2(ad116) mutant worms expressing a dominant negative wwp-1(C762A) have significantly shorter lifespans than control worms expressing GFP. Lifespan values are given in Supplementary Table 1.
UBC-18 is a putative E2 that regulates pharyngeal morphogenesis during early embryonic development19-21. UBC-18 is homologous to human UbcH7, and similar to S. cerevisiae Ubc5p and Ubc4p21. Recently a two-hybrid screen using UBC-18 as a bait identified WWP-1 and the RING finger E3 ligases, ARI-1 and F56D2.2 as UBC-18 interactors19. Unlike ubc-18 and ari-1 dsRNA, inactivation of wwp-1 by dsRNA treatment failed to produce a pharynx unattached phenotype in pha-1(e2123) animals, suggesting that WWP-1 may function with UBC-18 to ubiquitinate targets not involved in pharyngeal development19. Consistent with this, RNAi of either ari-1 or F56D2.2 did not affect the long lifespan of eat-2 mutant animals (Supplementary Fig. 10).
We found that UBC-18 is indeed a functional E2. UBC-18 formed a thiol ester bond with ubiquitin (Fig. 3a), and recombinant WWP-1 ubiquitin ligase activity required UBC-18 and E1 in vitro (Fig. 3b). Extracts prepared from worms mutant for ubc-18, ubc-18(ku354)21, greatly reduced WWP-1-dependent ubiquitin ligase activity, which was restored by the addition of recombinant UBC-18 (Fig. 3b). Finally, we confirmed that UBC-18 and WWP-1 associate in vitro (Fig. 3c).
Figure 3. WWP-1 exhibits ubiquitin ligase activity in a UBC-18 dependent manner in vitro.

a, UBC-18 forms thiol sensitive adducts with ubiquitin. In vitro ubiquitin conjugation reaction in which samples were subjected to SDS/PAGE with or without β-mercaptoethanol (β-mercap). b, UBC-18 is essential for ubiquitin ligase activity in vitro. Left panel: In vitro ubiquitination assay of WT WWP-1 using N2 or ubc-18 mutant [ubc-18(ku354)] embryo extract. Recombinant UBC-18 was added in the last lane. Right panel: In vitro ubiquitination assay using purified components. c, GST pulldown assay in which GST WWP-1 (or GST alone) bound to glutathione-agarose beads was incubated with cell lysates expressing ubc-18.
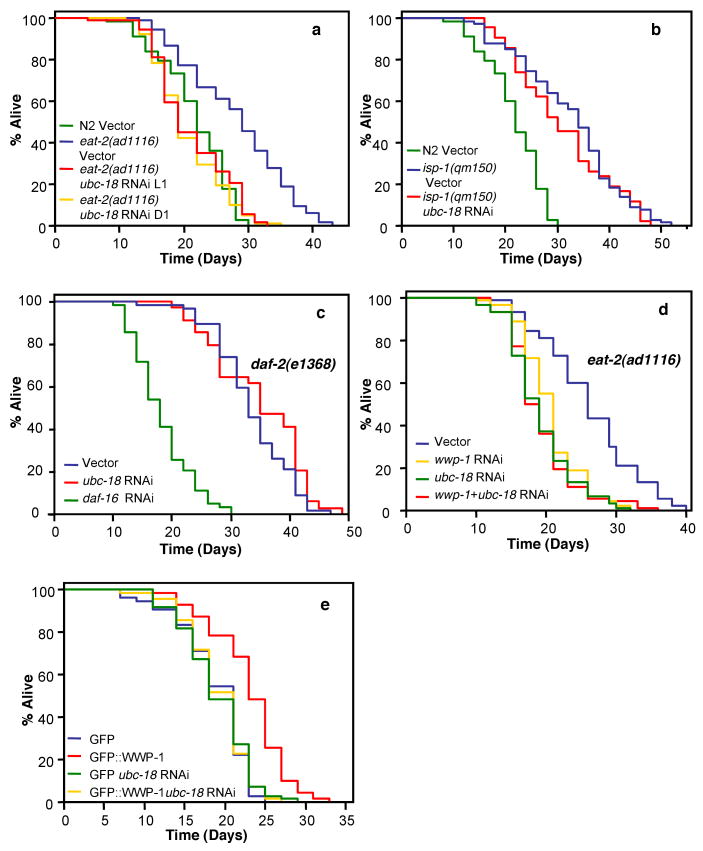
We tested whether ubc-18 was essential for DR-induced longevity. Like wwp-1, ubc-18 plays a role in stress resistance in C. elegans (Supplementary Fig. 1c-e). However, we found that overexpression of ubc-18 was unable to extend lifespan in C. elegans (Supplementary Fig. 11). Possibly, UBC-18 is not limiting for WWP-1 function in lifespan. Loss of ubc-18 function reduced lifespan at 25°C (Supplementary Fig. 2c), but only slightly at 20°C (Supplementary Fig. 3c-d). However, RNAi depletion of ubc-18 completely suppressed the increased longevity of eat-2 mutants (Fig. 4a). This decreased lifespan is unlikely to be due to impaired pharynx function, since RNAi was initiated at the L1 stage when the pharynx is completely developed, and ubc-18 RNAi initiated at the first day of adulthood, also suppressed the increased longevity of eat-2(ad1116) animals. Like wwp-1 depletion, we did not see a difference in pharyngeal pumping rates with loss of ubc-18 (Supplementary Table 4), and RNAi depletion of ubc-18 had no effect on the long lifespan of isp-1(qm150) (Fig. 4b) or daf-2 mutant animals (Fig. 4c and Supplementary Fig. 9b). In addition, epistasis analysis of wwp-1 and ubc-18 indicated that combined knockdown of both genes by RNAi in eat-2(ad1116) animals did not shorten lifespan any further than RNAi of either single gene (Fig. 4d). Finally, knockdown of ubc-18 suppressed the extended lifespan of wwp-1 overexpressing animals (Fig. 4e).
Figure 4. WWP-1 and UBC-18 function together to regulate DR induced longevity.
Lifespan values are given in Supplementary Table 1. Two-way ANOVA analysis is presented in Supplementary Table 7. Knockdown of ubc-18 expression by RNAi is shown in Supplementary Fig. 15d. a, Lifespan analysis of eat-2(ad1116) mutant worms fed bacteria expressing ubc-18 dsRNA or control vector initiated upon hatching of eggs (L1) or day 1 adults (D1). b,c, Lifespan analysis of isp-1(qm150) (b) and daf-2(e1368) (c) fed bacteria expressing wwp-1 dsRNA or control vector. d, Lifespan analysis of eat-2(ad1116) mutant animals fed bacteria expressing both wwp-1 dsRNA and vector (wwp-1 RNAi), ubc-18 dsRNA and vector (ubc-18 RNAi) or wwp-1 and ubc-18 dsRNA (wwp-1 + ubc-18 RNAi). e, Lifespan analysis of wwp-1 overexpressing worms (GFP∷WWP-1) or control worms (GFP) fed bacteria expressing ubc-18 dsRNA or control vector.
In summary, the UBC-18/WWP-1 complex functions to specify the longevity response of DR animals. Because E2s often function with multiple E3s, it is surprising to find that ubc-18 was not only essential, but also specific for the response to DR. M7.1 (UBC-2/LET-70) is most homologous to UbcH5, a mammalian E2 that associates with HECT ubiquitin ligases. Unlike ubc-18, loss of ubc-2 did not specifically suppress the extended longevity of eat-2 mutants and resulted in general sickness of animals (Supplementary Fig. 12). The other E3s that interact with UBC-18 may be dedicated instead to the developmental function of UBC-18, as is the case for ARI-119. It is interesting that WWP-1 and UBC-18 expression is observed in several neurons localized in the head and tail of adult animals (Supplementary Fig. 13), since many recent studies in C. elegans and Drosophila suggest that signals derived from the nervous system can control longevity22-25. Although it is intriguing to speculate that a few key neuronal cells in the nervous system are the site of action of WWP-1/UBC-18 to regulate longevity, expression is not confined to a few neurons, as is the case for the DR regulator, SKN-1B7. Furthermore, expression of WWP-1/UBC-18 is found in intestinal cells, another site where longevity cues are expressed in the worm26.
Since several transcription factors have been identified as targets for the mammalian orthologues of wwp-117, we investigated whether WWP-1 may target one of the two transcription factors essential for DR longevity in the worm: PHA-4 and SKN-1B7,8. The genetic epistasis analysis of ubc-18/wwp-1 suggested that PHA-4 may be a target for ubiquitination. Using our in vitro ubiquitination assay for WWP-1, we were unable to detect ubiquitinated conjugates for either PHA-4 or SKN-1B in a purified system (Supplementary Fig. 14). Recently it has been shown that pha-4 and the CeTor pathway antagonize one another to regulate longevity in adults27. Our results might suggest that wwp-1 may feed into the CeTor pathway as well. The identification of the targets of UBC-18/WWP-1 is needed to allow precise placement of this complex in the DR pathway.
Our study uncovers for the first time a role of the ubiquitin pathway in longevity in response to dietary restriction. Given the strong conservation of wwp-1 with mouse and human WWP1, an attractive hypothesis is that the mammalian orthologue will also be critical for DR induced longevity. A detailed understanding of the pathways that mediate the benefits of DR may lead to novel therapies for age-related diseases.
Methods Summary
C. elegans methods
The wwp-1 mutant strain was generated by backcrossing RB1178 [wwp-1(ok1102)] to N2 three times (Supplementary Fig. 15b). Nematodes were handled using standard methods 28.
Lifespan analysis
Lifespan analyses were performed as described 29. Bacterial dilution DR lifespans were performed as described8 with the following modifications: synchronized populations of eggs were hatched and grown at 20°C on NG agar plates containing OP50 E. coli until the L4 larval stage when they were transferred to plates of OP50 containing 100 μg/ml FUDR. At day 1 adulthood, worms were transferred into liquid culture. All lifespans were performed at 20°C unless noted.
Protein extraction for in vitro ubiquitination assay
C. elegans embryos were isolated using an alkaline hypochlorite solution from gravid N2 worms grown at 20°C 30. The embryos were resuspended in lysis buffer [50 mM Tris/HCl (pH 7.5), 0.75 mM EDTA, 1.5 mM DTT, 2.5 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A] and homogenized with 30 strokes in a Dounce homogenizer. The extract was centrifuged 15,000 × g at 4°C and stored at −70°C.
In vitro ubiquitination assay
The ubiquitination assay was carried out by incubating 1 μg Flag-tagged ubiquitin, 0.5 μg GST-WWP-1 (WT or C762A mutant), 0.1 μg UBC-18 and 15-20 μg embryo extract in 30 μl reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT and 5 mM ATP) for 1 hour at 30°C. The reaction was stopped with sample buffer and run on denaturing protein gels. Ubiquitinated substrates were identified by anti-Flag (M2, Sigma) immunoblotting. For in vitro ubiquitination assays using purified components, similar conditions were administered except 0.5 μg UBC-18 and 1 μg E1 were used. To measure UBC-18 ubiquitin conjugation, a similar reaction was performed and the reaction was stopped with sample buffer lacking β-mercaptoethanol.
Supplementary Material
Acknowledgments
We thank members of the Dillin laboratory for discussion, Philip Marks and Huaiyu Sun for their help. We thank Anne Brunet for the solid plate DR protocol. wwp-1(ok1102) was generated by the C. elegans Gene Knockout Consortium, and some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by USPHS grants CA54418 and CA82683 (to T.H.), NIDDK 070696 and NIA AG027463 grants, Ellison Medical and Glenn Medical Foundations (to A.D.). A.C.C was supported by an American Cancer Society Postdoctoral Fellowship and The Rossi Endowment. T.H. is a Frank and Else Schilling American Cancer Society Research Professor.
Footnotes
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 2.Pirozzi G, et al. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 3.Huang K, et al. A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene. 2000;252:137–145. doi: 10.1016/s0378-1119(00)00216-x. [DOI] [PubMed] [Google Scholar]
- 4.Astin JW, O'Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair (Amst) 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 8.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 9.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 10.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 13.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 15.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 16.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 17.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Hoppe T, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 19.Qiu X, Fay DS. ARI-1, an RBR family ubiquitin-ligase, functions with UBC-18 to regulate pharyngeal development in C. elegans. Dev Biol. 2006;291:239–252. doi: 10.1016/j.ydbio.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 20.Fay DS, et al. The coordinate regulation of pharyngeal development in C. elegans by lin-35/Rb, pha-1, and ubc-18. Dev Biol. 2004;271:11–25. doi: 10.1016/j.ydbio.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Fay DS, Large E, Han M, Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development. 2003;130:3319–3330. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- 22.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 23.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 24.Parkes TL, et al. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 25.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 26.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 27.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 30.Hope I. C elegans: A practical approach Oxford. Oxford University Press; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.