Abstract
Background
Single nucleotide polymorphisms (SNPs) in the progesterone receptor (PGR) gene have been associated with the risk of endometrial cancer. However, no study has systematically evaluated the role of the PGR gene in endometrial carcinogenesis.
Methods
Exposure information and DNA samples collected in the Shanghai Endometrial Cancer Study, a population-based case-control study of 1,204 incident cases and 1,212 age frequency-matched population controls, were used in this study. Seven tag SNPs were identified for the PGR gene plus the 5 kb flanking regions using the Han Chinese data from the HapMap project with a pairwise r2 ≥ 0.90. These 7 SNPs captured 92% of SNPs in the region with a pairwise r2 ≥ 0.90 or 100% of SNPs with a pairwise r2 ≥ 0.80. Genotyping of polymorphisms was performed by using the Affymetrix MegAllele Targeted Genotyping System. A logistic regression model was employed to compute adjusted odds ratios (ORs) and 95% confidence intervals (CIs).
Results
Of seven tag SNPs assessed, two polymorphisms in the 3’ flanking region of the PGR gene, rs11224561 and rs471767, were associated with the risk of endometrial cancer. Genotype CC of SNP rs11224561 was associated with decreased risk (OR=0.68, 95% CI=0.50-0.92) compared to the TT genotype. Carrying the G allele of the rs471767 SNP was also associated with decreased risk, although the association was not statistically significant (OR=0.78, 95%CI=0.59-1.04 and OR=0.32, 95%CI=0.03-3.05 for the AG and GG genotypes, respectively, compared with the homozygote AA).
Conclusion
Our findings suggest that polymorphisms in the 3’ flanking region of the PGR gene may be associated with the risk of endometrial cancer.
Keywords: progesterone receptor gene, single nucleotide polymorphism, endometrial cancer
Introduction
Endometrial cancer is a hormone-related disease. It is well recognized that excessive estrogen stimulation unopposed by progesterone plays a central role in the development of endometrial cancer across all ethnic populations (1, 2). Progesterone opposes estrogen-induced proliferation by interacting with its receptor (PGR) (3, 4), primarily through two functionally distinct PGR isoforms, PRA and PRB. These isoforms are produced by a single-copy PGR gene from two alternative promoters and translational start sites (5).
Genetic variations may result in alteration of the biological function of PGR (6, 7), thus altering progesterone-mediated tumor suppression and contributing to an individual’s susceptibility to endometrial cancer in Western populations (7, 8). For example, the PROGINS Alu insertion, which is in complete linkage disequilibrium (LD) with the Val660Leu polymorphism (rs1042838), was associated with an increased risk of endometrial cancer in a Brazilian population (8). Single nucleotide polymorphism (SNP) rs10895068 (+331G/A), a SNP related to increased expression of the PRB isoform (9), was found to predispose women to endometrial cancer in a US population (7). A recent study, however, observed a null association between endometrial cancer risk and +331G/A polymorphisms among Swedish women (10). These polymorphisms are very rare in the Chinese population (http://www.ncbi.nlm.nih.gov/SNP), a population with a low risk of endometrial cancer. This suggests that genetic polymorphisms in the PGR gene may play an important role in the development of endometrial cancer. To our knowledge, this hypothesis has not yet been comprehensively evaluated.
The implementation of the International HapMap Project has enabled rapid acquisition of data on common SNPs in an entire gene and exploration of disease-associated genetic variants in that gene using a comprehensive approach (11). In this study, we evaluated whether genetic variants in the PGR gene confer susceptibility to endometrial cancer by using a SNP tagging approach using data from the Shanghai Endometrial Cancer Study (SECS), a large, population-based case-control study conducted in urban Shanghai, China.
Materials and Methods
Study Subjects
Details of the SECS have been described elsewhere (12). Briefly, 1,454 newly-diagnosed endometrial cancer cases aged 30 to 69 years were identified between 1997 and 2003 through the population-based Shanghai Cancer Registry, of which 1,204 cases (82.8%) participated in the study. Controls were randomly selected from the general population of urban Shanghai using the Shanghai Resident Registry according to the age distribution of endometrial cancer cases in 1996. Women with a history of any cancer or hysterectomy were not eligible. Of the 1,629 eligible women contacted, 1,212 (74.4%) participated in the study. The study protocols were approved by the Institutional Review Boards of all institutes involved in the study, and written, informed consent was obtained from all participants prior to interview.
Study participants were interviewed in person by trained retired medical professionals using a structured questionnaire. Detailed information on demographic factors, menstrual and reproductive history, hormone use, prior disease history, physical activity, tobacco and alcohol use, diet, weight history, and family history of cancer was collected for all participants. Body weight, height, and circumferences of the waist and hips were measured according to a standardized protocol at the time of interview. Menopause was defined as the cessation of the menstrual period for at least 12 months before diagnosis for cases and interview for controls, excluding those lapses caused by pregnancy, breastfeeding or estrogen hormone use. Body mass index (BMI, weight in kilograms/height in meters2) and waist-to-hip circumference ratio (WHR) were calculated using measured anthropometrics.
Of the study participants who completed an in-person interview, 857 cases and 856 controls donated a blood sample and 282 cases and 286 controls provided a buccal cell sample. 189 cases and 198 controls provided samples using a mouthwash method; and 93 cases and 88 controls provided samples using a buccal swab method. Due to the very low DNA yield of the buccal swab method, we did not include buccal swab DNA samples in the genotyping. In addition, there were 9 cases and 37 controls whose samples contained very little DNA; they were not included in this project. DNA samples from 1,037 cases (86.1%, 856 blood and 181 buccal cell) and 1,018 controls (84.0%, 835 blood and 183 buccal cell) were included in this study. We and others have previously compared the genotyping results derived using DNA isolated from mouthwash samples and from blood and found that buccal cell DNA provides valid genotyping results (13, 14). All these buccal and blood DNA samples were genotyped for an additional 23 SNPs by using Taqman with 14 blind duplicate pairs for each SNP. The average concordance rate for these 23 SNPs was 99.6%.
SNP selection, identification and genotyping
Tag SNPs were selected by searching Han Chinese data from the HapMap project (www.hapmap.org) using the Tagger program (15). The following criteria were used to identify tag SNPs: 1) SNPs were located in the PGR gene or within the flanking 5 kb regions, 2) had a minor allele frequency (MAF) ≥ 0.05, and 3) the other unselected SNPs could be captured by one of the tag SNP with a LD of r2 ≥ 0.90. SNP selection was completed in December 2005. As a result, a total of seven tag SNPs, rs11224561, rs471767, rs12223699, rs11571234, rs547378, rs11224579, and rs11224598, were identified, as listed in Appendix 1. Genotype distributions for all SNPs were consistent with Hardy-Weinberg equilibrium among both cases and controls (Appendix I).
Appendix 1.
Primary information of genotyped SNPs of PGR gene, the Shanghai Endometrial Cancer Study, 1997-2003.
| Chromosome position a | rs# in dbSNP | Location | Allele b | MAF in cases | MAF in controls | PHWE for cases | PHWE for controls | Call rate (%) |
|---|---|---|---|---|---|---|---|---|
| PGR gene: | rs11224561 | 3’ flanking | C/T | 0.29 | 0.32 | 0.47 | 0.16 | 99.51 |
| Chr11:100,414,312..100,506,464 | rs471767 | 3’ flanking | G/A | 0.05 | 0.06 | 0.40 | 0.76 | 99.85 |
| rs12223699 | intron 6 | C/T | 0.10 | 0.10 | 0.96 | 0.29 | 99.95 | |
| rs11571234 | intron 4 | A/T | 0.16 | 0.16 | 0.46 | 0.96 | 100.00 | |
| rs547378 | intron 4 | A/G | 0.22 | 0.22 | 0.41 | 0.31 | 99.90 | |
| rs11224579 | intron 3 | C/T | 0.26 | 0.26 | 0.32 | 0.99 | 99.95 | |
| rs11224598 | intron 2 | C/T | 0.20 | 0.20 | 0.17 | 0.71 | 100.00 |
Version of NCBI Build 36.
Bolded alleles are minor alleles.
These SNPs were genotyped using the Affymetrix MegAllele Targeted Genotyping System with the Molecular Inversion Probe (MIP) method (16) as part of large-scale genotyping efforts that included 1,737 SNPs. Genotyping was conducted at the Vanderbilt Microarray Shared Resource following the manufacturer’s protocol. Briefly, 2.01 ug of genomic DNA was annealed to the assay panel overnight at 58°C. After annealing, the samples were split into 4 equal aliquots. Each aliquot was gap filled with 4 different aliquots receiving a different dNTP. The dNTP was ligated to produce a padlocked probe and then digested with exonucleases. The padlocked probe was then cleaved at a specific cleavage site and inverted. The inverted probe was the substrate for two rounds of PCR. After passing quality control (QC) tests, samples were hybridized to the arrays. Arrays were then washed, stained, and detected via the scanner and analyzed by using the Affymetrix protocol.
As a QC procedure, we included 39 blinded QC samples and 12 HapMap DNA samples in the genotyping. The consistency rates for these samples were ≥ 97.4% for all SNPs. The genotyping of PGR SNPs was highly successful, with call rates of 99.5-100% (median: 99.95%). Finally, the laboratory staff remained blind to the case-control status and identity of all samples.
Statistical Analyses
Chi-squared statistics and the t test were used to evaluate case-control differences in the distribution of risk factors and genotypes of the PGR gene. Logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (95% CIs). Interactive effects between a dichotomous risk factor and genotypes were evaluated by introducing the products of two dummy variables describing the 3 genotypes (two homozygous and one heterozygous) and the non-genetic risk factor in the logistic model along with the main effect terms. A likelihood ratio test was conducted by comparing the model including the main effects only with that including both the main effects and the interaction terms to derive the P-value for the multiplicative interaction test. We also examined the joint effects of multiple SNPs that were found to be associated with disease risk by grouping women together according to the number of minor alleles they carried at the two polymorphic sites. LD between polymorphisms was assessed by HaploView software (17), and haplotype blocks were defined using the methods of Gabriel et al (18). Haplotypes were constructed and associations between haplotypes and endometrial cancer risk were analyzed using HAPSTAT software (19). Five common haplotypes (frequency > 5%) for the four polymorphic sites in the haplotype block were constructed in the order of rs471767, rs12223699, rs11571234, and rs547378. All statistical tests were based on two-tailed probability.
Results
Of 1,037 eligible endometrial cancer cases in this study, 967 (93.2%) were adenocarcinoma, 12 (1.2%) were papillary serous carcinoma, 12 (1.2%) were clear-cell carcinoma, 33 (3.2%) were with other pathological types or 13 unknown (1.3%). Presented in Table 1 are selected demographic and risk factor characteristics of the subjects genotyped for PGR polymorphisms in this study. 1,037 cases and 1,018 controls were similar with respect to age, educational status, cigarette smoking, and use of hormone replacement therapy. Compared with controls, cases were more likely to have a family history of cancer, an earlier age at menarche, a later age at menopause, fewer live births, a higher BMI and WHR, and were less likely to have ever used oral contraceptives or to have engaged in regular physical activity. There were no appreciable differences seen in the distribution of demographic or risk factors between the entire study population (data not shown) and those with genotyping data.
Table 1.
Comparison of cases and controls with genotyping data on demographic characteristic and selected risk factors for endometrial cancer, the Shanghai Endometrial Cancer Study, 1997-2003.
| Subject Characteristics | Cases (n=1037) | Controls (n=1018) | P-value a |
|---|---|---|---|
| Age (years, x̄ ± sd) | 54.3 ± 8.5 | 54.5 ± 8.5 | 0.66 |
| ≥ Middle school education (%) | 78.5 | 77.6 | 0.45 |
| Regular smoker (%) | 3.1 | 3.5 | 0.57 |
| Regular alcohol consumption (%) | 3.1 | 5.3 | 0.01 |
| 1st degree relative with cancer (%) | 35.1 | 29.1 | <0.01 |
| Age at menarche (x̄ ± sd) | 14.6 ± 2.6 | 14.8 ± 2.3 | <0.01 |
| Number of pregnancies (x̄ ± sd) | 2.6 ± 1.5 | 2.9 ± 1.5 | <0.01 |
| Postmenopausal (%) | 56.4 | 61.9 | 0.01 |
| Age at menopause b (x̄ ± sd) | 50.2 ± 3.6 | 49.0 ± 3.6 | <0.01 |
| Years of menstruation (x̄ ± sd) | 32.7 ± 5.1 | 30.7 ± 5.3 | <0.01 |
| Ever used oral contraceptives (%) | 18.2 | 25.3 | <0.01 |
| Ever used HRT (%) | 4.7 | 4.3 | 0.66 |
| Diagnosis of diabetes (%) | 15.0 | 3.5 | <0.01 |
| Body mass index (x̄ ± sd) | 25.8 ± 4.1 | 23.8 ± 3.5 | <0.01 |
| Waist-to-hip ratio (x̄ ± sd) | 0.84 ± 0.05 | 0.82 ± 0.06 | <0.01 |
| Engaged in regular physical activity (%) | 28.1 | 34.1 | <0.01 |
For χ2 test (categorical variables) or non parameter Wilcoxon test (continuous variables).
Only among postmenopausal women.
As shown in Table 2, the CC genotype of the rs11224561 polymorphism was significantly associated with a reduced risk of endometrial cancer (OR=0.68, 95%CI: 0.50-0.92) as compared to the homozygous major genotype TT. The G allele of SNP rs471767 was associated with a marginally reduced risk of endometrial cancer (OR per allele=0.77, 95%CI=0.58-1.01). No significant associations were observed between endometrial cancer and the other five SNPs.
Table 2.
Association of the tag SNPs in the PGR gene with endometrial cancer risk, the Shanghai Endometrial Cancer Study, 1997-2003.
| Genotype | Cases (%) | Controls (%) | P for χ2 test | Age-adjusted OR (95%CI) | Age-adjusted OR for per allele | P for trend |
|---|---|---|---|---|---|---|
| rs11224561 | 1032 | 1013 | ||||
| TT | 513 (49.7) | 477 (47.1) | 0.04 | 1.00 | ||
| CT | 436 (42.3) | 422 (41.7) | 0.96(0.80-1.15) | |||
| CC | 83 (8.0) | 114 (11.3) | 0.68(0.50-0.92) | 0.88(0.77-1.00) | 0.05 | |
| CT/CC | 519 (50.3) | 536 (52.9) | 0.24 | 0.90(0.76-1.07) | ||
| rs471767 | 1035 | 1017 | ||||
| AA | 941 (90.9) | 900 (88.5) | 0.06 | 1.00 | ||
| AG | 93 (9.0) | 114 (11.2) | 0.78(0.59-1.04) | |||
| GG | 1 (0.1) | 3 (0.3) | 0.32(0.03-3.05) | 0.77(0.58-1.01) | 0.06 | |
| AG/GG | 94 (9.1) | 117 (11.5) | 0.07 | 0.77(0.58-1.03) | ||
| rs12223699 | 1036 | 1018 | ||||
| TT | 832 (80.3) | 813 (79.9) | 0.93 | 1.00 | ||
| CT | 193 (18.6) | 197 (19.4) | 0.95(0.76-1.18) | |||
| CC | 11 (1.1) | 8 (0.8) | 1.33(0.53-3.33) | 0.99(0.81-1.21) | 0.90 | |
| CT/CC | 204 (19.7) | 205 (20.1) | 0.80 | 0.97(0.78-1.20) | ||
| rs11571234 | 1037 | 1018 | ||||
| AA | 725 (69.9) | 712 (69.9) | 0.89 | 1.00 | ||
| AT | 288 (27.8) | 279 (27.4) | 1.01(0.83-1.23) | |||
| TT | 24 (2.3) | 27 (2.7) | 0.88(0.50-1.53) | 0.99(0.84-1.17) | 0.90 | |
| AT/TT | 312 (30.1) | 306 (30.1) | 0.99 | 1.00(0.83-1.21) | ||
| rs547378 | 1035 | 1018 | ||||
| GG | 627 (60.6) | 621 (61.0) | 0.78 | 1.00 | ||
| AG | 363 (35.1) | 341 (33.5) | 1.05(0.88-1.27) | |||
| AA | 45 (4.4) | 56 (5.5) | 0.80(0.53-1.20) | 0.98(0.85-1.14) | 0.78 | |
| AG/AA | 408 (39.4) | 397 (39.0) | 0.84 | 1.02(0.85-1.22) | ||
| rs11224579 | 1036 | 1018 | ||||
| TT | 555 (53.6) | 554 (54.4) | 0.90 | 1.00 | ||
| CT | 415 (40.1) | 394 (38.7) | 1.05(0.88-1.26) | |||
| CC | 66 (6.4) | 70 (6.9) | 0.94(0.66-1.35) | 1.01(0.88-1.16) | 0.90 | |
| CT/CC | 481 (46.4) | 464 (45.5) | 0.70 | 1.03(0.87-1.23) | ||
| rs11224598 | 1037 | 1018 | ||||
| TT | 663 (63.9) | 656 (64.4) | 0.86 | 1.00 | ||
| CT | 341 (32.9) | 320 (31.4) | 1.05(0.87-1.27) | |||
| CC | 33 (3.2) | 42 (4.1) | 0.78(0.49-1.25) | 0.99(0.84-1.15) | 0.86 | |
| CT/CC | 374 (36.1) | 362 (35.5) | 0.81 | 1.02(0.85-1.22) |
We further examined the joint effects of two suggested risk SNPs, rs11224561 and rs471767, by grouping women together according to the number of minor alleles they carried at the two polymorphic sites. These two SNPs were moderately correlated (r2=0.11). Compared to women with no minor allele at the two polymorphic sites, the risk of endometrial cancer decreased with an increasing number of minor alleles (P for trend=0.02) (Table 3).
Table 3.
Combined effect of SNPs in the PGR gene in endometrial cancer risk, the Shanghai Endometrial Cancer Study, 1997-2003.
| Cases (%) | Controls (%) | OR (95% CI) | P value | |||||
|---|---|---|---|---|---|---|---|---|
| Number of minor alleles at rs11224561 and rs471767 | ||||||||
| 0 | 508 (49.3) | 477 (47.1) | ref (1.00) | |||||
| 1 | 378 (36.7) | 345 (34.1) | 1.02(0.85-1.25) | 0.77 | ||||
| 2 | 115 (11.2) | 149 (14.7) | 0.73(0.55-0.96) | 0.02 | ||||
| 3-4 | 29 (2.8) | 41 (4.1) | 0.67(0.41-1.09) | 0.10 | ||||
| P for trend=0.02 | ||||||||
| Haplotypes in the PGR gene † | Log-additive model (OR and P value) | Dominant model (OR and P value) | Recessive model (OR and P value) | |||||
| ATAG | 63.1 | 61.4 | ref (1.00) | ref (1.00) | ref (1.00) | |||
| ATTA | 16.2 | 16.4 | 0.96 (0.81-1.14) | 0.66 | 0.99 (0.82-1.19) | 0.93 | 0.86 (0.55-1.32) | 0.48 |
| ACAG | 10.4 | 10.5 | 0.97 (0.79-1.19) | 0.78 | 0.98 (0.79-1.21) | 0.82 | 0.98 (0.52-1.82) | 0.94 |
| ATAA | 5.8 | 5.9 | 0.96 (0.74-1.25) | 0.77 | 0.95 (0.72-1.25) | 0.70 | 1.14 (0.42-3.17) | 0.79 |
| GTAG | 4.6 | 5.9 | 0.76 (0.57-1.00) | 0.05 | 0.78 (0.59-1.04) | 0.09 | 0.34 (0.05-2.46) | 0.28 |
In the order of SNPs rs471767, rs12223699, rs11571234 and rs547378 based on their chromosome position.
ORs were age-adjusted, P value was 0.42 for the test of overall difference between cases and controls in haplotype frequencies and 0.39 for the global test for entire set of haplotypes.
Presented in Figure 1 is the LD structure of the PGR gene. Four SNPs, one in the 3’ flanking region, one in intron 7, and the other two in intron 4, comprised one LD block. Within this block, haplotype GTAG, the only common haplotype containing the variant G allele of SNP rs471767, was associated with a marginally significant, reduced risk of endometrial cancer under the log-additive model (OR=0.76, 95%CI: 0.57-1.00) and dominant model (OR=0.78, 95%CI: 0.59-1.04) compared with the most common haplotype ATAG (Table 3). When haplotypes were created using 7 tag SNPs, four common haplotypes (TATAGTT, CATTACC, TACAGTT and CGTAGTT) were reconstructed. Similarly, only one haplotype, the haplotype that contained the variant G allele of SNP rs471767 (CGTAGTT), had a slight inverse association with the risk of endometrial cancer (OR=0.80, 95%CI: 0.50-1.10) under the dominant model (data not shown in the table).
Figure 1.
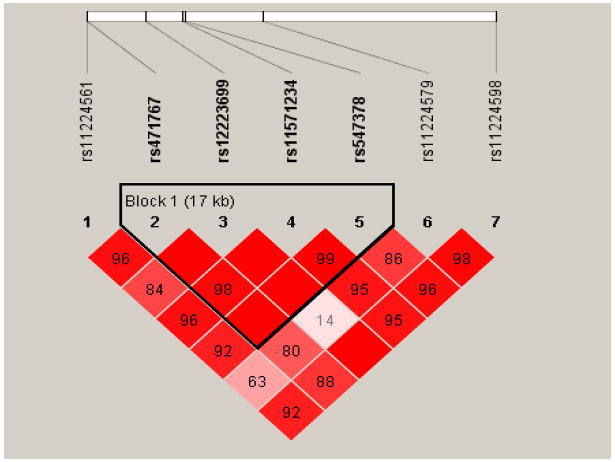
Pairwise LD (D’) between tag SNPs at PGR gene. Diamonds without a number correspond to D’=1. The block was defined using the method of confidence intervals (18).
Because menstrual status, oral contraceptive use, and body size may influence the sex hormone milieu, we further evaluated the possible modifying effect of these factors. The inverse association between SNP rs11224561 and cancer risk appeared to be more evident among pre-menopausal women and women with a higher WHR, but none of the tests for multiplicative interaction were statistically significant (data not shown in the table). A similar association pattern was observed for SNP rs471767. Further analysis showed that neither the number of minor alleles at rs11224561 or rs471767 nor haplotype ATAG interacted with any sex hormone related factors (data not shown in the table).
Discussion
Three previous studies have evaluated genetic variations in the PGR gene in relation to endometrial cancer risk (7, 8, 10). A case-control study nested within the Nurses’ Health Study found that the +331G/A PGR gene polymorphism (rs10895068) was linked to endometrial cancer risk, possibly by altering expression of the PRB isoform (7). It was suggested that the PRB isoform acts as a stronger transcription factor than the PRA isoform in vitro (20). However, no association between this polymorphism and endometrial cancer was observed in a population-based case-control study conducted in Sweden (10). Recently, a case-control study from Brazil found that the PROGINS polymorphism (homozygotes of the insertion allele) was associated with an increased risk of endometrial cancer (8), consistent with the observation of an increased risk of endometriosis for this allele (21-23). The PROGINS polymorphism is a 306 bp Alu insertion in intron 7 of the PGR gene and is also marked by a missense SNP in exon 4 (rs1042838) and a silent SNP in exon 5 (rs1042839) (7). Because the minor allele frequencies for both rs1042838 and rs10895068 are less than 2% in the Chinese population (http://www.hapmap.org), they were not selected for the current study. However, because of their low prevalence in Chinese population, it is unlikely that these potential functional polymorphisms play an important role in cancer risk in our population.
In the current study, by using a SNP tagging approach, we identified and evaluated seven tag SNPs which covered 92% of SNPs in the region with a pairwise r2 ≥ 0.90 or 100% of SNPs with a pairwise r2 ≥ 0.80. To our knowledge, this is the first study to use such a comprehensive approach to investigate the role of the PGR gene in endometrial cancer risk. The two SNPs, rs11224561 and rs471767, for which we observed an association with endometrial cancer, have not been investigated in the past. Neither of these SNPs were in LD with rs1042838, the marker SNP of the PROGINS polymorphism, in either the Caucasian or Chinese/Japanese populations (R2 < 0.10) according to Hapmap data. These SNPs and SNP rs10895068 are located more than 95 kb apart, although the R2s are not available for the Hapmap data. Therefore, the observed association with rs11224561 and rs471767 is unlikely to be accounted for by known functional polymorphisms. In addition, the risk of endometrial cancer decreased with an increasing number of minor alleles at these two polymorphic sites, suggesting a cumulative effect of the polymorphism. Haplotype analysis also showed a significant effect for the two SNPs. Given that both of these SNPs are located in the 3’ flanking region of the PGR gene, it is possible that polymorphisms in this region may regulate the translation of the PGR gene and thus increase the anti-proliferative activity of progesterone. It is of note that the frequency of the minor allele (G) of rs471767 was 6.0% in our population controls and HapMap Chinese, but is 32.5% among HapMap Caucasians. On the other hand, the frequency of the minor allele (C) of SNP rs11224561, which was 32.0% in our controls and 33.3% in HapMap Chinese, was the most common allele (87.5%) among HapMap Caucasians (http://www.ncbi.nlm.nih.gov/SNP). Because the incidence of endometrial cancer among Chinese women is much lower than among Caucasian women, these data do not appear to support a causal effect for these two SNPs in endometrial cancer etiology. Furthermore, due to the multiple comparisons made in this study, we cannot rule out the possibility that the significant associations were caused by chance. Further investigation is needed to ascertain the nature of the SNP-disease association and to identify the underlying causal polymorphisms.
This study has a number of strengths, including the population-based study design, the relatively high participation rate, the relatively homogeneous ethnic background (>98% Han Chinese), low HRT use, and the low frequency of hysterectomy (5.1%) in the study population. In addition, the application of the SNP tagging approach in SNP selection made it possible to systematically evaluate the genetic markers of the PGR gene. However, the sample size was not sufficiently large for testing interactions. Chance findings cannot be excluded.
In summary, we found that two tag SNPs in the 3’ flanking region of the PGR gene, rs11224561 and rs471767, were associated with the risk of endometrial cancer among Chinese women. Our findings will need to be validated in future studies.
Acknowledgments
We would like to thank Dr. Fan Jin for her contributions to implementing the study in Shanghai, Ms. Regina Courtney, Dr. Shawn Levy, and the Vanderbilt Microarray Shared Resource for their contributions to the genotyping, and Ms. Bethanie Hull for her assistance in the preparation of this manuscript. All microarray experiments were performed at the Vanderbilt Microarray Shared Resource. The Vanderbilt Microarray Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Diabetes Research and Training Center (P60 DK20593), the Vanderbilt Digestive Disease Center (P30 DK58404) and the Vanderbilt Vision Center (P30 EY08126). The study would not have been possible without the support of the study participants and research staff of the Shanghai Endometrial Cancer Study. This work was supported by USPHS grant R01 CA92585 from the National Cancer Institute.
Footnotes
Conflicts of Interest: None to declare.
References
- 1.Siiteri PK. Steroid hormones and endometrial cancer. Cancer Res. 1978;38:4360–6. [PubMed] [Google Scholar]
- 2.Key TJ, Pike MC. The dose-effect relationship between ‘unopposed’ oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. Br J Cancer. 1988;57:205–12. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gielen SC, Hanekamp EE, Hanifi-Moghaddam P, Sijbers AM, van Gool AJ, Burger CW, Blok LJ, Huikeshoven FJ. Growth regulation and transcriptional activities of estrogen and progesterone in human endometrial cancer cells. Int J Gynecol Cancer. 2006;16:110–20. doi: 10.1111/j.1525-1438.2006.00279.x. [DOI] [PubMed] [Google Scholar]
- 4.Smid-Koopman E, Kuhne LC, Hanekamp EE, Gielen SC, De Ruiter PE, Grootegoed JA, Helmerhorst TJ, Burger CW, Brinkmann AO, Huikeshoven FJ, Blok LJ. Progesterone-induced inhibition of growth and differential regulation of gene expression in PRA- and/or PRB-expressing endometrial cancer cell lines. J Soc Gynecol Investig. 2005;12:285–92. doi: 10.1016/j.jsgi.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–14. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vegeto E, Allan GF, Schrader WT, Tsai MJ, McDonnell DP, O’Malley BW. The mechanism of RU486 antagonism is dependent on the conformation of the carboxy-terminal tail of the human progesterone receptor. Cell. 1992;69:703–13. doi: 10.1016/0092-8674(92)90234-4. [DOI] [PubMed] [Google Scholar]
- 7.De Vivo I, Huggins GS, Hankinson SE, Lescault PJ, Boezen M, Colditz GA, Hunter DJ. A functional polymorphism in the promoter of the progesterone receptor gene associated with endometrial cancer risk. Proc Natl Acad Sci U S A. 2002;99:12263–8. doi: 10.1073/pnas.192172299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junqueira MG, da Silva ID, Nogueira-de-Souza NC, Carvalho CV, Leite DB, Gomes MT, Baracat EC, Lopes LA, Nicolau SM, Goncalves WJ. Progesterone receptor (PROGINS) polymorphism and the risk of endometrial cancer development. Int J Gynecol Cancer. 2007;17:229–32. doi: 10.1111/j.1525-1438.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 9.Sartorius CA, Melville MY, Hovland AR, Tung L, Takimoto GS, Horwitz KB. A third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoform. Mol Endocrinol. 1994;8:1347–60. doi: 10.1210/mend.8.10.7854352. [DOI] [PubMed] [Google Scholar]
- 10.Dossus L, Canzian F, Kaaks R, Boumertit A, Weiderpass E. No association between progesterone receptor gene +331G/A polymorphism and endometrial cancer. Cancer Epidemiol Biomarker & Prev. 2006;15:1415–6. doi: 10.1158/1055-9965.EPI-06-0215. [DOI] [PubMed] [Google Scholar]
- 11.Couzin J. Genomics. The Hapmap gold rush: researchers mine a rich deposit. Science. 2006;312:1131. doi: 10.1126/science.312.5777.1131. [DOI] [PubMed] [Google Scholar]
- 12.Xu WH, Shrubsole MJ, Xiang YB, Cai Q, Zhao GM, Ruan ZX, Cheng JR, Zheng W, Shu XO. Dietary Folate intake, MTHFR genetic polymorphisms and the risk of endometrial cancer among Chinese women. Cancer Epidemiol Biomarker & Prev. 2007;16:281–7. doi: 10.1158/1055-9965.EPI-06-0798. [DOI] [PubMed] [Google Scholar]
- 13.Cheng JR, Guan SF, Wang XL, Han LH, Gao YT. Feasibility of genetic polymorphisms analysis using genomic DNA obtained from human buccal cells. Ai Zheng. 2005;24(7):893–7. [PubMed] [Google Scholar]
- 14.Laine ML, Farre MA, Crusius JB, van Winkelhoff AJ, Pena AS. The mouthwash: a non-invasive sampling method to study cytokine gene polymorphisms. J Periodontol. 2000;71(8):1315–8. doi: 10.1902/jop.2000.71.8.1315. [DOI] [PubMed] [Google Scholar]
- 15.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 16.Hardenbol P, Yu F, Belmont J, Mackenzie J, Bruckner C, Brundage T, Boudreau A, Chow S, Eberle J, Erbilgin A, Falkowski M, Fitzgerald R, Ghose S, Iartchouk O, Jain M, Karlin-Neumann G, Lu X, Miao X, Moore B, Moorhead M, Namsaraev E, Pasternak S, Prakash E, Tran K, Wang Z, Jones HB, Davis RW, Willis TD, Gibbs RA. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–75. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 19.Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29(4):299–312. doi: 10.1002/gepi.20098. [DOI] [PubMed] [Google Scholar]
- 20.Kumar NS, Richer J, Owen G, Litman E, Horwitz KB, Leslie KK. Selective down-regulation of progesterone receptor isoform B in poorly differentiated human endometrial cancer cells: implications for unopposed estrogen action. Cancer Res. 1998;58:1860–5. [PubMed] [Google Scholar]
- 21.Lattuada D, Somigliana E, Vigano P, Candiani M, Pardi G, Di Blasio AM. Genetics of endometriosis: a role for the progesterone receptor gene polymorphism PROGINS? Clin Endocrinol (Oxf) 2004;61:190–4. doi: 10.1111/j.1365-2265.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- 22.Wieser F, Schneeberger C, Tong D, Tempfer C, Huber JC, Wenzl R. PROGINS receptor gene polymorphism is associated with endometriosis. Fertil Steril. 2002;77:309–12. doi: 10.1016/s0015-0282(01)02984-3. [DOI] [PubMed] [Google Scholar]
- 23.De Carvalho CV, Nogueira-De-Souza NC, Costa AM, Baracat EC, Girão MJ, D’Amora P, Schor E, da Silva ID. Genetic polymorphisms of cytochrome P450cl7alpha (CYP17) and progesterone receptor genes (PROGINS) in the assessment of endometriosis risk. Gynecol Endocrinol. 2007;23:29–33. doi: 10.1080/09513590601024707. [DOI] [PubMed] [Google Scholar]


