Abstract
Current yeast interactome network maps contain several hundred molecular complexes with limited and somewhat controversial representation of direct binary interactions. We carried out a comparative quality assessment of current yeast interactome datasets, demonstrating that high-throughput yeast two-hybrid (Y2H) provides high-quality binary interaction information. As a large fraction of the yeast binary interactome remains to be mapped, we developed an empirically-controlled mapping framework to produce a “second-generation” high-quality high-throughput Y2H dataset covering ~20% of all yeast binary interactions. Both Y2H and affinity-purification followed by mass spectrometry (AP/MS) data are of equally high quality but of a fundamentally different and complementary nature resulting in networks with different topological and biological properties. Compared to co-complex interactome models, this binary map is enriched for transient signaling interactions and inter-complex connections with a highly significant clustering between essential proteins. Rather than correlating with essentiality, protein connectivity correlates with genetic pleiotropy.
A crucial step towards understanding cellular systems properties is mapping networks of physical DNA-, RNA- and protein-protein interactions, the “interactome network”, of an organism of interest as completely and accurately as possible. One approach consists in systematically testing all pairwise combinations of predicted proteins to derive the “binary” interactome. Early attempts at binary interactome mapping used high-throughput yeast two-hybrid (Y2H), in which a protein interaction reconstitutes a transcription factor that activates expression of reporter genes. High-throughput Y2H maps have been generated for Saccharomyces cerevisiae (1-3), Caenorhabditis elegans (4-6), Drosophila melanogaster (7), and human (8-10). An alternative approach consists in generating “co-complex” interactome maps, achievable by high-throughput co-affinity purification followed by mass spectrometry (AP/MS) to identify proteins bound to tagged baits, as done for Escherichia coli (11, 12), S. cerevisiae (13-16), and human (17).
To investigate fundamental questions of interactome network structure and function, it is necessary to understand how the size and quality of currently available maps, including thorough evaluation of differences between binary and co-complex maps, might have affected conclusions about global and local properties of interactome networks (18, 19). Here, we address these issues using the yeast S. cerevisiae as a model system.
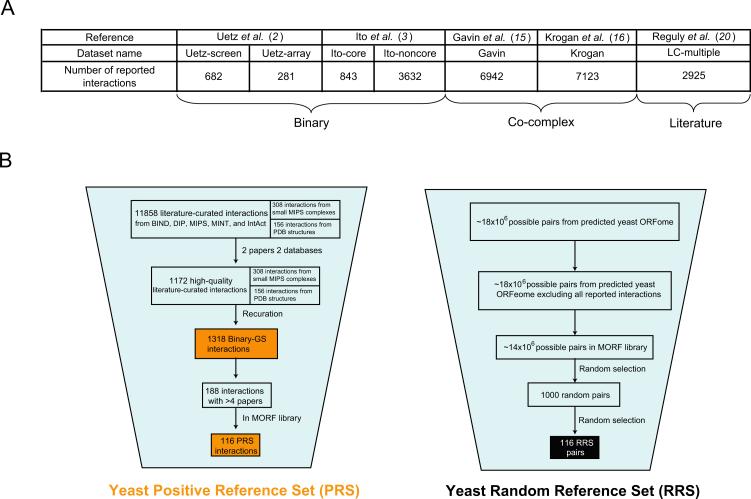
First, we compared the quality of existing high-throughput binary and co-complex datasets to information obtained from curating low-throughput experiments described in the literature (Fig. 1A). For binary interactions we examined: (i) the subset found by Uetz et al. in a proteome-scale all-by-all screen (“Uetz-screen”), excluding the pairs found in a focused, potentially biased experiment involving only 193 baits (“Uetz-array”) (2); and (ii) the Ito et al. interactions found three times or more (“Ito-core”), independently from those found one or two times (“Ito-noncore”), a distinction recommended by the authors but seldom applied in the literature (3). For co-complex associations, we investigated two high-throughput AP/MS datasets referred to as “Gavin” (15) and “Krogan” (16). For literature-curated interactions, we only considered those curated from two or more publications (“LC-multiple”) (20), which we considered of higher quality than those curated from a single publication.
Fig. 1.
Evaluation of S. cerevisiae protein-protein interaction datasets. (A) Number of interactions reported in various large-scale S. cerevisiae protein-protein interaction datasets. (B) Schema of pipeline used to assemble binary positive and random reference sets. (C) Fraction of a random sample of interactions from each dataset confirmed by Y2H and PCA. (D) Fraction of positives in each dataset calculated using MIPS and Binary-GS. (Error bars indicate standard error).
To experimentally compare the quality of these datasets, we selected a representative sample of ~200 protein interaction pairs from each one and tested them by means of two independent interaction assays, Y2H and a yellow fluorescent protein complementation assay (PCA) (21) [Supporting Online Material (SOM) I]. In PCA, bait and prey proteins are fused to non-fluorescent fragments of yellow fluorescent protein that, when brought in close proximity by interacting proteins, reconstitute a fluorescent protein in mammalian cells. In contrast, reconstitution of a transcription factor in Y2H experiments takes place in the nucleus of yeast cells. In terms of assay designs, Y2H and PCA can be considered as orthogonal assays and can be used to validate each other's results.
No single assay is expected to detect 100% of genuine interactions, and the actual fraction of positives detected is inherently linked to the stringency at which the assay is implemented. To identify the optimal scoring condition of each assay we selected a set of ~100 well-documented yeast protein-protein interaction pairs [“positive reference set” (PRS)] and a set of ~100 random pairs [“random reference set” (RRS)] (Fig. 1B; SOM II). Because RRS pairs were picked uniformly from the 14×106 possible pairings of proteins within our yeast ORFeome collection (22) (excluding those reported as interacting), these pairs are extremely unlikely to be interacting.
Sampled pairs from binary Uetz-screen and Ito-core datasets tested positive at levels as high as the positive control PRS, demonstrating their high quality (Fig. 1C). A sample of literature-curated LC-multiple interactions tested slightly lower with Y2H, while being indistinguishable by PCA (Fig. 1C), demonstrating that high-throughput Y2H datasets can be comparable in quality to literature-curated information. In striking contrast, sampled pairs from Ito-noncore tested at levels similar to the negative control RRS, confirming the low quality of this particular dataset (Fig. 1C).
Sampled pairs from Gavin and Krogan high-throughput AP/MS datasets tested poorly in our two binary interaction assays (Fig. 1C), albeit at levels similar to Munich Information Center for Protein Sequences (MIPS) complexes, a widely-used gold standard (23). This observation demonstrates that, at least for detecting binary interactions, Y2H performs better than AP/MS
Our experimental data quality assessment shows that binary Uetz-screen, Ito-core, and LC-multiple datasets are of high quality, while Ito-noncore should not be used. AP/MS datasets, although of intrinsically good quality (15, 16), should be used with caution when binary interaction information is needed.
Our experimental results contrast strikingly with computational analyses that suggested that high-throughput Y2H datasets contain more false positives than literature-curated or high-throughput AP/MS datasets (24, 25). In computational analyses, the quality of a dataset is often determined by the fraction of interactions also present in a pre-defined gold standard set (24). Generally, MIPS complexes have been considered as gold standard with all proteins constituting a given complex modeled as interacting with each other. Such modeling results in limited and biased sampling issues against binary interactions since not all proteins in a complex contact each other directly (fig. S1), and not all direct physical interactions occur within complexes (fig. S2; SOM III). Hence, while MIPS complexes are appropriate for benchmarking co-complex membership datasets, they are not for binary interaction datasets. This distinction is corroborated by the poor experimental confirmation rate of pairs from MIPS complexes using binary assays (Fig. 1C).
To computationally re-examine the quality of existing yeast interactome datasets we assembled a binary gold standard set (“Binary-GS”) of 1,318 high-confidence physical binary interactions (Fig. 1B; SOM III). Binary-GS includes direct physical interactions within well-established complexes as well as conditional interactions (e.g., dependent on posttranslational modifications) and thus represents well-documented direct physical interactions in the yeast interactome (26). When measured against Binary-GS, the quality of high-throughput Y2H datasets (with the exception of Ito-noncore) was substantially better (SOM IV and V) than that of high-throughput AP/MS datasets (Fig. 1D). Our results demonstrate the distinct nature of binary and co-complex data. Generally, Y2H datasets contain high quality direct binary interactions, whereas AP/MS co-complex datasets are composed of direct interactions mixed with preponderant indirect associations (SOM VI).
The proteome-wide binary datasets, Uetz-screen and Ito-core, contain 682 and 843 interactions, respectively (2, 3). The overlap between these two datasets appears low (3, 24): 19% of Uetz-screen and 15% of Ito-core interactions were detected in the other dataset. Given our demonstration of high quality for these datasets (Fig. 1C, D), we conclude that the small overlap stems primarily from low sensitivity (i.e., many false negatives) rather than from low specificity (i.e., many false positives as previously suggested).
Several factors might affect sensitivity. First, the space of pair-wise protein combinations actually tested in each dataset might have been considerably different. We refer to the fraction of all possible pairs tested in a given screen as the “completeness”. For example, missing 10% of ORFs in each mapping project could reduce the common tested space down to 66% [(0.9×0.9) × (0.9×0.9)] of all possible pair-wise combinations. Second, different protein interaction assays or even different versions of the same assay detect different subsets of pairs out of all possible interactions, explaining partly the limited overlap between datasets obtained with different Y2H versions. For any assay, the “assay-sensitivity” is estimated as the fraction of PRS interactions detected, which for our Y2H assay was determined empirically to be ~20% (Fig. 1C). Finally, when screening tens if not hundreds of millions of protein pairs in any tested space, that search space might need to be sampled multiple times to report all or nearly all interactions detectable by the assay used. The fraction of all theoretically detectable interactions by a particular assay found in a given experiment is its “sampling-sensitivity”. These three parameters fully account for the seemingly small overlap between Ito-core and Uetz-screen (SOM VII), demonstrating that a large fraction of the S. cerevisiae binary interactome remains to be mapped. Therefore, we carried out a new proteome-scale yeast high-throughput Y2H screen (fig. S3).
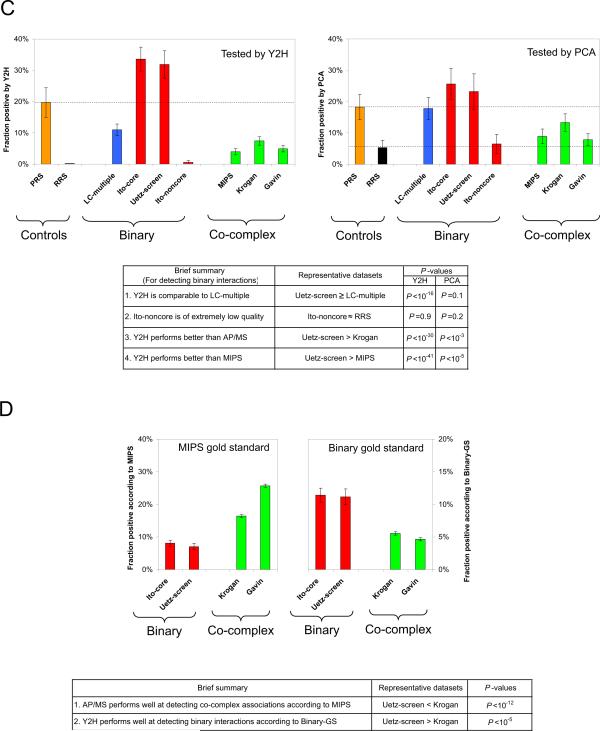
We used 5,796 Gateway-cloned ORFs available in the yeast MORF collection (22). After subcloning these ORFs into Y2H vectors and removing auto-activators (27, 28), our search space became 3,917 DB-Xs against 5,246 AD-Ys, representing a completeness of 77% (Fig. 2A; SOM VI), comparable to that of recent AP/MS datasets (15) (~78%; SOM VI).
Fig. 2.
Large-scale Y2H interactome screen. (A) Completeness of the Y2H screen. (B) Sampling-sensitivity of CCSB Y2H screens measured by screening a subspace multiple times. (C) Fraction of protein pairs in PRS, RRS, and CCSBYI1 that test positive by PCA, MAPPIT or Y2H. (D) Overlaps between three high-quality large-scale S. cerevisiae Y2H datasets (*P<10-7).
To address sampling-sensitivity, we determined what fraction of all detectable interactions is found in each pass after eight trials in a search space of 658 DB-X and 1,249 AD-Y ORFs. A single trial identified about 60% of all possible interactions that can be detected with our high-throughput Y2H, whereas three to five repeats were required to obtain 80-90% (Fig. 2B; SOM VI). We decided to screen the whole search space three times independently to yield an estimated sampling-sensitivity of 85% (Fig. 2B). In total ~88,000 colonies were picked, of which 21,432 scored positive upon more detailed phenotyping (SOM I). After identifying all putative interaction pairs by sequencing, phenotypically retesting them using fresh cultures from archival stocks, and eliminating de novo auto-activators (28), we obtained a final dataset, “CCSBYI1”, of 1,809 interactions among 1,278 proteins.
To validate the overall quality of CCSB-YI1, we tested 94 randomly-chosen interactions by PCA and mammalian protein-protein interaction trap (MAPPIT; SOM I) (21, 29). MAPPIT takes place at the mammalian cell membrane and measures interactions via activation of STAT3-dependent reporter expression. Using both PCA and MAPPIT the confirmation rate of CCSB-YI1 was similar to those of Ito-core and Uetz-screen (Fig. 1C). The precision [i.e., fraction of true positives in the dataset (30)] of CCSB-YI1 is estimated at 94-100% (Fig. 2C; fig. S4; SOM VI). Additionally, the performance of our high-throughput Y2H approach was confirmed via a larger RRS of 1,000 random pairs (30) (Fig. 1B), none of which tested positive (SOM II).
The overlaps of Uetz-screen (27%) and Ito-core (35%) with CCSB-YI1 (Fig. 2D) can be explained by the completeness, assay- and sampling-sensitivity of the three experiments (SOM VII) and agree well with the results of the pairwise confirmation of those two datasets (Fig. 1C). Similar principles apply to other large-scale experiments such as AP/MS, likely accounting for the low overlap between Krogan and Gavin (~25%; fig. S5B).
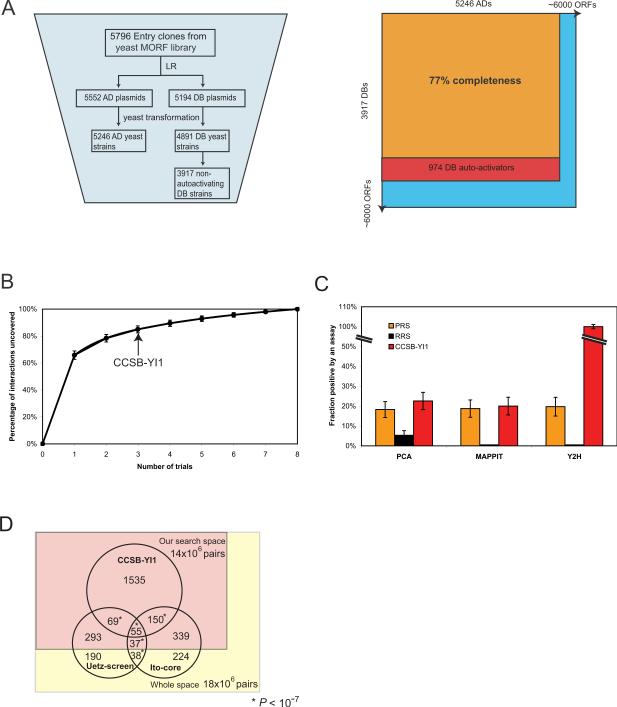
Factoring in completeness, precision, assay-, and sampling-sensitivity, we estimated that the yeast binary interactome consists of ~18,000±4,500 interactions (SOM VI), experimentally validating previous computational estimates of 17,000 to 25,000 interactions (31, 32). To obtain a more comprehensive map of the binary yeast interactome we combined the three available high-quality proteome-scale Y2H datasets (SOM VII). The union of Uetz-screen, Ito-core, and CCSB-YI1, “Y2H-union”, contains 2,930 binary interactions among 2,018 proteins, which, according to our empirical estimate of the interactome size, represents ~20% of the whole yeast binary interactome (Fig 3A).
Fig. 3.
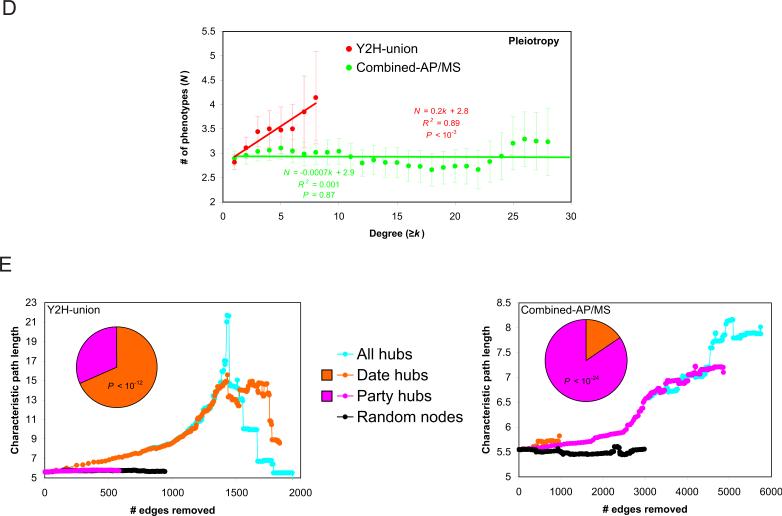
Network analysis of Y2H-union, Combined-AP/MS and LC-multiple datasets. (A) Network representations. Shown are relationships between increasing degree of a gene product and (B) the fraction of essential genes with the corresponding degree, (C) the fraction of essential genes with the corresponding degree for Y2H datasets, (D) the number of phenotypes associated with deletion of the encoding gene. (E) Contribution of date hubs and party hubs as measured by change in the characteristic path length after simulated removal of edges by deleting the indicated types of nodes. Inset: fraction of date hubs and party hubs for each dataset.
We re-examined global topological features of this new yeast interactome network, facing lower risk of over-interpreting properties due to limited sampling and various biases in the data (18). To contrast topological properties of the binary Y2H-union network with that of the co-complex network, we used an integrated AP/MS dataset (33), which was generated by combining raw high-throughput AP/MS data (15, 16). This “Combined-AP/MS” dataset, composed of 9,070 co-complex membership associations between 1,622 proteins, attempts to model binary interactions from co-complex data (Fig. 3A).
As found previously for other macromolecular networks, the connectivity or “degree” distribution of all three datasets is best approximated by a power-law (34) (fig. S6; SOM VIII). Highly connected proteins, or “hubs”, are reportedly more likely encoded by essential genes than less connected proteins (35). Surprisingly, Y2H-union lacked any correlation between degree and essentiality (Fig. 3B). This discrepancy might stem from biases in the datasets available at the time of the original observation: interactions reported in Uetz et al. (Uetz-array and Uetz-screen) and literature-curated interactions. Although Uetz-array is of high quality (fig. S7), its experimental design could negatively influence network analyses. Most hub proteins in Uetz-array were found as baits (fig. S8) and the percentage of essential proteins in the 193 bait proteins is two times higher (34.7%) than that of all protein-encoding ORFs in the yeast genome (18.4%), explaining the high correlation between degree and essentiality (Fig. 3C). Likewise, literature-curated interactions seem prone to sociological and other inspection biases (SOM VII). Thus, we refrain heretofore from using LC-multiple in our further topological and biological analyses. No significant correlation between degree of connectedness and essentiality was observed in any of the three proteome-wide high-throughput binary datasets available today (i.e., Ito-core, Uetz-screen, and CCSB-YI1; Fig. 3C), as well as new versions of our C. elegans and human interactome maps (fig. S9; SOM IX).
Hub proteins instead relate to pleiotropy, the number of phenotypes observed as a consequence of gene knock-out (SOM I). There was a significant correlation in Y2H-union between connectivity and the number of phenotypes observed in global phenotypic profiling analyses of yeast genes (36) (Fig. 3D). Thus the number of binary physical interactions mediated by a protein seems to better correlate with the number of cellular processes in which it participates than its essentiality. The correlation between degree and number of phenotypes is not observed in Combined-AP/MS, likely because co-complex associations reflect the size of protein complexes more than the number of processes they might be involved in.
We confirmed the concept of modularity in the yeast interactome network, whereby date hubs that dynamically interact with their partners appear particularly central to global connectivity while static party hubs appear to function locally in specific biological modules (37). The proportion of date and party hubs is strikingly different between Y2H-union and Combined-AP/MS (Fig. 3E). There are significantly more date hubs in the binary network, whereas party hubs are prevalent in the co-complex network. In the binary network, date hubs are crucial to the topological integrity of the network, while party hubs have minimal effects. However, in the co-complex network, date and party hubs affect the topological integrity of the network equally, likely because most hubs in Combined-AP/MS reside in large stable complexes, while hubs in Y2H-union preferentially connect diverse cellular processes.
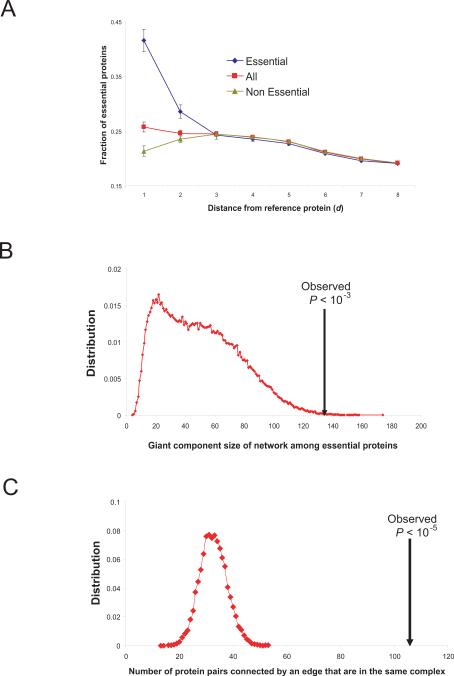
Surprisingly, essential proteins strongly tended to interact with each other (Fig. 4A; SOM IX). Concentrating on the subnetwork formed by interactions mediated by and among essential proteins (fig. S10), we found a giant component whose size is much larger than expected by chance (Fig. 4B). To better understand the clustering of essential proteins, we examined the interacting essential protein pairs that are also reported to be in the same complex, finding 106 interacting essential protein pairs, a greater number than expected by chance (Fig. 4C; SOM IX).
Fig. 4.
Clustering of essential proteins. (A) Average fraction of essential proteins among proteins whose distance are equal to d from a protein selected from essential, non-essential and all proteins. (B) Giant component size of network formed by essential proteins (arrow) compared to 100,000 random networks of same topological properties. (C) The number of interacting essential proteins that are also found in the same complex compared to 10,000 random selections of proteins of the same number as essential proteins (SOM IX).
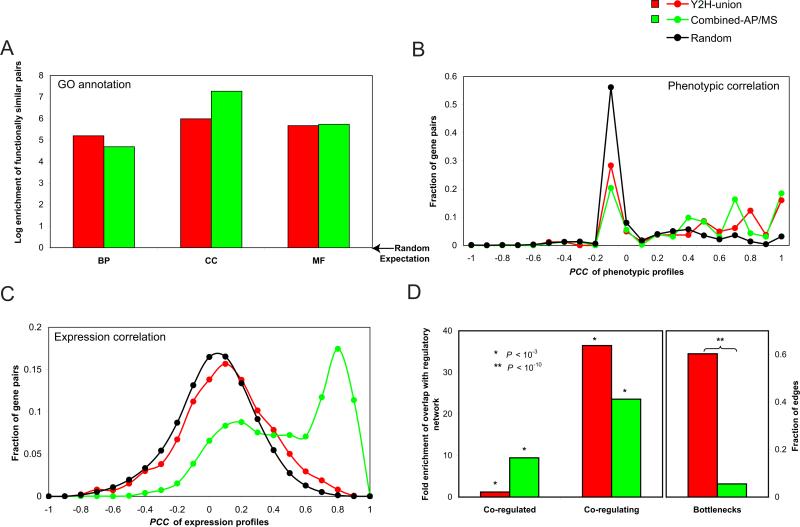
We investigated the overall relationships between Y2H-union and Gene Ontology (GO) attributes (38), phenotypic and expression profiling similarities (39), and transcriptional regulatory networks (40). Both Y2H-union and Combined-AP/MS show significant enrichment (all P<10-10) for functionally similar pairs in all three GO branches (Fig. 5A) (41). There is also significant enrichment of positive correlations of phenotypic profiles (36) between interacting pairs in both datasets (Fig. 5B; fig. S11). Such interactions supported by strong phenotypic information constitute likely possibilities of functional relationships. Lastly, both datasets are significantly enriched with pairs co-expressed across many conditions (fig. S12), although Combined-AP/MS shows higher enrichment (Fig. 5C), agreeing well with the different nature of the two assays: AP/MS aims at detecting stable complexes whereas Y2H tends to detect more transient and condition specific protein interactions. This observation is further supported by enrichment of kinase-substrate pairs in Y2H-union (SOM X; fig. S13).
Fig. 5.
Biological features of yeast interactome datasets. (A) Enrichment of interacting protein pairs (relative to random) that share GO annotations in the biological process, cellular component and molecular function branches of GO ontology. (B) Pearson correlation coefficient (PCC) of phenotypic profiles between interacting pairs in different datasets. (C) Co-expression correlation between interacting pairs. (D) Left panel: enrichment of interacting proteins as targets of a common TF (co-regulated), and enrichment of interacting TFs in a common MIM (co-regulating) (*P<10-3). Right panel: fraction of bottlenecks from each dataset in the combined network (SOM XI). Top 10% of edges with the highest betweenness are defined as “bottlenecks” (45).
To explore the mechanisms behind co-expression of interacting protein pairs we combined transcriptional regulatory networks with interactome network information (40). Interacting proteins in both networks showed a tendency to be co-regulated by common transcription factors (TFs; Fig. 5D). Similarly to what we observed in the co-expression correlation analysis (Fig. 5C), the enrichment for interacting pairs in Combined-AP/MS was significantly higher than that of Y2H-union. Strikingly, we observed a significant enrichment of protein-protein interactions between TFs involved in a common “multi-input motif” (42, 43) (MIM, where multiple TFs co-regulate a given set of genes; Fig. 5D; SOM ×). The fraction of co-regulating TF pairs is much higher in the binary interactome than in the co-complex network, suggesting that various TFs function together to form transient complexes to differentially regulate transcriptional targets (44).
These observations suggest that our binary interactome dataset is enriched in transient or condition-specific interactions linking different subcellular processes and molecular machines. To further explore this possibility we calculated “edge-betweenness” for each interaction in a merged network of all available interactions (SOM XI), measuring the number of shortest paths between all protein pairs that traverse a given edge. The higher edge-betweenness of interactions from Y2H-union shows the tendency of Y2H to detect key interactions outside of complexes, significantly more often than AP/MS (Fig. 5D). Several examples of such complex-to-complex connectivity are evident in a complete map of MIPS complexes connected by Y2H interactions (fig. S14).
Overall, we infer that Y2H interrogates a different subspace within the whole interactome than AP/MS, and Y2H interactions represent key connections between different complexes and pathways. Y2H and AP/MS provide orthogonal information about the interactome and are both vital to obtain a complete picture of cellular protein-protein interaction networks.
Acknowledgments
Supported by funds from the W.M. Keck Foundation awarded to M.V. and F.P.R.; by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative awarded to M.V. and CCSB; by National Institutes of Health (NIH) grant (R01-HG001715) awarded to M.V. and F.P.R.; by National Institutes of Health (NIH) grants (U01-A1070499-01 and U56-CA113004) awarded to A.-L.B.; by grants from the University of Ghent (GOA12051401) and the Fund for Scientific Research Flanders (FWO-V G.0031.06) awarded to J.T.; by a grant from the National Cancer Institute of Canada awarded to C.B.; and by an NIH grant (HG003224) awarded to F.P.R. I.L. is a postdoctoral fellow with the FWO-V. M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium). We thank members of our laboratories for helpful discussions, and Agencourt Biosciences for sequencing assistance.
Footnotes
Supporting Online Material SOM Text Figs. S1-35 Tables S1-5 References
References and Notes
- 1.Fromont-Racine M, Rain JC, Legrain P. Nat. Genet. 1997;16:277. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 2.Uetz P, et al. Nature. 2000;403:623. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, et al. Proc. Natl. Acad. Sci. USA. 2001;98:4569. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walhout AJ, et al. Science. 2000;287:116. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 5.Reboul J, et al. Nat. Genet. 2003;34:35. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 6.Li S, et al. Science. 2004;303:540. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giot L, et al. Science. 2003;302:1727. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 8.Colland F, et al. Genome Res. 2004;14:1324. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual JF, et al. Nature. 2005;437:1173. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Stelzl U, et al. Cell. 2005;122:957. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Butland G, et al. Nature. 2005;433:531. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 12.Arifuzzaman M, et al. Genome Res. 2006;16:686. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavin AC, et al. Nature. 2002;415:141. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 14.Ho Y, et al. Nature. 2002;415:180. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 15.Gavin AC, et al. Nature. 2006;440:631. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 16.Krogan NJ, et al. Nature. 2006;440:637. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 17.Ewing RM, et al. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han JD, Dupuy D, Bertin N, Cusick ME, Vidal M. Nat. Biotechnol. 2005;23:839. doi: 10.1038/nbt1116. [DOI] [PubMed] [Google Scholar]
- 19.Scholtens D, Vidal M, Gentleman R. Bioinformatics. 2005;21:3548. doi: 10.1093/bioinformatics/bti567. [DOI] [PubMed] [Google Scholar]
- 20.Reguly T, et al. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remy I, Michnick SW. Methods Mol. Biol. 2004;261:411. doi: 10.1385/1-59259-762-9:411. [DOI] [PubMed] [Google Scholar]
- 22.Gelperin DM, et al. Genes Dev. 2005;19:2816. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mewes HW, et al. Nucleic Acids Res. 2006;34:D169. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, et al. Nature. 2002;417:399. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 25.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Nat. Biotechnol. 2004;22:78. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, et al. Genome Res. 2004;14:1107. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidalain PO, Boxem M, Ge H, Li S, Vidal M. Methods. 2004;32:363. doi: 10.1016/j.ymeth.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Walhout AJ, Vidal M. Genome Res. 1999;9:1128. doi: 10.1101/gr.9.11.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyckerman S, et al. Nat. Cell Biol. 2001;3:1114. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 30.Jansen R, Gerstein M. Curr. Opin. Microbiol. 2004;7:535. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Grigoriev A. Nucleic Acids Res. 2003;31:4157. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen R, et al. Science. 2003;302:449. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 33.Collins SR, et al. Mol. Cell. Proteomics. 2007;6:439. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Barabási AL, Albert R. Science. 1999;286:509. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 35.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Nature. 2001;411:41. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 36.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. Mol. Syst. Biol. 2005;1:0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han JD, et al. Nature. 2004;430:88. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 38.Gene Ontology Consortium Nucleic Acids Res. 2008;36:D440. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal M. Cell. 2001;104:333. doi: 10.1016/s0092-8674(01)00221-5. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Gerstein M. Proc. Natl. Acad. Sci. USA. 2006;103:14724. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H, Jansen R, Stolovitzky G, Gerstein M. Bioinformatics. 2007;23:2163. doi: 10.1093/bioinformatics/btm291. [DOI] [PubMed] [Google Scholar]
- 42.Lee TI, et al. Science. 2002;298:799. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 43.Milo R, et al. Science. 2002;298:824. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 44.Luscombe NM, et al. Nature. 2004;431:308. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. PLoS Comput. Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]