Abstract
New habitat-based models for spread of hantavirus are developed which account for interspecies interaction. Existing habitat-based models do not consider interspecies pathogen transmission, a primary route for emergence of new infectious diseases and reservoirs in wildlife and man. The modeling of interspecies transmission has the potential to provide more accurate predictions of disease persistence and emergence dynamics. The new models are motivated by our recent work on hantavirus in rodent communities in Paraguay. Our Paraguayan data illustrate the spatial and temporal overlaps among rodent species, one of which is the reservoir species for Jabora virus and others which are spillover species. Disease transmission occurs when their habitats overlap. Two mathematical models, a system of ordinary differential equations (ODE) and a continuous-time Markov chain (CTMC) model, are developed for spread of hantavirus between a reservoir and a spillover species. Analysis of a special case of the ODE model provides an explicit expression for the basic reproduction number, , such that if , then the pathogen does not persist in either population but if , pathogen outbreaks or persistence may occur. Numerical simulations of the CTMC model display sporadic disease incidence, a new behavior of our habitat-based model, not present in other models, but which is a prominent feature of the seroprevalence data from Paraguay. Environmental changes that result in greater habitat overlap result in more encounters among various species that may lead to pathogen outbreaks and pathogen establishment in a new host.
Keywords: Hantavirus, Interspecies pathogen transmission, Basic reproduction number
1. Introduction
Successful transmission of a directly transmitted pathogen requires opportunities for contact between species. These opportunities often occur when the preferred habitat of a species overlaps or is invaded by a second species. Interspecies interactions, especially among species competitively utilizing the same resources, often result in aggressive encounters. If a pathogen is present in a reservoir host, the encounter may result in pathogen transmission to a naive host or adaptation of the pathogen to create a new reservoir. The reservoir population, the carrier of the pathogen and the long-term host, often does not exhibit disease symptoms or experience any additional mortality.
In this investigation, we develop, analyze, and numerically simulate solutions to two new habitat-based models for the spread of a directly transmitted pathogen between two species. Our goal is to model the process of interspecies pathogen transmission based on species habitat preferences. The motivation for the models comes from our recent study of hantavirus in Paraguay. Hantavirus (Family Bunyaviridae) is a genus of viruses, each generally associated with a specific rodent species (i.e., mice and rats). Approximately 30 different hantaviruses exist throughout the world, some of which cause human infection (Mills et al., 1999). Human infection is incidental, generally due to indirect transmission from contact with infectious rodent excreta, but may result in hantavirus pulmonary syndrome with a mortality rate as high as 37% (CDC, 2002). One of the reservoir species for hantavirus in Paraguay is Akodon montensis (Montane Akodont found in Eastern Paraguay, Northeastern Argentina and Southeastern Brazil) carrier of Jabora virus (JABV, GenBank # EF492471). Our empirical data show that although these species exhibit different habitat preferences, the combination of partial habitat flexibility and temporally variable climatic and soil and vegetation conditions, results in periodic microgeographic sympatry of Akodon with one or both of the spillover species.
Mathematical models for the spread of hantavirus in rodents have concentrated primarily on the dynamics of the reservoir population (Abramson and Kenkre, 2002, Abramson et al., 2003; Allen et al., 2003, Allen et al., 2006a, Allen et al., 2006b; Sauvage et al., 2003, Sauvage et al., 2007; Wesley, 2008, Wesley et al., 2009, Wolf et al., 2006). A multi-species epizootic model for susceptible and infected hosts was formulated and analyzed by McCormack and Allen (2007) but this model was not spatially explicit and did not account for differences in epizootiology of reservoir and spillover species. Our new models take into account habitat partitioning and important differences in the epizootiology of the reservoir and spillover populations. The role of the spillover species in pathogen and disease emergence is not well understood. It has been speculated that the spillover species may contribute to maintenance of the pathogen in the wild, provided there is spillback infection (McCormack and Allen, 2007) or the spillover species may be instrumental in the evolution of new hantaviruses (Chu et al., 2006). Spillover infections occur in hantavirus (Delfraro et al., 2008, Palma et al., 2009, Klingstrom et al., 2002; Torrez-Martinez et al., 1998, Weidmann et al., 2005) but are not unique to hantaviruses (Daszak et al., 2000); they have been documented in other zoonotic diseases including rabies (Nadin-Davis and Loza-Rubio, 2006; Nel et al., 1997), Nipah virus (Chua, 2003), canine distemper, parvovirus (Fiorello et al., 2006), and the SARS coronavirus (Holmes, 2003).
The habitat-based models consist of three regions: a preferred habitat for each of the reservoir and the spillover populations, and a third region of overlap (or boundary region) where interspecies encounters and pathogen transmission may occur. We formulate two models, the first model is a deterministic model, a system of ordinary differential equations (ODE), whereas the second model is a stochastic model, a continuous-time Markov chain (CTMC) model. The ODE system is analytically tractable in the case that encounters in the boundary region are brief. In this case an explicit expression for the basic reproduction number, , can be calculated, the threshold for disease outbreaks. The basic reproduction number is one of the most important parameters in the study of disease ecology. Specifically, it is the number of secondary infections caused by introduction of one infectious individual into an entirely susceptible population (see Anderson and May, 1991, Hethcote, 2000). The ODE system does not capture the few cases that occur due to interspecies interactions in the region of overlap. Therefore, we formulate a CTMC model for this purpose. Numerical solutions of the stochastic model illustrate sporadic infection in the spillover species when habitats overlap, a prominent feature of the seroprevalence data from Paraguay. Analysis and simulation of our new models show that as the number of encounters in the overlap region and the time spent in the overlap region increase (which may be triggered by habitat change), there is greater likelihood of pathogen outbreaks and disease persistence in the reservoir and spillover populations (through increase in ). Interspecies encounters and pathogen transmission in the region of overlap may be the first step in the evolution of a new hantavirus strain.
2. Empirical data and motivation for the model
Recent data collected in Paraguay (2005–2007) have shown cases of hantaviral infection in A. montensis, the reservoir species for JABV. Spillover infection, presumably of JABV, has been found in several other mouse species, including Necromys lasiurus (Hairy-tailed Akodont in Central Brazil, Southeastern Peru, Eastern Paraguay and Northeastern Argentina) and Oxymycterus delator (Paraguayan Hocicudo in Eastern Paraguay and South Central Brazil).
2.1. Habitat characteristics
Our field work was conducted in the Mbaracayú Biosphere Reserve in eastern Paraguay, which lies in the western-most portion of the interior Atlantic Forest. Vegetation composition in this area is typical of the mixture of intact, disturbed, and deforested areas found in eastern Paraguay (Fernández Soto and Mata Olmo, 2001).
Within this landscape of mixed habitat types, rodents were sampled on two mark-recapture grids, R3A and R3B, representing contrasting potential habitat for A. montensis (see Fig. 1 ). Site R3A is largely deforested, with its natural cover replaced by native and exotic graminoids and forbs. This site is highly disturbed by human activities and is intensively managed for pasturing and grazing. Reforestation is suppressed and graminoid cover maintained by frequent prescribed burning. Large ungulate grazers, primarily domestic cattle (Bos taurus) are present on this site year-round. Vegetation in the site is dominated by species of the genus Andropogon, warm temperature/tropical grasses used as grazing forage. Other common vegetation genera include Merostachys (bamboo) along the fringes of pastures and Xyris (a forb) in lower, wetter areas. Although dominated by herbaceous species, R3A retains a few islands of woody vegetation and trees, especially along the edges of the deforested areas. These forest remnants are better microhabitats for Akodon, and most captures of this species were along the northwest and southeast corners of the grid, the edges closest to the woodlands.
Fig. 1.
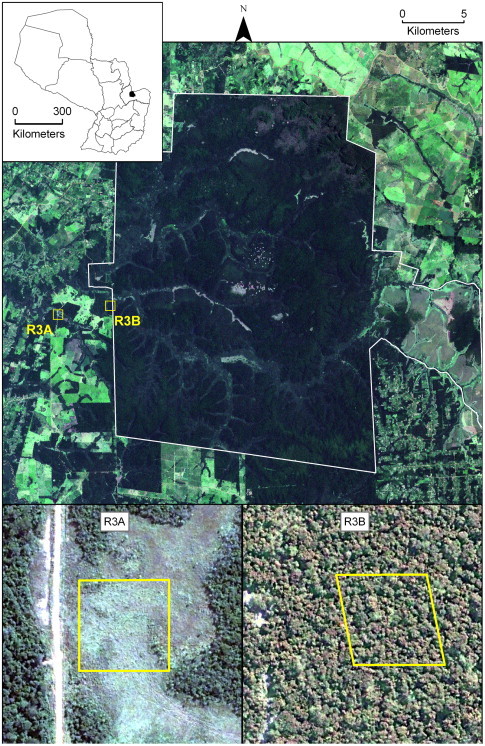
Natural color satellite imagery showing location and general land cover characteristics of the two data collection grids. Larger image (acquired February 2003 from Landsat ETM+) shows the general setting of the collection grids within and near the Mbaracayú forest reserve (reserve boundaries are indicated by the white outline). Smaller images (acquired May 2005 from DigitalGlobe Quickbird) show detailed views of the forested (R3B) and deforested (R3A) sites. Boxes indicate the exact boundaries of the 1 ha trapping grids. Location of the study site within Paraguay is indicated by the inset map.
Site R3B contrasts with R3A in that its dominant cover consists of native forest and its associated vegetation community. Although native cover remains, the site shows evidence of recent human disturbance, especially selective logging and nearby road construction. These disturbances have resulted in fragmentation of the native vegetation cover, producing numerous internal edges and gaps in the forest canopy. These edges and gaps are associated with a dense understory at the forest floor, favorable habitat for A. montensis (Pardiñas and D’Elía, 2003). Dominant vegetation genera in areas of intact or mostly intact forest include Cedrela and Balfourodendron. In disturbed areas, a lower canopy dominated by Sorocea bonplandii (a lower, woody shrub-like tree) is common, with herbaceous species and Bromelia common in the understory, and Merostachys frequently in forest gaps.
2.2. Empirical data
Each sampling grid consisted of an square array of trap stations, set 10 m apart. One standard Sherman Live Trap was set on the ground at each station. Where vegetation structure permitted (R3B only), another trap was placed 2–3 m above ground, in branches or vines, to sample other more arboreal species. However, although A. montensis is preferentially a forest-dweller, it only rarely climbs in the vegetation (651 of 661, 98.5%, of our captures of this species on R3B were on the ground). Both grids were sampled in nine sessions from February 2005 to May 2007, using standard mark-recapture techniques for small mammals (Wilson et al., 1996). Each session included eight consecutive nights (seven in February 2005). Animals were individually marked with a subdermally- implanted passive integrated transponder (PIT) tag, which can be read by passing the electronic reader near the animal's body. Date, grid, trap station, and the PIT tag number were recorded, animals were identified to species, and sex and weight were recorded. Blood, saliva, urine, and feces were collected the first time an animal was captured during each sampling session, for assay of hantavirus antibody and viral RNA presence. Animals were then released at the site of capture. All field protocols were approved by the Texas Tech University Animal Care and Use Committee.
A total of 582 captures was recorded on R3B, and 332 on R3A. Species captured on R3B included A. montensis, Calomys callosus, Oligoryzomys fornesi, O. nigripes, and Oryzomys megacephalus; on R3A, A. montensis, C. callosus, C. tener, N. lasiurus, O. nigripes, and O. delator were encountered. In this report we consider only the populations of A. montensis, N. lasiurus, and O. delator, as exemplifying the scenario being modeled. A. montensis has been described as the primary reservoir of JABV. RNA-positive individuals of Akodon were encountered on R3B and R3A, and seropositive individuals of N. lasiurus and O. delator were trapped and identified on R3A. Each of these species is widely distributed in the central Southern Cone of South America (Carleton and Musser, 2005), and may be locally abundant in their preferred habitat. Habitat preferences differ somewhat among the three, which is critical to this field situation and to the model which we present herein. N. lasiurus and O. delator prefer grasslands, with Necromys preferring dry soil and Oxymycterus preferring wet or even saturated soils. A. montensis preferentially inhabits disturbed woodlands, also venturing into old-fields and grasslands which include forbs and brushy growth (Redford and Eisenberg, 1992, Goodin et al., manuscript). Our data from nine sampling sessions through 27 months support these descriptions of habitat preferences, and further indicate that the microgeographic separation among these three species is partial and temporally variable and often incomplete, with overlap (microsympatry) occurring sporadically between A. montensis and one or both of the spillover species. Fig. 2 illustrates this situation on site R3A, and also indicates the presence of seropositive individuals for each of these species in close proximity to the others.
Fig. 2.
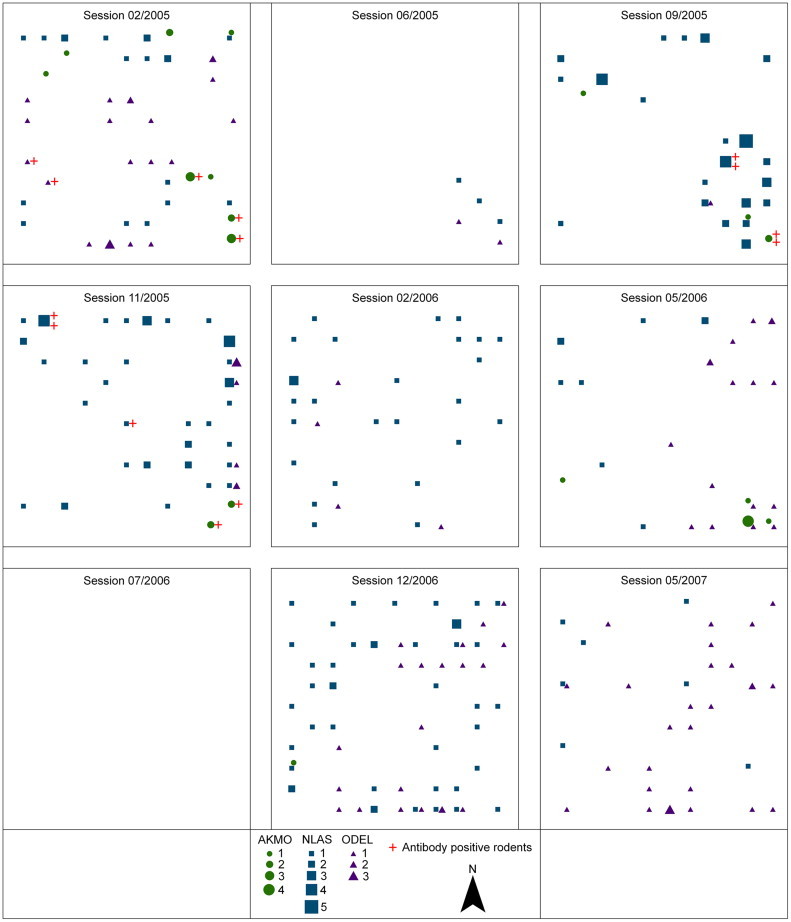
Data for nine trapping sessions in 2005–2007 for three species A. montensis (AKMO), N. lasiurus (NLAS) and O. delator (ODEL) at site R3A. Symbols indicate number of animals captured at each station during eight nights of trapping, and the number of these which were seropositive.
At site R3B, A. montensis was captured but neither N. lasiurus nor O. delator. Based on 2005–2007 data, 21 out of 84 A. montensis males (25%) tested for hantavirus showed positive titers for antibodies or RNA, including 12 (14.3%) that were only antibody-positive, eight (9.5%) that were antibody-positive and RNA-positive, and one (1.2%) that was only RNA-positive. Only four out of 68 A. montensis females (5.9%) tested for hantavirus showed positive titers for antibodies with no detectable viral RNA. In other studies of hantavirus ecology, male seroprevalence was higher than female seroprevalence (Bernshtein et al., 1999, Childs et al., 1994, Glass et al., 1998, Klein et al., 2001, McIntyre et al., 2005, Mills et al., 1997, Yahnke et al., 2001).
3. Model derivation
Based on the empirical data, we model only male rodents in the spread of hantavirus and use two infectious stages for the reservoir host, a highly infectious stage and a persistent stage, I and P. The highly infectious stage represents animals that are RNA-positive and may or may not have antibodies, and the persistent stage represents animals that are only antibody-positive. This latter persistent stage is less infectious than the highly infectious stage. Two infectious stages were assumed in models for Puumala hantavirus in bank voles (Sauvage et al., 2003, Sauvage et al., 2007, Wolf et al., 2006).
First, two basic models for the reservoir and the spillover species, each within their own habitat, are formulated. Then these two models are merged into a habitat-based model, where interspecies pathogen transmission may occur in a region of overlap of the two habitats (such as R3A).
3.1. Basic model
The disease stages for the reservoir species include susceptible, , exposed or latently infected, , highly infectious, , and persistently infectious, . The subscript refers to the reservoir species. The total population density is . The per capita birth rate and survival to the adult reproductive stage is . There is no vertical transmission. Disease-related deaths are not known to occur in the reservoir host (Mills et al., 1997). The natural death rate depends on population density, a strictly increasing function of the population density, and . The transmission coefficients for the two infectious stages are and , respectively. The models described below assume density-dependent transmission (pseudo-mass action incidence) as in other hantavirus models (Abramson and Kenkre, 2002; Abramson et al., 2003; Allen et al., 2006a, Allen et al., 2006b). The models and results can be easily generalized to frequency-dependent transmission which in some cases may provide a better fit to data (Begon et al., 1999). In stable environments, frequency-dependent transmission may be appropriate, but in the overlap region, where rodents occupy the region for only a short period of time, encounters are most likely density-dependent. The average length of the exposed and highly infectious periods are and , respectively. All parameters are assumed to be positive unless noted otherwise. The model for the reservoir population takes the following form:
| (3.1) |
The total population density satisfies the following differential equation:
| (3.2) |
From the assumptions on , it follows that there exists a unique positive constant , the carrying capacity, such that and . With frequency-dependent transmission the terms are replaced with .
The spillover species responds differently to hantavirus infection. Presumably, the infection is only short-term, an acute stage, A, and therefore, we assume no disease-related deaths occur. But this assumption can be modified. The model is an SEAR model, where animals pass through the stages of being susceptible, latent, infectious, and finally recovered. A subscript s is used to identify the spillover species and distinguish it from the reservoir species. The differential equations for hantaviral infection in the spillover species are similar to the reservoir species but transmission of hantavirus occurs only from the infectious stage and is the recovery rate. The model for the spillover species takes the following form:
| (3.3) |
We assume satisfies similar assumptions as , so that the total male population density for the spillover species satisfies a differential equation similar to Eq. (3.2). Likewise, there exists a unique positive constant , the carrying capacity of the spillover population, such that .
3.2. A habitat-based epizootic model
The reservoir and spillover species generally have preferred habitats as shown by the data. The spillover species is rarely found in the habitat where the reservoir species is dominant and vice versa (see Fig. 2). However, contact between these two species occurs in a boundary or overlap region adjacent to their habitats, where densities of the two species may be relatively low. Encounters between infectious and susceptible animals in this boundary region may result in interspecies transmission of hantavirus. Fig. 3 is a schematic of the three regions, the preferred habitats for the reservoir and the spillover species and the boundary region.
Fig. 3.
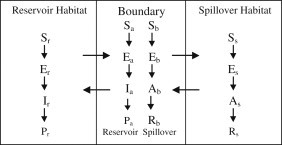
A schematic of the three regions representing preferred habitats for the reservoir and spillover species and the boundary or overlap region adjacent to the two habitats.
These habitats are connected via movement to and from the preferred habitat and the boundary region. Time spent in this boundary region is short for both species. The majority of the population is susceptible, especially in the case of the spillover species.
Suppose the per capita rate of movement into the boundary region is low, i.e., is small, and that the per capita rate of movement out of this boundary region is high, i.e., is large. The same movement rates are assumed for each species. Thus, for each of the differential equations (3.1) and (3.3) movement into and out of the boundary region is included. Subscripts a and b on the differential equations denote the reservoir and spillover species, respectively, in the boundary region.
3.2.1. ODE model
Based on the preceding assumptions, the differential equations for the reservoir species in its preferred habitat take the following form:
| (3.4) |
Similar differential equations apply to the spillover species, where terms for movement into and out of the preferred habitat are added to the differential equations (3.3).
Because rodents are in the boundary region for a short period of time, on the order of days, no births nor deaths occur in this region. We assume that the carrying capacity in the preferred habitat remains constant. That is, the stable preferred habitat density for the reservoir species is which can be thought of as a population source for the boundary region. The carrying capacity in the boundary region may increase or decrease relative to if increases or decreases, respectively. The differential equations for the reservoir species in the boundary region are
| (3.5) |
and for the spillover species they are
| (3.6) |
Rodents may change their disease status while in the boundary, e.g., or . An inherent assumption in the models is that the time spent in each of the disease states is exponentially distributed. For example, the probability that an initially exposed reservoir host transitions to the infectious stage while in the boundary region is
| (3.7) |
Other models based on more general probability distributions such as the gamma distribution provide alternative formulations (Feng et al., 2007; Lloyd, 2001a, Lloyd, 2001b). More data are required to determine the form of the distributions. In this investigation, we consider the simplest form, an exponential distribution. If and if the number of rodents exposed to the infection in the boundary region is relatively small, then transitions between disease stages in the boundary region may have little impact on the disease dynamics. In Section 4, an explicit expression for the basic reproduction number is derived when these transition rates are set to zero: and .
The total male population densities in the preferred habitat and in the boundary region are and for the reservoir species and and for the spillover species. Thus, the differential equations for the total male population densities are
| (3.8) |
Initial conditions are nonnegative and strictly positive in the preferred habitat; and .
3.2.2. CTMC model
The ODE model can be easily extended to a CTMC model which includes variability in the birth, death, transmission, and movement processes. In the CTMC model, the 16 random variables are integer-valued taking on values in the set . There are 38 different events, that include births, deaths, transmission, and movement. Let be a vector of 16 discrete random variables associated with the CTMC process:
Based on the 38 events, the infinitesimal transition probabilities can be defined, , where for sufficiently small (Allen, 2003, Karlin and Taylor, 1975). For example, the probability of a birth in the reservoir population is
where the time step is chosen so that the possibility of more than one transition or change in units of time is negligible. The 38 events and their corresponding transition probabilities are described in the Appendix.
Due to climatic variations within the year—dry, wet, and transitional (D, W, T) periods—there may be greater overlap of the habitats during certain periods of the year. One way of modeling this variability in the overlap region is to modify the rate of movement into or out of the boundary region for each species, depending on their habitat preferences during each of these periods. For example, A. montensis is seen in the overlap region more frequently in period T than in periods D or W, when densities of O. delator are high. In the model, we do not specifically include this seasonal variability but we do consider the effects of changes in and on the basic reproduction number , the threshold for disease outbreaks. More data are required to predict whether certain periods are more likely to result in spillover infection.
4. Model analysis
There are three types of equilibria for the ODE habitat-based model: an extinction equilibrium, where the population density is zero; a unique disease-free equilibrium (DFE), where all the infectious and recovered states are zero but the susceptible states are positive; and enzootic equilibria (EE), where some infectious states have positive values. It can be easily shown that the extinction equilibrium is unstable; the population persists. For example, it follows from Eqs. (3.8) that in the preferred habitats, the population densities approach a constant value, their respective carrying capacities,
The preferred habitats serve as a population source for the boundary region. The densities in the boundary region depend on the densities of these source populations and the movement rates into and out of this region. In particular, in the boundary region, the reservoir and spillover population densities are
respectively. As the ratio increases, so do the population densities in the boundary region, whereas the densities in the preferred habitats will approach their respective carrying capacities. Hence, the total reservoir population density (preferred boundary) is and the total spillover population density is . The equilibrium values for the unique DFE are , , , and ; all of the other equilibrium values are zero. We assume initial densities are less than or equal to their respective carrying capacities, , with and . Whether the DFE is stable depends on the basic reproduction number for the habitat-based model.
We calculate reproduction numbers for each of the preferred habitats and an approximation to the overall basic reproduction number for the habitat-based model (3.4)–(3.6), . If , then it is likely that the disease persists in the reservoir and spillover populations. The reproduction numbers can be calculated using the next generation matrix approach (van den Driessche and Watmough, 2002). For the general system (3.4)–(3.6), it is possible to show that the basic reproduction number is a positive root of a fourth degree polynomial but it is difficult to obtain a simple analytical expression for . The simplifying assumption
| (4.1) |
leads to an explicit expression for (shown below). This explicit expression is a close approximation to the overall basic reproduction number, if the time spent in the boundary is short relative to the time spent in each of the disease states. This expression is very useful in interpreting the contributions to disease outbreaks by the reservoir and the spillover species and in making comparisons to other reproduction numbers.
Assume condition (4.1) holds. First, the reproduction number for the reservoir species (assuming the spillover species is not present) is
Second, the reproduction number for the spillover species (assuming the reservoir species is not present) is
If there are no interspecies interactions so that , then . Each of the reproduction numbers simplifies to well-known reproduction numbers for SEIP or SEAR models (3.1) or (3.3), respectively. In this case, it is straightforward to calculate the enzootic equilibrium for the reservoir host,
| (4.2) |
whenever the reproduction number for the SEIP model (3.1) is
At equilibrium, the proportion of animals that are RNA- or antibody-positive, , is
| (4.3) |
To derive an expression for the basic reproduction number for the habitat-based model when (4.1) holds, we first define an expression which depends on interspecies or crossover transmission. That is, let the intraspecies transmission parameters be zero, and , and the interspecies transmission parameters, , , and , be nonzero. The reproduction number for interspecies pathogen transmission is defined as
Note that depends on the ratio directly and indirectly through the carrying capacities in the overlap region, and . The preceding definition can be used to define the basic reproduction number for the habitat-based model (3.4)–(3.6):
| (4.4) |
(Derivation of this formula is given in the Appendix.) It follows that ; interspecies pathogen transmission increases the basic reproduction number. A similar relationship was shown in a multi-species SI model of McCormack and Allen (2007).
The local stability of the DFE follows directly from the results of van den Driessche and Watmough (2002). Global stability of the DFE when and condition (4.1) holds can be verified by construction of a Liapunov function. The full system (3.4)–(3.6) consists of 16 differential equations which makes it difficult to find an explicit closed form solution for an enzootic equilibrium (EE). However, existence and uniqueness of a positive EE can be verified when and condition (4.1) holds. It is shown, in the Appendix, that the EE for the full system (3.4)–(3.6) is a fixed point of and . Then the existence of a unique positive EE follows by applying a theorem (Hethcote and Thieme, 1985) on existence and uniqueness of a positive fixed point. The following theorem summarizes the preceding results.
Theorem 4.1
A basic reproduction number exists for system (3.4)–(3.6) such that if , the DFE is locally asymptotically stable. If condition (4.1) holds for system (3.4)–(3.6), then
- (i)
has the form given in (4.4),
- (ii)
if , then the DFE is globally asymptotically stable, and
- (iii)
if , then the DFE is unstable and there exists a unique positive enzootic equilibrium.
5. Numerical examples
Selection of parameters values is based on estimates from the literature for hantavirus ( and , ) and on trapping and demographic data for the reservoir species A. montensis and for the spillover species N. lasiurus or O. delator (, , and equilibrium ratios). The reservoir species A. montensis and one spillover species (N. lasiurus) are widely distributed in many of our study sites sampled in Paraguay, whereas the other spillover species (O. delator) is less widespread and generally less abundant. We choose carrying capacities of and for the reservoir and spillover species in their respective habitats. Although the values for and are not known, the selected values are close to the estimates for minimal number known alive based on data from the trapping regions. The basic time unit in the model is one year. We assume total number of births per female per year that survive to the adult reproductive stage is six for the reservoir and the spillover species (several litters per year). Assuming an equal sex ratio, for the male population the number of males that survive to reproductive age is . Population growth is assumed to satisfy a logistic growth assumption so that for the reservoir and spillover species,
so that and . We assume an average duration of two weeks for the latent period and for the spillover infectious stage , and an average duration of three months for the highly infectious stage for the reservoir species (Bernshtein et al., 1999, Lee et al., 1981, Padula et al., 2004). Hence, , , and , e.g., weeks. In addition, , , , and . Animals enter the boundary region several times per year and stay only a short time. The average length of time in the boundary region is less than the average length of time for the latent period or acute infectious period, and , . In the numerical examples, we let and which means, on average, each animal may make eight visits per year to the boundary region, spending about one week in the boundary region. The probability there is a transition from an exposed to an infectious state while in the boundary region is 1/3 (see Eq. (3.7)). Even though this probability is not small, the basic reproduction number given by (4.4) is a good approximation to the overall basic reproduction number for our parameter values (shown below). The parameter values are reasonable but are chosen for illustrative purposes (a range of values, and , are considered later in this section). These parameter choices give population densities in the boundary region of and .
The transmission parameters cannot be estimated directly. Instead, we make some reasonable assumptions about their relationship to disease transmission in the infectious stages, and . The product is the number of infectious contacts that result in infection by a highly infectious reservoir animal per year (at equilibrium). Based on the summary data for proportion of animals RNA- or antibody-positive, 0.25 at site R3B, if we equate formula (4.3) to 0.25 and let and , this leads to (when ). Thus, the value of is chosen so that (which is close to ) is between one and two. We assume that the highly infectious stage of the reservoir host (RNA-positive and/or antibody-positive) is three times as infectious as the persistently infectious stage (only antibody-positive), i.e., (Bernshtein et al., 1999, Lee et al., 1981, Padula et al., 2004). In addition, we assume the transmissibility of the pathogen in the acute infectious stage of the spillover host is the same as for the persistent stage in the reservoir host, . This leads to . In the boundary region, intraspecific transmissibility remains the same as in the preferred habitats, but interspecies transmissibility is doubled due to aggressive encounters. In particular,
| (5.1) |
| (5.2) |
The basic parameter values are given in Table 1 .
Table 1.
Basic parameter values for the ODE and the CTMC models for the reservoir and the spillover species.
| Reservoir parameter | Value | Spillover parameter | Value |
|---|---|---|---|
| 100 | 50 | ||
| 3 | 3 | ||
| 26 | 26 | ||
| 4 | 26 | ||
| 26 | 26 | ||
| 4 | 26 | ||
| 0.075 | 0.025 | ||
| 0.025 | 0.025 | ||
| 0.075 | 0.15 | ||
| 0.025 | 0.05 | ||
| 0.05 |
If (4.1) holds and the remaining parameter values are as in Table 1 with and , we obtain , so that . The approximate reproduction numbers based on the analysis in Section 4 are
The overall basic reproduction number for the parameters in Table 1 is which is close to the approximation 1.42. The disease persists in the habitat-based model. For the parameter values in Table 1, one sample path of the CTMC model and the solution to the ODE model are graphed for the two infectious stages of the reservoir species (see Fig. 4 ).
Fig. 4.
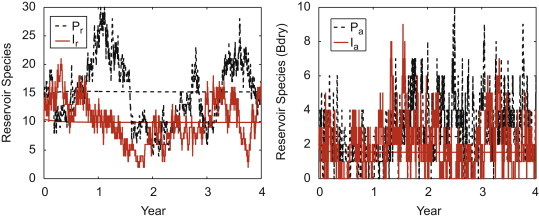
Solution to the ODE model (straight lines) and one sample path of the CTMC model (highly variable curves) for the reservoir species in its preferred habitat ( and ) and in the overlap region ( and ). The parameter values are given in Table 1 with and Initial values are at the equilibrium values given in (4.2), rounded to the nearest integer.
Although the pathogen persists, the infection in the spillover population is very low (straight line is the ODE equilibrium value); only sporadic infection occurs in the sample path for the spillover species in the preferred habitat and in the boundary region (Fig. 5 ).
Fig. 5.
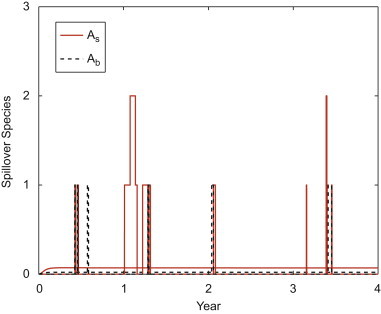
Solution to the ODE model and one sample path of the CTMC model for the spillover species in its preferred habitat () and in the boundary region (). In the ODE model, and , whereas, in the CTMC model, the values for the random variables and lie in the set . Initial values and parameter values are the same as those in Fig. 4.
For the ODE model with parameter values given in Table 1, , and , there is a unique enzootic equilibrium which is locally asymptotically stable:
With no interspecies transmission and no overlap region () the equilibrium values for the reservoir host, based on the formulas given in (4.2), are . These latter equilibrium values show that the percentage of highly infectious and persistently infectious rodents are in close agreement with the summary data for A. montensis at site R3B, i.e., 25% are infected.
The CTMC simulation with interspecies transmission illustrates the sporadic infection in the spillover population (as in site R3A) and provides information about the variability in number of cases. A quasistationary probability distribution is reached in the CTMC model (conditional on nonextinction). Approximations (estimated from 10,000 sample paths) to the quasistationary probability distributions for the two infectious stages in the reservoir species are graphed in Fig. 6 . The mean values for and are and . In the absence of interspecies transmission and , the mean values are and .
Fig. 6.
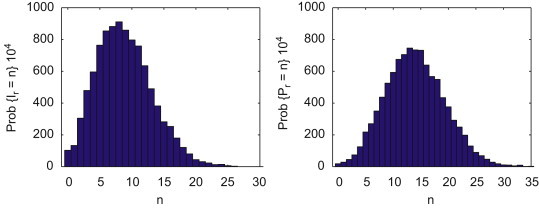
Probability histograms for the reservoir species in the preferred habitat for the two infectious stages, and (10,000 sample paths of the CTMC model). Parameter values are given in Table 1. The mean and standard deviation of the distributions for are and and for they are and .
Encounters that lead to interspecies pathogen transmission can be measured by the magnitude of . The greater the habitat overlap, the greater the number of interspecies and intraspecies encounters which in turn increase the likelihood of pathogen outbreaks and disease persistence. Changes that affect the overlap region will have the greatest impact on the parameters and rather than the parameters affecting transmission, births, or deaths. As more animals enter and stay in the boundary region, that is, if increases and decreases, then increases and consequently, increases. This increase can be seen in Fig. 7 . The value of , computed from formula (4.4), is compared to the exact value of based on the parameter values in Table 1. Both reproduction numbers show similar increases with (average number of visits/year) and (average number of days in the boundary region). The difference between these two reproduction numbers is also computed (Fig. 7(c)); the largest relative difference is 0.12, when and .
Fig. 7.
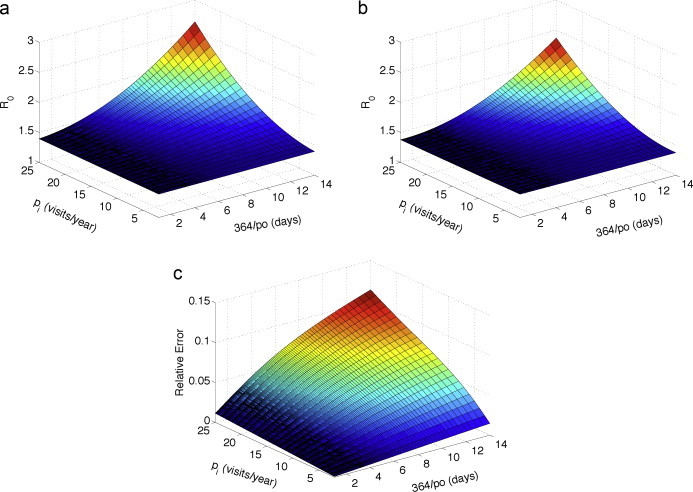
The basic reproduction number for the habitat-based model (3.4)–(3.6) as a function of the average number of visits to boundary region and average length of time (days) spent in the boundary region , (a) based on formula (4.4), [Approx], and (b) based on the parameter values in Table 1, [Exact]. The graph in (c) is the relative difference or relative error in these two values: ([Approx]−[Exact])/[Exact].
6. Discussion
Biologically-motivated models for pathogen spread between two species were formulated, an ODE model (3.4)–(3.6) and a CTMC model. The models are based on the fact that spatial overlap of habitats leads to greater numbers of interspecies encounters. From the ODE model, an explicit expression for the basic reproduction number was calculated based on assumption (4.1), as well as reproduction numbers for the preferred habitats, and , and for crossover or interspecies transmission, . In this case, we showed global stability of the disease-free equilibrium when and existence of an enzootic equilibrium when . Greater number of interactions among species allow the pathogen to be transmitted more frequently from an infectious host to a susceptible host. This, in turn, increases so that it exceeds the reproduction number in the preferred habitat, (Fig. 7), which ultimately results in greater likelihood of outbreaks and disease persistence. As illustrated in Fig. 2, the overlap region is spatially- and temporally-dependent. We did not consider temporal variability of this overlap region which may depend on seasonal variations. But we did include demographic variability due to births, deaths, transmission, and movement in the CTMC model. Seasonal variations, in general, will cause additional variability in the solution behavior (e.g., Allen et al., 2006a). As more data are collected, the effects of seasonal and climatic variations on the reservoir and spillover species will be studied. In addition, controlled studies are needed to obtain data on the duration and shape of the rodents’ disease stage distributions.
Interspecies pathogen transmission, where a known virus “jumps” into a new host, is one of the primary reasons for the large increase in emerging diseases in wildlife in recent years (Daszak et al., 2000, Parrish et al., 2008, Richomme et al., 2006). Our mathematical models illustrate the first step in this emergence and the role that the spillover species may play in emerging diseases. Our models were developed for spread of hantavirus in rodents but can be modified and applied to other species, where spatial spread results in spillover infection.
Acknowledgments
This research was supported by a Grant from the Fogarty International Center #R01TW006986-02 under the NIH NSF Ecology of Infectious Diseases initiative. We thank R.K. McCormack for preliminary discussions on this work, the Fundación Moises Bertoni for facilitating access to the field sites, and the Vendramini family for allowing us to work in Estancia Rama III. The Secretaría de Ambiente provided necessary permits for working with wildlife. In addition, we thank the referees for their helpful suggestions.
Appendix A.
A.1. Basic reproduction number
The basic reproduction number for system (3.4)–(3.6) is obtained via the next generation matrix approach (van den Driessche and Watmough, 2002). Reorder the vector of 16 state variables as follows:
Then , where is the changes due to new infections and is the other transitions. The Jacobian matrix of and , evaluated at the DFE, is
where F and V are matrices, corresponding to the Jacobian matrices for the first 10 variables in . The symbol represents a zero matrix. Matrix , a matrix, has eigenvalues with negative real part. The basic reproduction number, , is the spectral radius of
where matrix is a nonnegative matrix. Thus,
the largest positive root of the characteristic polynomial of matrix , a fourth degree polynomial. Under condition (4.1),
The elements of matrix are
| (A.1) |
where and .
The characteristic polynomial of matrix is
Note that , , and . It follows that the largest positive root of the quadratic equation is , given by Eq. (4.4). Local stability of the DFE if and instability if follows directly from van den Driessche and Watmough (2002). This verifies the first part of Theorem 4.1.
A.2. Proof of Theorem 4.1(ii)–(iii)
Before we give the proof of Theorem 4.1(ii)–(iii), some preliminary facts are stated in the two cases, and . When , to verify global stability, we use an expression equivalent to ,
It can be easily shown that iff (. Let
It is clear that is forward invariant. As , the total population density of each patch approaches their respective carrying capacities and . We assume solutions are initially in . Then the -limit set of solutions to (3.4)–(3.6) is a subset of
which is contained in . We will show for that solutions to the limiting system (beginning in ) approach the DFE. Then global asymptotic stability of the DFE will follow from the theory of asymptotically autonomous systems.
The EE can be can be written as a fixed point problem. It can be easily verified that at an EE, the values of the variables and can be expressed in terms of and that the variables and can be expressed in terms of . Also, due to these relationships, , , and can be expressed in term of and . If the solutions to and are positive, then all of these other variables are also positive. Using these relationships and some algebraic manipulation lead to the following expressions for and :
| (A.2) |
and
| (A.3) |
where
and are defined in (A.1).
Proof of Theorem 4.1(ii)–(iii)
Assume . Consider the limiting system of (3.4)–(3.6), where the total population densities are at their respective carrying capacities. Define the Liapunov function for the limiting system as follows:
where
The coefficients , , are positive if and . But implies and . Therefore, if , is nonnegative and equals zero only if
equals the zero vector. Differentiating with respect to along solution trajectories for the limiting system leads to
where . A computer algebra system can be used to verify that for , . It follows that for (or equivalently . The Liapunov–Lasalle extension theorem (LaSalle, 1976) implies solutions to the limiting system in approach the largest positively invariant subset of the set where . Hence, solutions to the limiting system converge to the DFE.
As noted previously, the -limit set of solutions to (3.4)–(3.6) is contained in . The preceding argument shows that all solutions beginning in converge to the DFE. It follows from the theory of asymptotically autonomous systems (Thieme, 1992, Theorem 4.1 or Castillo-Chavez and Thieme, 1995, p. 39) that if , then the -limit set of (3.4)–(3.6) is the DFE.
Next, assume . To prove existence and uniqueness of a positive EE, denote the vector-valued function , where and are defined in (A.2) and (A.3), respectively. The function is positive, continuous, and bounded (each of the component functions is bounded by and ). In addition, . The derivative of evaluated at the origin is
where are defined in (A.1). It is clear that is irreducible with spectral radius equal to . With the aid of a computer algebra system, it can be shown that F is a monotone increasing function in each of its variables. These properties of F are sufficient to show the existence of a positive EE (Hethcote and Thieme, 1985, p. 209). To ensure a unique EE exists, a sufficient condition is that the function F is strictly sublinear: A vector-valued function from into itself is called strictly sublinear if for fixed in and fixed in , there exists an such that , for .
We define positive constants and so that and for a fixed and fixed in , then choose . The positive constants
and
where
and
Hence, the function F defined in (A.2) and (A.3) is strictly sublinear. All of the conditions of Theorem 2.1 in Hethcote and Thieme (1985, p. 209) are satisfied. The function F has a unique fixed point which implies that a unique positive EE exists for the full system (3.4)–(3.6). □
A.3. CTMC model
A CTMC model can be derived based on the ODE model (3.4)–(3.6), where variability is included in the birth, death, transmission, and movement processes (Allen, 2003). There are 38 events in the CTMC model, where a change occurs. Let be a vector of 16 discrete random variables
where , etc., and . The 38 events are divided into events for birth, death, and transmission (22 events) and for movement (16 events). If there is a change in the th random variable at time t we denote it by , ; the variable t is omitted for simplicity. There is no change in the variable if . The 22 transition probabilities based on birth, death, and transmission are as follows (some ):
where and . The 16 transition probabilities associated with movement between the preferred habitat and the boundary region are
The probability of no change is one minus the sum of the 38 probabilities defined above and the probability of any other change is .
Differential equations for the joint probability function of X follow from these infinitesimal transition probabilities. The joint probability function is a solution of the forward Kolmogorov differential equations, an infinite system of differential equations. For multivariate processes, the form of these differential equations is often too complicated for analytical purposes. This is the case for our CTMC model with 16 random variables. The general form of the forward Kolmogorov differential equations depends on the set , where . Let denote the rate at which events occur for , where the values of are defined above. For example, for the 38th event, , and for so that . There are 38 events for which . The general form for the forward Kolmogorov differential equations is
where , (see e.g., Bailey, 1990).
References
- Abramson, G., Kenkre, V.M., 2002. Spatiotemporal patterns in hantavirus infection. Phys. Rev. E 66, 0011912-1-5. [DOI] [PubMed]
- Abramson G., Kenkre V.M., Yates T.L., Parmenter R.R. Traveling waves of infection in the hantavirus epidemics. Bull. Math. Biol. 2003;65:519–534. doi: 10.1016/S0092-8240(03)00013-2. [DOI] [PubMed] [Google Scholar]
- Allen L.J.S. Prentice-Hall; Upper Saddle River, NJ: 2003. An Introduction to Stochastic Processes with Applications to Biology. [Google Scholar]
- Allen L.J.S., Langlais M., Phillips C.J. The dynamics of two viral infections in a single host population with applications to hantavirus. Math. Biosci. 2003;186:191–217. doi: 10.1016/j.mbs.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Allen, L.J.S., Allen, E.J., Jonsson, C.B., 2006a. The impact of environmental variation on hantavirus infection in rodents. In: Gumel, A.B., Castillo-Chavez, C., Mickens, R.E., Clemence, D.P. (Eds.), Proceedings of the Joint Summer Research Conference on Modeling the Dynamics of Human Diseases: Emerging Paradigms and Challenges, Contemporary Mathematics Series, vol. 410, AMS, Providence, RI, pp. 1–15.
- Allen L.J.S., McCormack R.K., Jonsson C.B. Mathematical models for hantavirus infection in rodents. Bull. Math. Biol. 2006;68:511–524. doi: 10.1007/s11538-005-9034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., May R.M. Oxford University Press; Oxford: 1991. Infectious Diseases of Humans, Dynamics and Control. [Google Scholar]
- Bailey N.T.J. Wiley; New York: 1990. The Elements of Stochastic Processes with Applications to the Natural Sciences. [Google Scholar]
- Begon M., Hazel S.M., Baxby D., Bown K., Cavanagh R., Chantrey J., Jones T., Bennett M. Transmission dynamics of a zoonotic pathogen within and between wildlife host species. Proc. R. Soc. London Ser. B. 1999;266:1939–1945. doi: 10.1098/rspb.1999.0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernshtein A.D., Apekina N.S., Mikhailova T.V., Myasnikov Y.A., Khlyap L.A., Korotkov Y.S., Gavrilovskaya I.N. Dynamics of Puumala hantavirus infection in naturally infected bank voles (Clethrinomys glareolus) Arch. Virol. 1999;144:2415–2428. doi: 10.1007/s007050050654. [DOI] [PubMed] [Google Scholar]
- Carleton, M.D., Musser, G.G., 2005. Order rodentia. In: Wilson, D.E., Reeder, D.M. (Eds.). Mammal Species of the World, third ed., vol. 2. Johns Hopkins University Press, Baltimore, MD, pp. 745–752.
- Castillo-Chavez, C., Thieme, H.R., 1995. Asymptotically autonomous epidemic models. In: Arino, O., Axelrod, D., Kimmel, M., Langlais, M. (Eds.), Mathematical Population Dynamics: Analysis of Heterogeneity, Theory of Epidemics, vol. 1, pp. 33–49.
- CDC MMWR, 2002. Hantavirus pulmonary syndrome—United States: updated recommendations for risk reduction, vol. 51(RR09), July 26, 2002, pp. 1–12. [PubMed]
- Childs J.E., Ksiazek T.G., Spiropoulou C.F., Krebs J.W., Morzunov S., Maupin G.O., Gage K.L., Rollin P.E., Sarisky J., Enscore R.E., Frey J.K., Peters C.J., Nichol S.T. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the Southwestern United States. J. Infect. Dis. 1994;169:1271–1280. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- Chu Y.-K., Milligan B., Owen R.D., Goodin D.G., Jonsson C.B. Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay. Am. J. Trop. Med. Hyg. 2006;75:1127–1134. [PMC free article] [PubMed] [Google Scholar]
- Chua K.B. Nipah virus outbreak in Malaysia. J. Clin. Virol. 2003;26:265–275. doi: 10.1016/s1386-6532(02)00268-8. [DOI] [PubMed] [Google Scholar]
- Daszak P., Cunningham A.A., Hyatt A.D. Emerging infectious disease of wildlife: threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- Delfraro A., Tomé L., D’Elía G., Clara M., Achával F., Russi J.C., Rodonz J.R. Juquitiba-like hantavirus from two nonrelated rodent species, Uruguay. Emerg. Infect. Dis. 2008;14:1447–1451. doi: 10.3201/eid1409.080455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Xu D., Zhao H. Epidemiological models for non-exponentially distributed disease states and applications to disease control. Bull. Math. Biol. 2007;69:1511–1536. doi: 10.1007/s11538-006-9174-9. [DOI] [PubMed] [Google Scholar]
- Fernández Soto, A., Mata Olmo, R., 2001. Deforestación y dinámica vegetal en un área de frontera agrícola del la Región Oriental del Paraguay. Revista Geonotas, Universidade Estadual de Maringá, vol. 5, p. 1.
- Fiorello C.V., Noss A.J., Deem S.L. Demography, ecology, and pathogen exposure of domestic dogs in the Izozog of Bolivia. Conserv. Biol. 2006;20:762–771. doi: 10.1111/j.1523-1739.2006.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass G.E., Livingston W., Mills J.N., Hlady W.G., Fine J.B., Biggler W., Coke T., Frazier D., Atherley S., Rollin P.E., Ksiazek T.G., Peters C.J., Childs J.E. Black Creek Canal Virus infection in Sigmodon hispidus in southern Florida. Am. J. Trop. Med. Hyg. 1998;59:699–703. doi: 10.4269/ajtmh.1998.59.699. [DOI] [PubMed] [Google Scholar]
- Goodin, D.G., Paige, R., Owen, R.D., Ghimire, K., Koch, D.E., Chu, Y.-K., Jonsson, C.B., Microhabitat characteristics of Akodon montensis, a reservoir for hantavirus, and hantaviral seroprevalence in an Atlantic forest site in eastern Paraguay. J. Vector Ecol., 34, 104–113 [DOI] [PubMed]
- Hethcote H.W. The mathematics of infectious diseases. SIAM Rev. 2000;42:599–653. [Google Scholar]
- Hethcote H.W., Thieme H.R. Stability of the endemic equilibrium in epidemic models with subpopulations. Math. Biosci. 1985;75:205–227. [Google Scholar]
- Holmes K. SARS-associated coronavirus. New Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Karlin S., Taylor H. second ed. Academic Press; NY: 1975. A First Course in Stochastic Processes. [Google Scholar]
- Klein S.L., Bird B.H., Glass G.E. Sex differences in immune responses and viral shedding following Seoul virus infection in Norway rats. Am. J. Trop. Med. Hyg. 2001;65:57–63. doi: 10.4269/ajtmh.2001.65.57. [DOI] [PubMed] [Google Scholar]
- Klingstrom J., Heyman P., Escutenaire S., Sjölander K.B., De Jaegere F., Henttonen H., Lundkvist A. Rodent host specificity of European hantaviruses: evidence of Puumala virus interspecific spillover. J. Med. Virol. 2002;68:581–588. doi: 10.1002/jmv.10232. [DOI] [PubMed] [Google Scholar]
- LaSalle J.P. SIAM; Philadelphia: 1976. The Stability of Dynamical Systems. [Google Scholar]
- Lee H.W., Lee P.W., Baek L.J., Song C.K., Seong I.W. Intraspecific transmission of Hantaan virus, etiological agent of Korean hemorrhagic fever, in the rodent Apodemus agrarius. Am. J. Trop. Med. Hyg. 1981;30:1106–1112. doi: 10.4269/ajtmh.1981.30.1106. [DOI] [PubMed] [Google Scholar]
- Lloyd A. Realistic distributions of infectious periods in epidemic models. Theor. Popul. Biol. 2001;60:59–71. doi: 10.1006/tpbi.2001.1525. [DOI] [PubMed] [Google Scholar]
- Lloyd A. Destabilization of epidemic models with the inclusion of realistic distributions of infectious periods. Proc. R. Soc. London Ser. B. 2001;268:985–993. doi: 10.1098/rspb.2001.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack R.K., Allen L.J.S. Disease emergence in multi-host epidemic models. Math. Med. Biol. 2007;24:17–34. doi: 10.1093/imammb/dql021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre N.E., Chu Y.-K., Owen R.D., Abuzeineh A., De La Sancha N., Dick C.W., Holsomback T., Nisbet R.A., Jonsson C. A longitudinal study of Bayou virus, hosts, and habitat. Am. J. Trop. Med. Hyg. 2005;73:1043–1049. [PubMed] [Google Scholar]
- Mills J.N., Ksiazek T.G., Ellis B.A., Rollin P.E., Nichol S.T., Yates T.L., Gannon W.L., Levy C.E., Engelthaler D.M., Davis T., Tanda D.T., Frampton J.W., Nichols C.R., Peters C.J., Childs J.E. Patterns of association with mammals in the major biotic communities of the southwestern United States. Am. J. Trop. Med. Hyg. 1997;56:273–284. doi: 10.4269/ajtmh.1997.56.273. [DOI] [PubMed] [Google Scholar]
- Mills J.N., Yates T.L., Ksiazek T.G., Peters C.J., Childs J.E. Long-term studies of hantavirus reservoir populations in the southwestern United States: rationale, potential and methods. Emerg. Infect. Dis. 1999;5:95–101. doi: 10.3201/eid0501.990111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis S.A., Loza-Rubio E. The molecular epidemiology of rabies associated with chiropteran hosts in Mexico. Virus Res. 2006;117:215–226. doi: 10.1016/j.virusres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Nel L., Jacobs J., Jaftha J., Meredith C. Natural spillover of a distinctly Canidae-associated biotype of rabies into an expanded wildlife host in southern Africa. Virus Genes. 1997;15:79–82. doi: 10.1023/a:1007979502754. [DOI] [PubMed] [Google Scholar]
- Padula P., Fogueroa R., Navarrette M., Pizarro E., Cadiz R., Bellomo C., Jofre C., Zaror L., Rodriguez E., Murúa R. Transmission study of Andes hantavirus infection in wild sigmodontine rodents. J. Virol. 2004;78:11972–11979. doi: 10.1128/JVI.78.21.11972-11979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma, R.E., Polop, J.J., Owen, R.D., 2009. Hantavirus-host ecology in the southern cone of South America: Argentina, Chile, Paraguay and Uruguay. Vector Borne Zoonotic Dis., in press. [DOI] [PubMed]
- Pardiñas U.F., D’Elía G. The genus Akodon (Muroidea: Sigmodontinae) in Misiones, Argentina. Mamm. Biol. 2003;68:129–143. [Google Scholar]
- Parrish C.R., Holmes E.C., Morens D.M., Park E.-C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford K.H., Eisenberg J.F. University of Chicago Press; Chicago: 1992. Mammals of the Neotropics: The Southern Cone. [Google Scholar]
- Richomme C., Gauthier D., Fromont E. Contact rates and exposure to inter-species disease transmission in mountain ungulates. Epidemiol. Infect. 2006;134:21–30. doi: 10.1017/S0950268805004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage F., Langlais M., Pontier D. Predicting the emergence of human hantavirus disease using a combination of viral dynamics and rodent demographic patterns. Epidemiol. Infect. 2007;135:46–56. doi: 10.1017/S0950268806006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage F., Langlais M., Yoccoz N.G., Pontier D. Modelling hantavirus in fluctuating populations of bank voles: the role of indirect transmission on virus persistence. J. Anim. Ecol. 2003;72:1–13. [Google Scholar]
- Thieme H.R. Convergence results and a Poincaré–Bendixson trichotomy for asymptotically autonomous differential equations. J. Math. Biol. 1992;30:755–763. [Google Scholar]
- Torrez-Martinez N., Bharadwaj M., Goade D., Delury J., Moran P., Hicks B., Nix B., Davis J.L., Hjelle B. Bayou virus-associated hantavirus pulmonary syndrome in Eastern Texas: identification of the rice rat, Oryzomys palustris as reservoir host. Emerg. Infect. Dis. 1998;4:105–111. doi: 10.3201/eid0401.980115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Driessche P., Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 2002;180:29–48. doi: 10.1016/s0025-5564(02)00108-6. [DOI] [PubMed] [Google Scholar]
- Weidmann M., Schmidt P., Vackova M., Krivanec K., Munclinger P., Hufert F.T. Identification of genetic evidence for dobrava virus spillover in rodents by nested reverse transcription (RT)-PCR and TaqMan RT-PCR. J. Clin. Microbiol. 2005;43:808–812. doi: 10.1128/JCM.43.2.808-812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, C.L., 2008. Discrete-time and continuous-time models with applications to the spread of hantavirus in wild rodents and human populations. Ph.D. Dissertation. Texas Tech University, Lubbock, Texas, USA.
- Wesley, C.L., Allen, L.J.S., Jonsson, C.B., Chu, Y.-K., Owen, R.D., 2009. A discrete-time rodent-hantavirus model structured by infection and developmental stages. In: International Conference on Difference Equations and Applications, July, 2006, Kyoto, Japan. Advanced Studies in Pure Mathematics, vol. 53, pp. 1–12.
- Wilson, D.E., Cole, F.R., Nichols, J.D., Rudran, R., Foster, M.S. (Eds.), 1996. Measuring and Monitoring Biological Diversity. Standard Methods for Mammals. Smithsonian Institution Press, Washington.
- Wolf C., Langlais M., Sauvage F., Pontier D. A multi-patch epidemic model with periodic demography, direct and indirect transmission and variable maturation rate. Math. Popul. Stud. 2006;13:153–177. [Google Scholar]
- Yahnke C.J., Meserve P.L., Ksiazek T.G., Mills J.N. Patterns of infection with Laguna Negra virus in wild populations of Calomys laucha in the central Paraguayan chaco. Am. J. Trop. Med. Hyg. 2001;65:768–776. doi: 10.4269/ajtmh.2001.65.768. [DOI] [PubMed] [Google Scholar]


