Abstract
A novel strategy for creating naturally-derived glycan microarrays has been developed. Glycosylamines are prepared from free reducing glycans and stabilized by reaction with acryloyl chloride to generate a glycosylamide in which the reducing monosaccharide has a closed ring structure. Ozonolysis of the protected glycan yields an active aldehyde, to which a bifunctional fluorescent linker is coupled by reductive amination. The fluorescent derivatives are easily coupled through a residual primary alkylamine to generate glycan microarrays. This strategy preserves structural features of glycans required for antibody recognition, and allows development of natural arrays of fluorescent glycans in which the cyclic pyranose structure of the reducing-end sugar residue is retained.
Keywords: Fluorescent labeling, Functional glycomics, Glycan array, Glycosylamine, Immobilization
Introduction
Functional glycomics has attracted great interest due to discoveries about the importance of complex glycoconjugates in biological processes, generally termed glycobiology (1). Protein-carbohydrate interactions are widely involved in many biological pathways, such as cell-cell adhesion (2-4), protein folding (5-8), disease pathogenesis (9), and others. Glycan microarrays, in which glycans are immobilized on activated glass surfaces and interrogated with proteins or pathogens, has been shown to be a successful tool for functional glycomics studies (10-12).
Solid-phase assays that involve either covalent or non-covalent glycan immobilization to various surfaces have been in use for decades (13, 14). As an early example, glycolipids have been separated on thin layer chromatography (TLC) and directly overlaid with proteins and antibodies (13, 14). A strategy was also developed to derivatize glycans to neoglycolipids (15, 16), which can be separated by TLC or immobilized directly onto nitrocellulose membranes for protein interaction assays. Biotin-streptavidin binding has also been utilized to prepare glycan microarrays (17), in which glycans are biotinylated and immobilized onto streptavidin-coated solid surfaces, either ELISA-type microtiter plates or glass chips.
Glycan microarray involving covalent immobilization has been developed based on derivatization of glycans with suitable functional groups, which are reactive with correspondingly activated solid surfaces. Thiol-maleimide (18, 19), azide-alkyne (20), and amino-NHS (21) or amino-epoxy (22, 23) reaction systems have all proved successful for glycan microarray purposes. The printed glycan array of the Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org) is comprised of >400 synthetic glycans coupled covalently through amino-NHS chemistry on a glass slide. This public glycan microarray has proved to be very successful for screening the binding specificity of glycan binding proteins (GBPs). It is anticipated that there are many thousands of different glycans, but expansion of the glycan library, however, is limited by the difficulty in synthesis of the complex naturally occurring glycan structures.
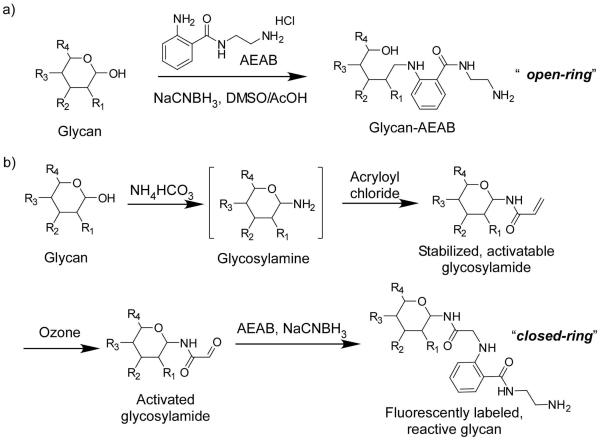
Natural glycan array development is a strategy in which glycans derived by enzymatic or chemical cleavage from natural sources, such as glycoproteins and glycolipids, are derivatized with a fluorescent linker, separated by multidimensional chromatography to obtain “tagged glycan libraries” or TGLs, and the purified tagged glycans can be printed as glycan microarrays. The TGLs, which are also more relevant to biological questions due to their natural origin, are not limited by complex syntheses and can be expanded quickly. We have successfully developed a novel bifunctional reagent, N-aminoethyl 2-aminobenzamide (AEAB), for preparing fluorescently labeled glycans by reductive amination for glycan microarray (24). As shown in Figure 1a this procedure results in glycan-AEAB derivatives that have a reduced or open-ring reducing end. Although most protein-carbohydrate interactions occur at the non-reducing end of glycans in glycoconjugates, this open-ring reducing end may in rare cases be a site of protein interaction. The current glycan microarray that is available through the CFG is populated with synthetic and semi-synthetic glycans having closed-ring glycans coupled to microscope slides. Bohorov et. al. (25) developed a method for derivatization of glycans using a modified hydroxylamine that retains a closed-ring form at the reducing end. However, the lack of spectroscopic properties in the linker limits its application in natural glycan array development, where microscale derivatization, characterization, and purification are essential due to the limited amounts of glycans available from natural sources. Here we report a microscale procedure, shown in Figure 1b, to fluorescently derivatize free glycans to glycosylamides, which retain a closed-ring reducing end.
Figure 1.
Design of bifunctional fluorescent derivatization of free reducing glycans via a) the common reductive amination approach and b) a novel approach that retains the full ring structure mimicking natural glycoconjugate linkages.
Results and Discussion
Fluorescent derivatization of free reducing sugars
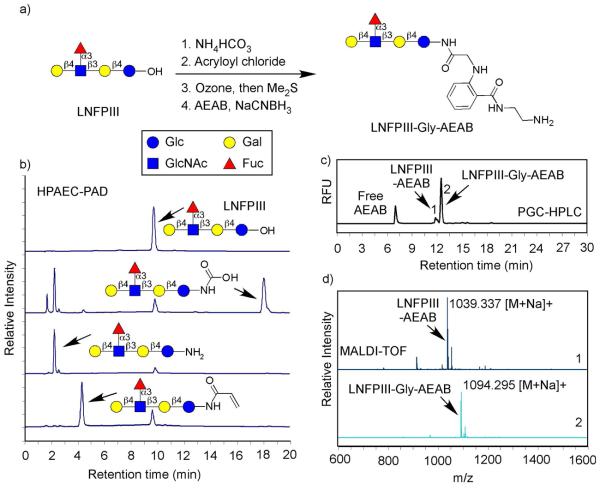
Figure 2a shows the derivatization procedure of a free reducing glycan (LNFPIII). We adopted the widely-used synthesis of a glycosylamine as the first step, where the reducing end selectively reacts with various acylation reagents. Glycans were mixed with water and excess ammonium bicarbonate and heated at 55°C for 1.5 h. This resulted in the carbamate of the glycosylamine, as shown by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis (Figure 2b). The resulting mixture was applied on either nonporous or porous carbon-based solid phase extraction (SPE) cartridges, i.e. carbograph or hypercarb cartridges. The cartridge was washed with dilute ammonium bicarbonate solution (10 mM) and eluted by 50% acetonitrile containing 10 mM ammonium bicarbonate. In this process, most of the glycan is transformed to glycosylamine with small amount of free reducing glycan (Figure 2b). Other researchers employed a low concentration ammonium bicarbonate solution during the size-exclusion chromatography of glycosylamines, presumably to prevent deamination of the unstable glycosylamine (26). This approach is compatible with our following chemical derivatizations and suitable for a small number of samples or small glycans (mono- and di-saccharides), which are only weakly retained by carbon SPE cartridge. Generally, we used the carbon SPE cartridge for desalting, which provides more consistency without the need of elution calibration or monitoring by chromatography. This procedure is also more practical for microgram level samples. The eluted solution was briefly evaporated in a Centra-vap to remove the organic solvent and lyophilized. To the lyophilized sample, cold saturated sodium bicarbonate solution and acryloyl chloride were added and the mixture was stirred vigorously for 10 min followed by incubation at room temperature for 1 h. The mixture was desalted by a carbon SPE cartridge and eluted by 50% acetonitrile with 0.1% trifluoroacetic acid (TFA). HPAEC-PAD analysis showed complete acylation (Figure 2b, bottom).
Figure 2.
The fluorescent derivatization of the free glycan LNFPIII: a) The conversion of LNFPIII (Galβ1,4(Fucα1,3)GlcNAcβ1,3Galβ1,4Glc) to LNFPIII-Gly-AEAB according to the reaction scheme in Figure 1b; b) HPAEC-PAD analysis of starting material, intermediates, and final product of the reactions (from top to bottom): free reducing LNFPIII, the glycosylamine carbamate after ammonium bicarbonate treatment, the glycosylamine after desalting with carbograph, and the glycosylamide after reaction with acryloyl chloride; c) The HPLC profile on PGC of the final product mixture using fluorescence detection; d) The MALDI-TOF analysis of peaks 1 and 2 in c), which were collected from preparative HPLC separation.
After evaporation, the glycosyl acrylamide was dissolved in methanol and treated with freshly generated ozone at −78°C and the reaction was terminated by addition of methyl sulfide. The solution was then evaporated to dryness and reacted with AEAB by reductive amination using the procedure previously described (24). The AEAB conjugated product, which is a fluorescent glycosylamide-glycyl-AEAB derivative, was applied to a porous graphitized carbon (PGC) column and the resulting HPLC profile is shown in Figure 2c. The peaks were collected and confirmed to be the glycan-AEAB conjugate (GAEAB, minor peak) generated from direct reductive amination and the glycan-gly-AEAB (GGAEAB, major peak) by MALDI-TOF analysis (Figure 2d). The fluorescently labeled glycan with a ring-closed structure, LNFPIII-gly-AEAB, was obtained in >80% relative yield (HPLC) (Figure 2c).
Parallel microscale derivatization of free reducing glycans
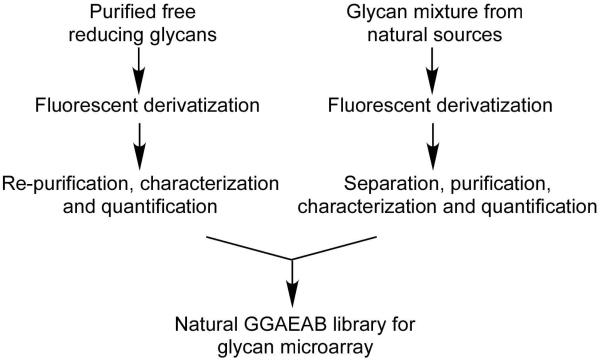
The utility of a glycan microarray is largely dependent on the size and versatility of the corresponding glycan library. Figure 3 shows the strategy of creating glycan microarray from both purified free reducing glycans, which may be commercially available, and glycan pools from natural sources. To demonstrate this principle, we carried out a parallel microscale derivatization of 26 commercially available free reducing glycans. While the experimental procedure is essentially the same as described above, a 96-well plate installed with hypercarb SPE cartridges was used to increase the synthetic throughput. The structures of glycans 1-26 are shown in Figure 4. All products were profiled by HPLC with generally > 50% relative yield (HPLC) and characterized individually by MALDI-TOF (Supporting Information). All products showed the expected mass spectra and were ready for microarray printing on amino-reactive glass surfaces.
Figure 3.
Two strategies for generating TGLs for the production of glycan microarray starting from either purified free glycans or complex mixtures of glycans released from glycoconjugates from natural sources.
Figure 4.
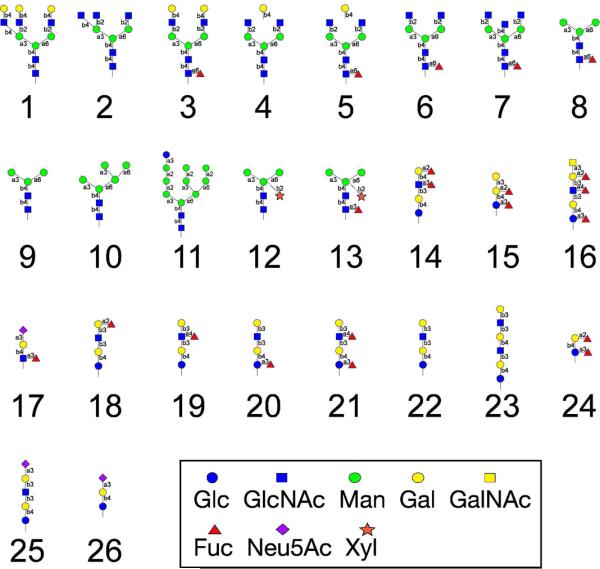
Structures of the defined glycans printed on the microarray. The symbol abbreviations for the glycans are indicated and are consistent with those used by the CFG.
Construction of a glycan microarray of GGAEAB conjugates
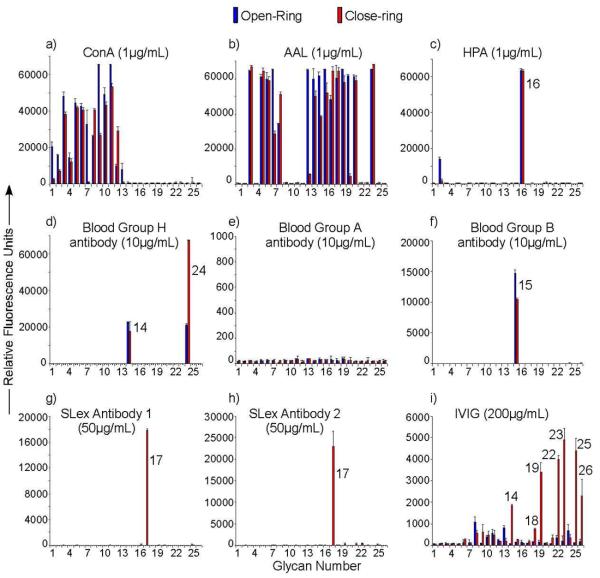
The GGAEABs synthesized from parallel microsynthesis were arrayed on NHS-activated glass slides to create a glycan microarray relatively easily as previously described (23, 24). To compare with the open-ring GAEAB conjugates, the free glycans were directly conjugated with AEAB by reductive amination in a separate process. The GAEAB conjugates, after purification, were printed side-by-side with the closed-ring GGAEABs. This fluorescent microarray was then interrogated by various lectins and antibodies to demonstrate its utility (Figure 5). ConA, which binds selectively to high mannose-, hybrid-, and biantennary complex-type N-glycans (27-29), showed expected binding towards N-glycans with varied affinities, which is generally true for both open-ring and closed-ring conjugates. Interestingly, for several N-glycans (glycans 1, 7, and 13), the closed-ring conjugates showed dramatically lower binding signals than the open-ring conjugates. Although glycans 1-13 have similar mannose moieties, their relative binding affinities are quite different. The triantennary glycans 1 and 7 and the pausi-mannose glycan 13 are known to be lower affinity ligands for ConA (27). It is possible that the more rigid presentation of closed-ring conjugates on microarray requires a tighter binding pocket than that of open-ring conjugates. This may affect ligands with relatively lower binding affinities more than those with higher binding affinities such as high-mannose glycans. This phenomenon is also shown in the recognition of glycans by AAL, which is a fucose-binding lectin (30). While most fucose-containing glycans showed strong binding by AAL, closed-ring conjugates of three glycans (7, 13 and 20) showed diminished binding compared to the open-ring conjugates. The fucose close to the reducing end in the closed-ring conjugates may be sterically restricted from binding by AAL, while it is more available in the open-ring conjugates. Although glycan 7 and 8 have similar fucose moieties, their non-reducing termini could finely tune the lectin-glycan interactions. Therefore, the decrease in binding was only shown for glycan 7. HPA, a lectin that binds terminal GalNAc residues (31, 32), showed expected interactions with a glycan containing terminal α-GalNAc (glycan 16). It also showed some interaction with a triantennary terminal GlcNAc-containing N-glycan (glycan 2), however, only in the open-ring form. Monoclonal antibodies to type-2 blood group H, A, and B showed extremely specific binding to glycans expressing such determinants. Anti-H antibody binds to type-2 H-antigens (glycans 14, 24), but not type-1 H-antigen (glycan 18). Interestingly, the closed-ring conjugates of glycan 24, which retained the type-2 structure, showed higher binding by anti-H antibody than the open-ring conjugates, in which the type-2 structure is incomplete due to the reduction of the reducing end. Anti-A antibody, which is specific for type-2 A-antigen, did not bind to type-1 A-antigen (glycan 16). Note that both the open and closed-ring forms of glycan 16 were bound by HPA, indicating the equivalent reactivity of the terminal α-GalNAc on these glycans (Figure 5). Anti-B showed strong binding to type-2 B-antigen (glycan 15), but not any other glycans. Two commercially available anti-sialyl Lewisx (SLex) antibodies showed specific binding only to the closed-ring SLex structure (glycan 17), and no binding was detectable to the open-ring SLex. This is apparently due to the ring-opening of the GlcNAc, which destroys the complete SLex epitope. Under such circumstances, only closed-ring conjugates can be effectively used as probes on microarrays. Recent studies have shown that commercial intravenous Immunoglobulin (IVIg) preparations, which represent pooled immunoglobulin G from several thousand presumably healthy donors and is used therapeutically, contains a large repertoire of anti-glycan antibodies (33). We tested whether antibodies in Sandoglobulin, a commercial IVIg preparation, differentially recognize glycans derivatized in the open-ring form versus closed-ring form. Interestingly, IVIg showed significant binding to closed-ring compared to open-ring derivatives of several glycans, most of which are human milk glycans larger than a tetrasaccharide. It is likely that antibodies in the complex mixture of IgG within IVIg recognize determinants in both the reducing and non-reducing ends of the glycans.
Figure 5.
Glycan array analysis of lectins binding to an array of 26 GGAEABs and the corresponding 26 GAEABs: Glycans from Figure 4 were prepared as open- (GAEABs as bars filled in blue) or closed-ring (GGAEABs as bars filled in red) derivatives and printed on NHS-slides as described in Methods. As described in the text with appropriate references, a) ConA recognizes high mannose-, hybrid-, and biantennary N-glycans; b) AAL recognizes Fuc-containing glycans; c) HPA binds to GalNAc residues; d) blood group H antibody recognizes Fucα1,2Galβ1-R; e) blood group A antibody recognizes type 2 chain GalNAcα1,3(Fucα1,2)Galβ1,4GlcNAcβ1-R but not type 1 chain GalNAcα1,3(Fucα1,2)Galβ1,2GlcNAcβ1-R (No. 16); f) blood group B antibody recognizes type 2 chain Galα1,3(Fucα1,2)Galβ1,4GlcNAcβ1-R; g) and h) anti-SLex 1 and 2, respectively, recognize NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAcβ1-R; i) IVIg is a product of pooled immunoglobulin G from thousands of individuals. Biotinylated lectins were detected by with cyanine 5-streptavidin (5 μg/mL). Binding of mouse antibodies (blood group antigen H, A, and B antibodies and anti-SLex antibodies) were detected by Alexa488 labeled goat anti-mouse IgM (5 μg/mL). Binding of Sandoglobulin IVIg was detected by Alexa488 labeled goat anti-human IgG (5 μg/mL).
Fluorescent derivatization of glycan mixtures from natural sources and HPLC separation
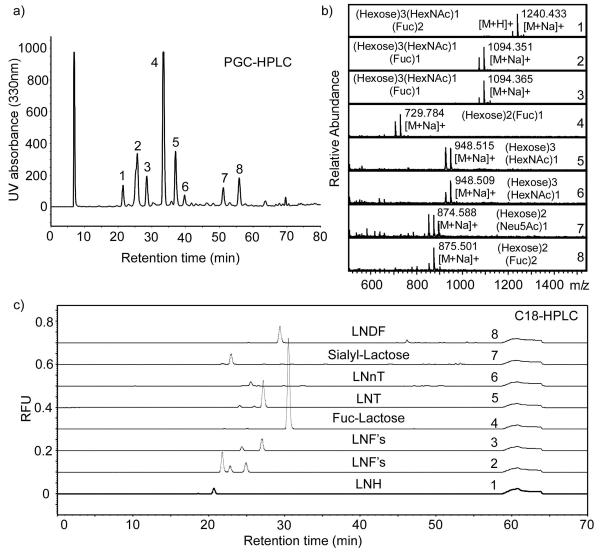
Although the microscale derivatization of individual free glycans enables a rapid route to building a natural GGAEAB library, the ability of derivatizing a glycan mixture from natural sources and separating them is more attractive. The expansion of the glycan library can be virtually unlimited due to the diversity of glycans in nature. To validate this approach, the Sephadex-G25 fraction C of human milk oligosaccharides (34), containing mostly glycans with <6 monosaccharide units were directly converted to glycosylamines, desalted, and acryloylated. Due to the incompleteness of this procedure, small amounts of free reducing glycans exist with the acryloylated glycan, which, after AEAB conjugation, might interfere with the separation of closed-ring GGAEAB conjugates. To solve this problem, the acryloylated product is treated with sodium borohydride before desalting to convert underivatized glycans to their corresponding alditols. The alditols are inert and can not be fluorescently labeled later. They can be easily separated from labeled GGAEAB later using C18 SPE cartridges. The purified acryloylated glycosylamines were treated with ozone and labeled by AEAB conjugation. The mixture was then separated by HPLC and GGAEAB fractions can be collected (Figure 6). Figure 6a showed the 1st dimension HPLC separation using a PGC column. Eight major peaks were collected and analyzed by mass spectrometry. The MALDI-TOF mass spectra confirmed that all the fractions are GGAEABs from human milk oligosaccharides (Figure 6b). Figure 6c showed the 2nd dimension HPLC profiles of all eight fractions on a C18 column, confirming that homogeneous fractions could be obtained through 2D-HPLC. This human milk TGL is a permanent library of fluorescently-labeled glycans possessing a closed ring reducing end that may be quantified, characterized by mass spectrometry and NMR for immediate definition of the glycome, or they may be immobilized onto various solid surfaces, and interrogated with glycan binding proteins to define potential functions prior to structural analysis. Thus, while the generating of such closed-ring GGAEAB derivatives may require several steps, as shown in Figure 1, an entire assortment of glycans can be processed through these steps as a mixture and then the fluorescently-labeled glycans may be purified by HPLC, thus reducing the overall time and effort required for glycan derivatization and purification by this approach.
Figure 6.
The HPLC separation of GGAEABs prepared from a fraction of human milk oligosaccharides containing low molecular weight oligosaccharides: a) The PGC-HPLC purification of 8 major fractions; b) MALDI-TOF spectra of collected fractions from PGC-HPLC separation of human milk GGAEABs; All spectra showed a single molecular masses as different adducts: [M+H]+, [M+Na]+ and [M+2Na]+, that correspond to compositions of known milk oligosaccharides ; c) The second dimensional C18-HPLC profile of the collected fractions in a) indicating the presence of isomers in fractions 2,3, and 5.
Glycan microarrays have been very successful in screening glycan binding proteins to provide valuable information on their specificity and binding properties (17, 21, 35). The most widely used and publicly available glycan microarray is the one provided by the CFG supported by the National Institute of General Medical Sciences of the National Institutes of Health (http://www.functionalglycomics.org/static/index.shtml). Most current glycan microarrays are based on glycans synthesized by chemical and/or enzymatic methods (36); thus, the expansion of such microarrays is limited due to the inherent complexity of glycan structures and difficulties in synthesis (37-42). We have previously developed a “natural glycan array” as an alternative solution for expanding functional glycomic research. Glycans from natural sources can be released and derivatized with bifunctional fluorescent linkers, such as 2, 6-diaminopyridine (DAP) (43) and N-aminoethyl 2-aminobenzamide (AEAB) (24). The labeled glycans can be separated, quantified, and arrayed onto surface-activated glass slides.
While derivatization of glycans by reductive amination is simple, quantitative, and sufficiently benign to preserve many labile glycan structural units, such as sialic acids and sulfates, direct reductive amination opens the ring of the reducing monosaccharide and compromises the intact glycan structure. In many cases, the open ring derivatives do not pose a concern, since many glycan-binding proteins recognize glycan determinants at the non-reducing end. However, to study the contribution of structure close to the reducing end such as human blood group A and B active trisaccharides, GalNAcα1-3(Fucα1-2)Gal and Galα1-3(Fucα1-2)Gal, respectively, we considered whether recognition may require retention of the closed-ring after derivatization, as occurs in all natural glycoconjugates.
Our results here show that for many proteins and antibodies, recognition of glycans with an open versus closed ring is not significantly different. However, for some glycan binding proteins, such as antibodies to SLex and some of the antibodies in IVIg, the open ring is clearly not significantly recognized in comparison to the closed ring structures. These results are consistent with studies by others on recognition of glycans by anti-Lex antibody (44) and selectins (45), in which open-ring forms of small glycans are not bound. Thus, the availability of this closed ring derivatization strategy using a fluorescent tag should enhance the power of glycan microarrays to detect glycan recognition. Other groups have also developed strategies to derivatize reducing glycans in closed-ring forms, using oxime chemistry (46) and hydroxylamine chemistry (25). Methods of generating glycan microarrays by directly immobilizing free glycans on the surface have also been developed (47-49). These approaches are also excellent, but they do not introduce a fluorescent tag, which is an essential feature for isolating and characterizing small amounts of glycans from natural sources.
This novel chemical derivatization strategy described here combined simple glycosylamine formation with the common reactions of N-acetylation, ozonolysis, and fluorescent AEAB conjugation, all of which can be accomplished using routine laboratory protocols. We chose the well-studied glycosylamine formation for several reasons. First, N-glycans are a major class of glycans with an amide linkage between the glycan and the peptide chain. Second, the glycosylamine is known to form a β-anomer preferentially upon N-acylation, which also matches many glycoconjugates linkages such as those in N-glycans. Third, the reaction is easy to perform, since the extremely high ammonium salt concentration in the product can be removed by simply using carbon-based SPE techniques. Thus, it is possible to extract the glycosylamines in a low salt concentration, and after lyophilization, the product can be directly used for N-acryloylation, which generates stabilized glycan conjugates for further derivatization. All steps except the glycosylamine formation are essentially quantitative. Obviously, it is important to have high yield reactions to avoid unnecessary purification steps. The last step of AEAB conjugation introduces a fluorescent label bearing a primary alkylamine onto the glycan. The final glycan derivative conserves the closed-ring structure of glycans and also introduces a fluorescent label to facilitate separation, purification, quantification, and characterization. Furthermore, the compound has a primary alkylamine group for solid phase immobilization, such as glycan microarray preparation. It is also worthwhile to note that due to its chemical nature, this new approach is not suitable for glycan structures with labile O-acetyl group, free amino group, or ozone-reactive double bonds, which only occur as a small fraction of natural glycans.
An essential feature for preparing natural glycan microarrays is to purify glycans from natural sources and microarray them. Our strategy here shows that closed-ring forms of natural glycans can be generated efficiently and the fluorescent nature of the GGAEABs allows them to be identified and purified from a glycan mixture by multi-dimensional HPLC, as demonstrated in Figure 6. Our procedure is amenable to both microscale parallel syntheses of derivatives from commercial glycans and synthesis and separation of glycan mixtures from natural sources. For our testing purposes we chose 26 glycans from commercial sources, which were easily derivatized in parallel, and the purified products were printed on NHS-activated glass slides. The lectin and antibody binding data matched the general expectation and validated the GGAEAB strategy for glycan microarray usage. In the future it will be important to further compare GBP recognition of open- versus closed-ring derivatives of glycans to explore the role of the reducing termini in recognition.
Methods
Materials
Free reducing glycans were purchased from V-labs and Glycoseparations and stored at −20°C until use. All chemicals were purchased from Sigma-Aldrich and used without further purification. Human milk was purchased from Mothers' Milk Bank, Austin, TX. Biotinylated lectins were purchased from Vector Labs. Antibodies against blood group A, B, and H were purchased from Santa Cruz Biotechnology, Inc. CSLEX1 antibody (CD15s) to SLex was purchased from BD Pharmingen. CHO131 antibody to SLex was purchased from R & D Systems. Sandoglobulin was a gift from CSL Behring AG, Bern Switzerland. HPLC solvents were purchased from Fisher Scientific. An Ultraflex-II TOF/TOF system from Bruker Daltonics was used for MALDI-TOF mass spectrometry analysis of glycan conjugates. HPAEC-PAD analysis was carried out with a Dionex ICS-3000 system using a Carbpac PA-100 column. A Shimazu HPLC CBM-20A system coupled with a UV detector SPD-20A and a fluorescence detector RF-10Axl was used for HPLC analysis and separation of GAEABs.
Glycosylamine formation, N-acetylation, reduction, ozone treatment, and AEAB conjugation
Free reducing glycans (0.1 mg – 10 mg) were dissolved in water (100 μL) in a 1.5 mL screw cap vial, and ammonium bicarbonate (200 mg) was added. The mixture was incubated at 55°C for 1.5 h, cooled, and diluted with 1 mL water. A 300 mg carbograph SPE column (8 mL), pre-conditioned with 1 column volume (c.v.) of 50% acetonitrile containing 10 mM ammonium bicarbonate and 3 c.v. of 10 mM ammonium bicarbonate. The sample was loaded on the carbograph column, washed with 10 mM ammonium bicarbonate (6 c.v.) and eluted with 50% acetonitrile with 10 mM ammonium bicarbonate (2 c.v.). The eluent was evaporated in a Speed-vac for 2 h and freeze-dried.
To the lyophilized powder in a 15 mL conical tube (sitting on ice), sodium bicarbonate was added (600 mg), followed by ice-cold saturated sodium bicarbonate solution (2 mL). Two hundred μL acryloyl chloride was added immediately and the mixture capped and vortexed for 5 min. The cap was slightly opened to release the pressure and the mixture was vortexed for another 5 min. The mixture was dissolved in water (5 mL) and applied on a 300 mg carbograph cartridge (precondition with 1 c.v. 50% acetonitrile and 0.1% TFA and 3 c.v. water). The reaction mixture was applied to the cartridge, washing with 6 c.v. water, and the glycosylamide was purified by elution with 50% acetonitrile and 0.1% TFA (2 c.v.). The eluent was evaporated by Speed-vac for 2 h to remove solvent and lyophilized.
Optional: For a complex mixture of glycans from human milk (or released from glycoconjugates), a sodium borohydride reduction step was used. To a glycan mixture dissolved in water (2 mL), an equal amount (by weight) of sodium borohydride (NaBH4) was directly added at 4°C. The reaction was vortexed with frequent cooling for 30 min. Acetic acid (twice the NaBH4 by weight) was added under cooling at 4°C and incubated for 10 min. The mixture was desalted using Carbograph as described above and lyophilized.
The lyophilized acryloyl derivatives were dissolved in methanol (2 mL) and cooled to −78°C (dry ice – ethanol). Ozone was bubbled through for ∼1 min (or until blue color stays). The samples were brought to room temperature, methyl sulfide (200 μL) was added, and the mixture was incubated at room temperature for 8 h.
The ozone treated glycan derivatives were dried in a Speed-vac for 2 h. Fresh solutions of AEAB (0.35 M) and sodium cyanoborohydride (1 M) in DMSO/AcOH (7/3) were prepared separately. AEAB solution (50–500 μL) and an equal volume of sodium cyanoborohydride solution were added to the dried sample and incubated at 65°C for 2 h. The resulting glycan derivatives were precipitated upon addition of 10 volumes of acetonitrile at −20°C for 2 h. The mixture was centrifuged and the supernatant discarded. The precipitate was reconstituted in water (100-500 μL) for HPLC analysis and purification.
For mono- and di-saccharides, which are not retained by carbograph very well, size exclusion chromatography can be used for the desalting and purification of the intermediates and products, as described in Supporting Information.
HPAEC-PAD and high performance liquid chromatography (HPLC) analysis of glycans and their derivatives
For HPAEC-PAD analysis, the linear gradient was set to 2.5-125 mM sodium acetate over 100 min in 100 mM sodium hydroxide. For HPLC analysis and preparation, UV absorption at 330 nm or fluorescence at 330 nm excitation and 420 nm emission was used to detected AEAB derivatives in HPLC analysis and separation. Both UV absorption and fluorescence intensity were used for the quantification of glycan derivatives using LNFPIII-AEAB as a standard.
For reverse phase HPLC with porous graphitized carbon (PGC) column, the mobile phase was acetonitrile and water with 0.1% TFA. For the glycan analysis, the concentration of acetonitrile increased from 15% to 45% in 30 min. For glycan separation of a glycan mixture, the concentration of acetonitrile increased from 15% to 45% in 90 min. For reverse phase HPLC with C18 column, a Vydec C18 column was used. The mobile phase is acetonitrile and water with 0.1% TFA. The concentration of acetonitrile increased from 1% to 5% in 60 min.
Printing, binding assay, and scanning
NHS-activated slides were purchased from Schott. Epoxy slides were purchased from Corning. Non-contact printing was performed using a Piezorray printer from Perkin Elmer. The average spot volume was within 10% variation of 1/3nL and spots were approximately 100 microns in diameter separated ∼200 microns center-to-center. All samples were printed in phosphate buffer (300 mM sodium phosphates, pH 8.5). After printing, the slides were boxed loosely and put in a high moisture chamber at 50°C and incubated for 1 h. The slides were then washed and blocked with 50mM ethanolamine in 0.1 M Tris buffer (pH 9.0) for 1 h. The slides can be dried by centrifugation and stored desiccated at −20°C for future use. Before assay, the slides were rehydrated for 5 min in TSM buffer (20 mM Tris-HCL, 150 mM sodium chloride (NaCl), 0.2 mM calcium chloride (CaCl2) and 0.2 mM magnesium chloride (MgCl2)). The slides were incubated with primary carbohydrate binding proteins (lectins and antibodies at concentrations indicated in Figures) for 1 h in TSM buffer (with 1% BSA). The bound proteins were detected by incubation with fluorescently labeled secondary antibodies. Biotinylated lectins were used in the binding assay and the bound lectins were detected by a secondary incubation with cyanine 5-streptavidin (5 μg/mL) in TSM buffer (with 1% BSA). Binding of mouse antibodies (blood group antigen H, A, and B antibodies and anti-SLex antibodies) were detected by incubation with Alexa488 labeled goat anti-mouse IgM (5 μg/mL). Binding of IVIg was detected by Alexa488 labeled goat anti-human IgG (5 μg/mL). For multi-panel experiments on a single slide, the array layout was designed using Piezorray software according to the dimension of a standard 16-chamber adaptor. The adaptor was applied on the slide to separate a single slide to 16 chambers sealed from each other during the assay.
The slides were scanned with a Perkin Elmer ProScanarray microarray scanner equipped with 4 lasers covering an excitation range from 488 nm to 637 nm. The scanned images were analyzed with the ScanArray Express software. For cyanine 5 fluorescence, 649 nm (Ex) and 670 nm (Em) were used. For Alexa488 fluorescence, 495 nm (Ex) and 519 nm (Em) were used. All the images obtained from the scanner were in grayscale and colored for easy discrimination.
Supplementary Material
Acknowledgements
This work was supported in part by a Bridge Grant to R.D.C. from the Consortium for Functional Glycomics under NIGMS, NIH Grant GM62116 and in part by RO1GM085448 to D.F.S. We thank Dr. Sylvia Miescher (Research and Development, CSL Behring AG, Bern Switzerland) for her kind gift of IVIg and Dr. Jamie Heimburg-Molinaro (Emory University School of Medicine) for manuscript editing and review.
Abbreviations
- AAL
Aleuria aurantia lectin
- AEAB
2-amino-N-(2-aminoethyl)-benzamide
- ConA
Concanavalin A
- DAP
2, 6-diaminopyridine
- GAEAB
Glycan-AEAB conjugate
- GBP
glycan binding protein
- GGAEAB
Glycosylamide-Gly-AEAB conjugate
- HPA
Helix pomatia agglutinin
- HPAEC-PAD
high performance anion-exchange chromatography with pulsed amperometric detection
- HPLC
high performance liquid chromatography
- LNFPIII
lacto-N-fucopentaose III
- NHS
N-hydroxysuccinimide
- PGC
porous graphitized carbon
- RFU
relative fluorescence unit
- SLex
Sialyl Lewis x
- SPE
solid phase extraction
- TLC
thin layer chromatography
- TGL
Tagged glycan library.
Footnotes
Supporting Information Available: This material is free via the Internet.
The authors declare they have no financial interest.
References
- 1.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2009. [PubMed] [Google Scholar]
- 2.Taylor ME, Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr Opin Cell Biol. 2007;19:572–7. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Lowe JB. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol. 2003;15:531–8. doi: 10.1016/j.ceb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Petrescu AJ, Wormald MR, Dwek RA. Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding. Curr Opin Struct Biol. 2006;16:600–7. doi: 10.1016/j.sbi.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31:156–63. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Moremen KW, Molinari M. N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr Opin Struct Biol. 2006;16:592–9. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruddock LW, Molinari M. N-glycan processing in ER quality control. J Cell Sci. 2006;119:4373–80. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- 9.Hooper LV, Gordon JI. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology. 2001;11:1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 10.Ratner DM, Adams EW, Disney MD, Seeberger PH. Tools for glycomics: mapping interactions of carbohydrates in biological systems. Chembiochem. 2004;5:1375–83. doi: 10.1002/cbic.200400106. [DOI] [PubMed] [Google Scholar]
- 11.Horlacher T, Seeberger PH. Carbohydrate arrays as tools for research and diagnostics. Chem Soc Rev. 2008;37:1414–22. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- 12.Park S, Lee MR, Shin I. Carbohydrate microarrays as powerful tools in studies of carbohydrate-mediated biological processes. Chem Commun (Camb) 2008:4389–99. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]
- 13.Magnani JL, Brockhaus M, Smith DF, Ginsburg V. Detection of glycolipid ligands by direct binding of carbohydrate-binding proteins to thin-layer chromatograms. Methods Enzymol. 1982;83:235–41. doi: 10.1016/0076-6879(82)83016-4. [DOI] [PubMed] [Google Scholar]
- 14.Lopez PH, Schnaar RL. Determination of glycolipid-protein interaction specificity. Methods Enzymol. 2006;417:205–20. doi: 10.1016/S0076-6879(06)17015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feizi T, Chai W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–8. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 16.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays -a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–45. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez RA, Blixt O. Identification of ligand specificities for glycan-binding proteins using glycan arrays. Methods Enzymol. 2006;415:292–310. doi: 10.1016/S0076-6879(06)15018-1. [DOI] [PubMed] [Google Scholar]
- 18.Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew Chem Int Ed Engl. 2002;41:3180–2. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 19.Ratner DM, Adams EW, Su J, O'Keefe BR, Mrksich M, Seeberger PH. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chembiochem. 2004;5:379–82. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 20.Bryan MC, Fazio F, Lee HK, Huang CY, Chang A, Best MD, Calarese DA, Blixt O, Paulson JC, Burton D, Wilson IA, Wong CH. Covalent display of oligosaccharide arrays in microtiter plates. J Am Chem Soc. 2004;126:8640–1. doi: 10.1021/ja048433f. [DOI] [PubMed] [Google Scholar]
- 21.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–8. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Boer AR, Hokke CH, Deelder AM, Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Anal Chem. 2007;79:8107–13. doi: 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- 23.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconj J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 24.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohorov O, Andersson-Sand H, Hoffmann J, Blixt O. Arraying glycomics: a novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology. 2006;16:21C–27C. doi: 10.1093/glycob/cwl044. [DOI] [PubMed] [Google Scholar]
- 26.Corradi Da Silva ML, Stubbs HJ, Tamura T, Rice KG. 1H NMR characterization of a hen ovalbumin tyrosinamide N-linked oligosaccharide library. Arch Biochem Biophys. 1995;318:465–75. doi: 10.1006/abbi.1995.1255. [DOI] [PubMed] [Google Scholar]
- 27.Kornfeld K, Reitman ML, Kornfeld R. The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J Biol Chem. 1981;256:6633–40. [PubMed] [Google Scholar]
- 28.Krusius T, Finne J, Rauvala H. The structural basis of the different affinities of two types of acidic N-glycosidic glycopeptides for concanavalin A--sepharose. FEBS Lett. 1976;72:117–20. doi: 10.1016/0014-5793(76)80911-8. [DOI] [PubMed] [Google Scholar]
- 29.Ogata S, Muramatsu T, Kobata A. Fractionation of glycopeptides by affinity column chromatography on concanavalin A-sepharose. J Biochem. 1975;78:687–96. doi: 10.1093/oxfordjournals.jbchem.a130956. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita K, Kochibe N, Ohkura T, Ueda I, Kobata A. Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. J Biol Chem. 1985;260:4688–93. [PubMed] [Google Scholar]
- 31.Hammarstrom S, Murphy LA, Goldstein IJ, Etzler ME. Carbohydrate binding specificity of four N-acetyl-D-galactosamine- “specific” lectins: Helix pomatia A hemagglutinin, soy bean agglutinin, lima bean lectin, and Dolichos biflorus lectin. Biochemistry. 1977;16:2750–5. doi: 10.1021/bi00631a025. [DOI] [PubMed] [Google Scholar]
- 32.Torres BV, McCrumb DK, Smith DF. Glycolipid-lectin interactions: reactivity of lectins from Helix pomatia, Wisteria floribunda, and Dolichos biflorus with glycolipids containing N-acetylgalactosamine. Arch Biochem Biophys. 1988;262:1–11. doi: 10.1016/0003-9861(88)90161-0. [DOI] [PubMed] [Google Scholar]
- 33.von Gunten S, Smith DF, Cummings RD, Riedel S, Miescher S, Schaub A, Hamilton RG, Bochner BS. IVIg Contains a Broad Repertoire of Anti-Carbohydrate Antibodis that is not Restricted to the IgG2 Subclass. J. Allergy Clin. Immunol. 2009 doi: 10.1016/j.jaci.2009.03.013. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobata A, Ginsburg V, Tsuda M. Oligosaccharides of human milk. I. Isolation and characterization. Arch Biochem Biophys. 1969;130:509–13. doi: 10.1016/0003-9861(69)90063-0. [DOI] [PubMed] [Google Scholar]
- 35.Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 2008;283:10109–23. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blixt O, Razi N. Chemoenzymatic synthesis of glycan libraries. Methods Enzymol. 2006;415:137–53. doi: 10.1016/S0076-6879(06)15009-0. [DOI] [PubMed] [Google Scholar]
- 37.Smith DF, Cummings RD. Deciphering Lectin Ligands through Glycan Arrays. In: Vasta GR, Ahmed H, editors. Animal Lectins: A Functional View. CRC Press; New York: 2008. pp. 49–62. [Google Scholar]
- 38.de Paz JL, Seeberger PH. Deciphering the glycosaminoglycan code with the help of microarrays. Mol Biosyst. 2008;4:707–11. doi: 10.1039/b802217h. [DOI] [PubMed] [Google Scholar]
- 39.Liang PH, Wu CY, Greenberg WA, Wong CH. Glycan arrays: biological and medical applications. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tateno H, Mori A, Uchiyama N, Yabe R, Iwaki J, Shikanai T, Angata T, Narimatsu H, Hirabayashi J. Glycoconjugate microarray based on an evanescent-field fluorescence-assisted detection principle for investigation of glycan-binding proteins. Glycobiology. 2008;18:789–98. doi: 10.1093/glycob/cwn068. [DOI] [PubMed] [Google Scholar]
- 41.Timmer MS, Stocker BL, Seeberger PH. Probing glycomics. Curr Opin Chem Biol. 2007;11:59–65. doi: 10.1016/j.cbpa.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Wang D. Carbohydrate microarrays. Proteomics. 2003;3:2167–75. doi: 10.1002/pmic.200300601. [DOI] [PubMed] [Google Scholar]
- 43.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–50. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 44.Streit A, Yuen CT, Loveless RW, Lawson AM, Finne J, Schmitz B, Feizi T, Stern CD. The Le(x) carbohydrate sequence is recognized by antibody to L5, a functional antigen in early neural development. J Neurochem. 1996;66:834–44. doi: 10.1046/j.1471-4159.1996.66020834.x. [DOI] [PubMed] [Google Scholar]
- 45.Leteux C, Stoll MS, Childs RA, Chai W, Vorozhaikina M, Feizi T. Influence of oligosaccharide presentation on the interactions of carbohydrate sequence-specific antibodies and the selectins. Observations with biotinylated oligosaccharides. J Immunol Methods. 1999;227:109–19. doi: 10.1016/s0022-1759(99)00077-0. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Chai W, Childs RA, Feizi T. Preparation of neoglycolipids with ring-closed cores via chemoselective oxime-ligation for microarray analysis of carbohydrate-protein interactions. Methods Enzymol. 2006;415:326–40. doi: 10.1016/S0076-6879(06)15020-X. [DOI] [PubMed] [Google Scholar]
- 47.Lee MR, Shin I. Facile preparation of carbohydrate microarrays by site-specific, covalent immobilization of unmodified carbohydrates on hydrazide-coated glass slides. Org Lett. 2005;7:4269–72. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 48.Park S, Lee MR, Shin I. Construction of carbohydrate microarrays by using one-step, direct immobilizations of diverse unmodified glycans on solid surfaces. Bioconjug Chem. 2009;20:155–62. doi: 10.1021/bc800442z. [DOI] [PubMed] [Google Scholar]
- 49.Zhi ZL, Powell AK, Turnbull JE. Fabrication of carbohydrate microarrays on gold surfaces: direct attachment of nonderivatized oligosaccharides to hydrazide-modified self-assembled monolayers. Anal Chem. 2006;78:4786–93. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.