Abstract
Brain metastases of breast cancer appear to be increasing in incidence as systemic therapy improves. Metastatic disease in the brain is associated with high morbidity and mortality. We present the first gene expression analysis of laser captured epithelial cells from resected human brain metastases of breast cancer compared to unlinked primary breast tumors. The tumors were matched for histology, TNM stage and hormone receptor status. Most differentially expressed genes were down-regulated in the brain metastases which included, surprisingly, many genes associated with metastasis. Q-PCR analysis confirmed statistically significant differences or strong trends in the expression of six genes: BMP1, PEDF, LAMγ3, SIAH, STHMN3 and TSPD2. Hexokinase 2 (HK2) was also of interest because of its increased expression in brain metastases. HK2 is important in glucose metabolism and apoptosis. In agreement with our microarray results, HK2 levels (both mRNA and protein) were elevated in a brain metastatic derivative (231-BR) of the human breast carcinoma cell line MDA-MB-231 relative to the parental cell line (231-P), in vitro. Knockdown of HK2 expression in 231-BR cells using shRNA reduced cell proliferation when cultures were maintained in glucose limiting conditions. Finally, HK2 expression was analyzed in a cohort of 123 resected brain metastases of breast cancer. High HK2 expression was significantly associated with poor patient survival post-craniotomy (P=0.028). The data suggest that HK2 overexpression is associated with metastasis to the brain in breast cancer and it may be a therapeutic target.
Introduction
Metastasis to the central nervous system (brain) is a contributor to breast cancer patient morbidity and mortality. Traditionally, brain metastases were diagnosed in ~15% of metastatic breast cancer patients late in the course of their disease and only palliative treatment was given. Improvements in breast cancer treatment have increased the number of patients with systemic responses and lengthened survival, however brain metastases have also increased as a “sanctuary” site (rev. in (1, 2)). This is best quantified in the Her-2+ subpopulation of breast cancers, where studies document that 25–40% of metastatic patients now develop brain metastases, often as the first site of relapse (rev. in (3) ). Other risk factors for brain metastases include “triple negative” or basal primary tumors, systemic metastases and young patient age (4–11). In order to understand the molecular events that underlie brain metastases, we initiated a gene expression profiling experiment using resected brain metastases of breast cancer.
Among the trends in gene expression, the overexpression of hexokinase 2 (HK2) was observed in brain metastases. HK2 is one of four members of the hexokinase family. The isoenzymes (HK1, HK2, HK3 and Glucokinase) are structurally similar however only HK1 and 2 are functionally similar. HK2, but not HK1, is overexpressed in several cancer types as compared to normal tissues; among primary tumors, high HK2 confers a poor prognosis (12–15). Ample evidence suggests that HK2 biochemical pathways are heightened in cancer and may constitute a potential therapeutic target. HK2 plays a key step early in glycolysis, phosphorylating glucose to produce glucose-6-phosphate. In some cell types, HK2 binds to the mitochondrial membrane to obtain ATP exported from oxidative phosphorylation. The Km for Mg-ATP bound to the mitochondrial membrane is approximately five times lower than that of free Mg-ATP (16). This efficient coupling of ATP from oxidative phosphorylation to the rate-limiting step of glycolysis may contribute to the Warburg effect, where tumor cells utilize glycolysis even in the presence of oxygen (16, 17). Not only does HK2 participate in bioenergetics, but its association with mitochondria has also been reported to prevent apoptosis (rev. in (18)). HK2 binds to the voltage-dependent anion channel (VDAC) on the mitochondrial membrane. In turn the VDAC/HK2 complex forms part of the permeability transition pore together with cyclophilin D and adenine nucleotide translocase (17, 19). How HK2 physically blocks apoptosis is still under investigation. One model suggests that the pro-apoptotic protein Bax also physically binds VDAC (in the absence of HK2) to promote cytochrome c release. Alternatively, HK2 may modulate binding of the anti-apoptotic protein Bcl-Xl to VDAC, with consequences for its downstream partnering with Bak and Bax (rev. in (20)). HK2 expression may also influence 18F 2-fluoro-2-deoxy-D-glucose (18F-FDG) uptake in tumors, thus impacting PET imaging for diagnosis and staging (13, 21, 22).
This study presents the first analysis of differentially expressed genes between resected brain metastases and primary breast carcinomas. A trend of elevated HK2 expression in brain metastases is demonstrated. In a brain metastasis cell line model, knock down of HK2 expression reduced cellular viability under conditions of limited glucose availability. Finally, the relevance of differential HK2 expression is demonstrated by the association of high HK2 expression in brain metastases with reduced patient survival.
Results
Microarray analysis of resected brain metastases and unlinked primary breast tumors
In order to identify potential molecular therapeutic targets for brain metastases of breast cancer, resected human brain metastases and unlinked primary tumors were subjected to a microarray analysis. Tissues were sectioned and epithelial cells procured by laser capture microdissection. Of 16 flash frozen brain metastases, 8 produced RNA of sufficient quality for hybridization, as did 9 of 20 primary breast tumors. Table 1 lists the characteristics of the tumors used. Most of the histological indicators were well balanced between the primary tumors and brain metastases. Tumors varied from T1 to T3, and N0 to N2 in both cohorts. Four of eight brain metastases and four of nine primary tumors were ER+. Most of the specimens were ductal in histology, with a lobular carcinoma in each cohort and a single brain metastasis from an inflammatory primary tumor. Her-2 amplification was over-represented in the brain metastases cohort (4/8) as compared to the primary tumors (1/9).
Table 1.
Histologic characteristics of microarray specimens.
| Specimen: | Site1: | TNM2: | ER3: | Her-24: | Histology: |
|---|---|---|---|---|---|
| 4-11-1 | BM | T1 N0 | Neg. | Amp. | Ductal |
| 4-11-2 | BM | T2 N1 M0 | Pos. | wt | Ductal |
| 4-11-3 | BM | T3 N2 | Pos. | wt | Lobular |
| 4-11-4 | BM | T3 N1 M0 | Neg. | wt | Inflammatory |
| 4-11-5 | BM | T3 N2 M0 | Pos. | Amp. | Ductal |
| 4-11-6 | BM | T2 N1 M0 | Neg. | Amp. | Ductal |
| 4-11-7 | BM | T2 N1 M0 | Neg. | Amp. | Ductal |
| 4-11-8 | BM | T1 N0 | Pos. | wt | Ductal |
| 4-11-9 | P | T2 N1 | Neg. | wt | Ductal |
| 4-11-10 | P | T1 N0 | Pos. | Amp. | Ductal |
| 4-11-11 | P | T1 | Pos. | wt | Ductal |
| 4-11-12 | P | T2 N0 | Neg. | wt | Ductal |
| 4-11-13 | P | T2 N1 | Neg. | wt | Lobular |
| 4-11-14 | P | T2 N1 | Neg. | wt | Ductal |
| 4-11-15 | P | T1 | Pos. | wt | Ductal |
| 4-11-16 | P | T2 | Pos. | wt | Ductal |
| 4-11-17 | P | T3 N2 Mx | Neg. | wt | Ductal |
BM, Brain metastasis; P, Primary tumor
Histology of brain metastases based on patient’s primary tumor.
Estrogen receptor: Pos., positive by immunohistochemistry; Neg., negative
Amp, amplified by FISH; wt, wild type DNA level
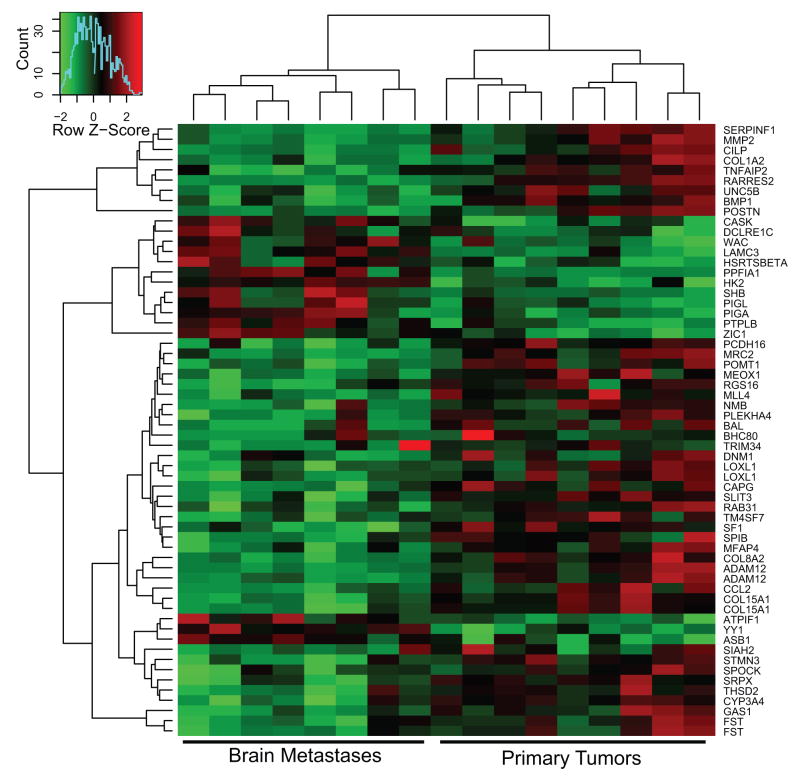
A heatmap displays the top 60 differentially expressed genes between the brain metastases and the primary tumors based upon supervised hierarchical clustering and elimination of expression signatures associated with common breast cancer pathways such as ER and Her-2 (Figure 1). The majority of differentially regulated genes were down-regulated in the brain metastases. Notably, metastasis associated genes were also down-regulated in the brain metastases including genes involved in extracellular matrix function (MMP2, COL1A2, LOXL1, MFAP4, COL8A2, ADAM12, COL15A1, THSD2), cell adhesion (PCDH16, TM4SF7), and motility (UNC5B, CAPG, CCL2, STMN3). Up-regulated genes included phosphatases (PPFIA1, PTPLB), phosphoinositide glycan anchor biosynthetic proteins which GPI anchor proteins to membranes (PIGL, PIGA), and signaling proteins (CASK, SHB). The only extracellular matrix gene up-regulated in the brain metastases was LAMγ3, a component of laminin 12.
Figure 1.
Microarray analysis of resected human brain metastases of breast cancer and unlinked primary breast tumors. Epithelial cells were procured by laser capture microdissection from brain metastases and primary tumors. Extracted and amplified RNAs were converted to Cy-dUTP labeled cDNA and hybridized to a 30K cDNA microarray. The control hybridization consisted of a mixture of RNAs from six human breast cancer cell lines, similarly processed. Supervised clustering of the data is shown where each sample is identified as a brain metastasis (left) or a primary tumor (right).
Because of the small number of specimens profiled and the rarity of resected brain metastases suitable for molecular analysis, validation experiments utilized 4 of the initially profiled specimens, in an attempt to repeat the gene expression trends observed on the microarray, supplemented with 8 independent samples each of unlinked brain metastases and primary tumors. Q-PCR was performed on amplified cDNA from laser capture microdissected epithelial cells. Table 2 lists the expression levels of nine genes from the microarray analysis. Significant differences or strong trends in the expression of several down-regulated genes were observed including: BMP1, which regulates cartilage formation (P=0.01), SIAH, seven in abstentia homolog 1 (P=0.001), STHMN3, stathmin-like 3 (P=0.009) and TSPD2, a thrombospondin derivative (P=0.0001). Two genes were up-regulated in brain metastases in the microarrary analysis and the validation cohort: LAMγ3, a component of laminin 12 (P=0.05) and HK2 (p=0.13). Because of the potential contributions of HK2 to tumor cell bioenergetics, growth and apoptosis, as well as HK2’s potential to be pharmacologically inhibited, functional studies of altered HK2 expression were initiated.
Table 2.
Validation of Differentially Expressed Genes.
| Gene: | Trend on Microarray1: | Mean ± SEM2 |
P value3 | |
|---|---|---|---|---|
| Brain Metastasis | Primary Tumor | |||
| BHC80 | Down in BM | 22.5 ± 4.9 | 63 ± 17.2 | 0.22 |
| BMP1 | Down in BM | 2.8 ± 0.5 | 7.1 ± 1.4 | 0.01 |
| HK2 | Up in BM | 0.9 ± 0.2 | 0.6 ± 0.1 | 0.13 |
| LAMγ3 | Up in BM | 0.7 +/− 0.2 | 0.3 +/− 0.1 | 0.05 |
| LOXL1 | Down in BM | 3.8 ± 0.6 | 9.0 ± 2.6 | 0.14 |
| PEDF | Down in BM | 5.4 ± 3.1 | 76.0 ± 36.5 | 0.001 |
| SIAH | Down in BM | 1.0 ± 0.3 | 6.6 ± 2.8 | 0.00011 |
| SLIT3 | Down in BM | 1.1 ± 0.3 | 2.4 ± 0.7 | 0.26 |
| STHMN3 | Down in BM | 0.6 ± 0.2 | 1.9 ± 0.6 | 0.0087 |
| TSPD2 | Down in BM | 0.1 ± 0.1 | 2.7 ± 0.8 | 0.00014 |
See Figure 1. BM, Brain metastasis
Adjusted for GAPDH levels
By exact two-tailed Wilcoxon rank sum test.
Knockdown of HK2 impairs proliferation under glucose limiting culture conditions
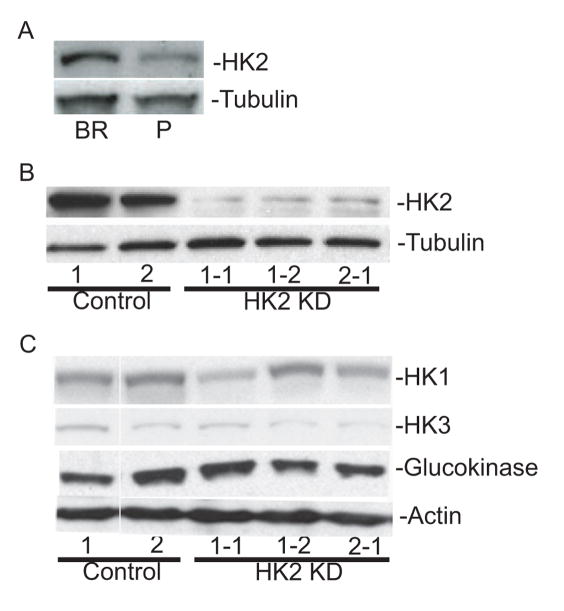
The human breast carcinoma cell line MDA-MB-231 was selected through six rounds of in vivo passage for a brain metastastic subline (231-BR) (23, 24). Several of the validated gene expression trends noted above between brain metastases and primary breast tumors were analyzed in the model system by comparing mRNA levels for a given gene between the parental MDA-MB-231 cell line and the 231-BR metastatic subclone. Of the 6 genes analyzed by Q-PCR only HK2 exhibited the same trend as seen in the tumor cohort. The 231-BR cells had a 2-fold increase in HK2 mRNA levels (16.37, 95% CI 9.24–23.5) compared to parental MDA-MB-231 cells (7.98, 95% CI 4.82–11.13). Increased HK2 protein expression was also noted in the 231-BR line compared to parental MDA-MB-231 cells, further suggesting that increased HK2 expression is coincident with brain metastatic colonization (Figure 2A). To test the functional consequences of altered HK2 expression, 231-BR cells were infected with a control shRNA or one of two HK2 shRNAs, and clones isolated. Three independent clones expressing HK2 shRNA exhibited a 56 to 65% reduction in HK2 protein levels relative to control clones as determined by western blot analysis (Figure 2B). The shRNA constructs used to knockdown HK2 are specific for this hexokinase family member as they did not appear to affect expression of the related isoenzyme family members, HK1, HK3, and Glucokinase (Figure 2C).
Figure 2.
Western blot analysis of HK2 expression in the 231-BR model system. A. HK2 protein levels in parental MDA-MB-231 human breast cancer cells (P) and a brain metastatic derivative (BR). Tubulin is shown as a loading control. B. Knockdown of HK2 expression in 231-BR cells is shown, including two independent clones of HK2 shRNA 1 (1-1, 1-2) and one clone of HK2 shRNA 2 (2-1). Two clonal lines expressing a control scrambled shRNA (Controls 1 and 2) are also shown. C. Expression levels of other hexokinase family members are not affected by HK2 knockdown. Protein levels for HK1, -3 and Glucokinase are shown for the clonal lines listed in panel B, with actin as a loading control.
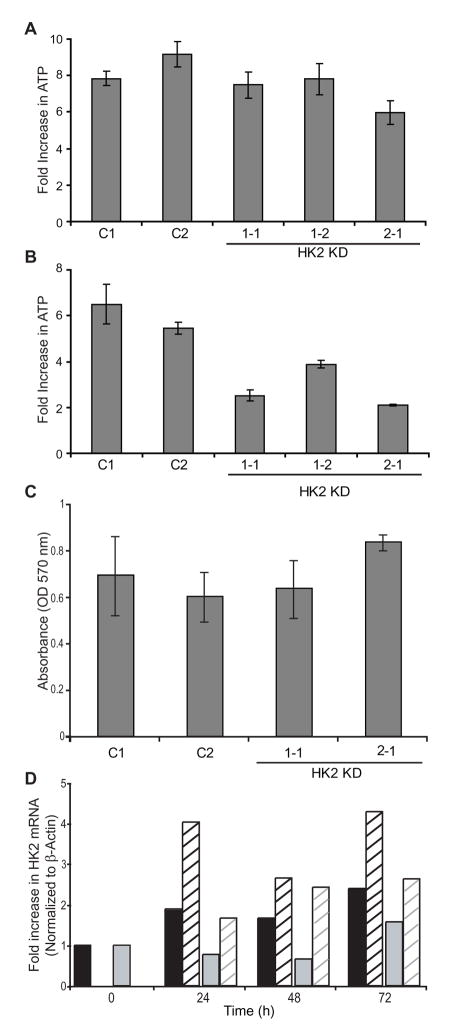
To determine if knockdown of HK2 altered cellular bioenergetics, intracellular ATP levels were determined over four days of culture (Figure 3). Under standard culture conditions intracellular ATP levels were unaffected by HK2 depletion (Figure 3A), possibly the result of redundant activities of Hexokinase homolog proteins, which are clearly present (Figure 2C). However, under these culture conditions, glucose is in excess as compared to physiological conditions (4.5 grams per liter in standard growth medium compared with human serum levels of 7–11 milligrams per liter). We therefore determined cellular ATP levels in limited glucose, 10% of standard culture conditions (Figure 3B). ATP levels in the two control clones increased 5 to 6-fold over time; a smaller rate of ATP increase was observed in each of the HK2 knockdown clones (2–4 fold, P<0.005 for each HK2 knockdown clone compared with the average of the controls, and P=0.003 for a comparison of the mean of the controls to the mean of the HK2 knockdown clones by one-way ANOVA). The amount of ATP is proportional to the number of viable cells and as such we confirmed these results using an MTT assay. No difference in viability was observed over 4 days of culture between the control and HK2 knockdown clones under standard culture conditions while in limiting glucose concentrations the HK2 knockdown clones exhibited a pronounced decrease in viability compared to controls (data not shown). This observation was also confirmed by trypan blue exclusion cell counting. An attempt was made to assess the effect of low oxygen conditions on ATP levels/viability; no difference was observed between control and HK2 knockdown clones when cultured in hypoxic conditions (Figure 3C). Further analysis showed that HK2 KD was overcome under hypoxic conditions by an increase in HK2 mRNA levels (Figure 3D).
Figure 3.
HK2 shRNA reduced cell viability, as measured by ATP concentration, of 231-BR cells in vitro under glucose limiting conditions. A. The control shRNA and HK2 shRNA clones were grown in normal culture medium (high glucose DMEM). B. The same clones were grown in medium diluted 10-fold into glucose free DMEM for four days (P<0.0001 by one-way ANOVA). Cellular ATP levels were measured by Cell Titer-Glo Luminescent cell viability assay at 24 and 96 hours of culture. The mean fold-increase ± standard deviation for n=3 experiments is shown for each graph. C. Knockdown of HK2 expression did affect the viability of cells grown in hypoxic conditions. The mean absorbance by MTT assay ± standard deviation for n=2 experiments is shown. D. HK2 mRNA knockdown was overcome when cells were grown in hypoxic conditions. The fold increase in HK2 mRNA compared to each clone at t=0 in normal growth conditions. HK2 mRNA was normalized to β-actin. Black bars; HK2 shRNA clone 1-1. Grey bars; HK2 shRNA clone 2-1. Hatched bars; clones grown in low oxygen conditions.
The effect of decreased HK2 expression on drug-induced apoptosis was examined to identify a role for HK2 in cell death in the 231-BR cells. Control and HK2 knockdown clones of 231-BR cells were incubated for 48 hours in the presence or absence of 0.5–1.0 μM doxorubicin or 1–50 nM paclitaxel in standard growth conditions. While both drugs showed a dose dependent increased in apoptosis as measured by Cell Death Detection ELISA+ for all cells there was no consistent difference in the amount of apoptosis between the control and HK2 knockdown clones. Similar experiments were performed using 5 Gy radiation as a relevant apoptosis inducer and again no significant difference was observed between control and knockdown clones (data not shown).
HK2 protein levels are associated with survival post-resection
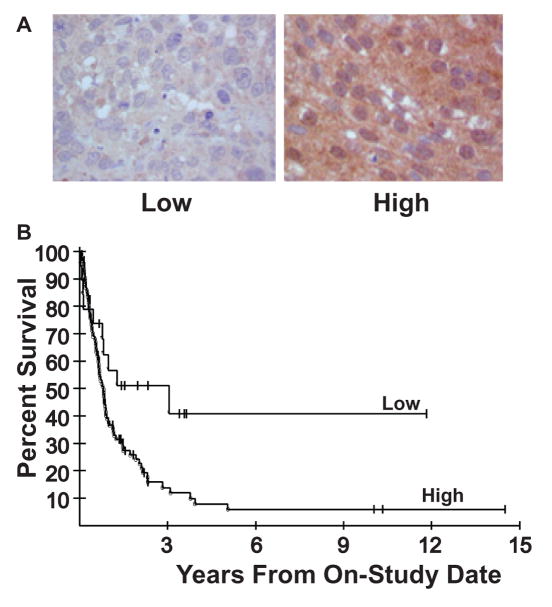
In order to further investigate HK2 as a potential molecular target for brain metastasis of breast cancer, we asked whether relative HK2 levels were associated with patient outcome post-resection. Immunohistochemistry (IHC) was performed on a cohort of 123 resected brain metastases of breast cancer. The cohort of resected brain metastases were scored as having no, low or high levels of HK2. Representative images of low and high HK2 staining levels are shown (Figure 5A). Ninety-five of the 123 tumors (77%) expressed high levels of HK2 staining and patient survival in this group was significantly reduced compared to the 19 of 123 (15%) patients low tumor HK2 levels (Figure 5B). The median survival for patients with high HK2 expression was 9.6 months while that for patients with low HK2 expression tumors was 17.5 months (P=0.028 for overall difference between the curves, by two-tailed log rank test). With regard to conventional histological markers, HK2 expression was not well associated with either Her-2 or ER expression (p=0.19 and 0.26, respectively, two-tailed Fisher’s exact test). Table 3 shows the results of a Cox proportional hazards model analysis of ER, Her-2 and HK2 staining data in this cohort. All three parameters were determined to be jointly statistically significant in this model. Furthermore, using a likelihood ratio test, HK2 expression provided a statistically significant increase in prognosis in the survival model, beyond that of ER and Her-2 together (P=0.0033). Thus, HK2 may be useful as a marker for prognosis in this population, if this finding were independently confirmed.
Table 3.
Cox proportional hazards model analysis of patient survival post-resection for brain metastasis of breast cancer1.
| Variable | Hazard Ratio (HR) | 95% Confidence Interval for HR | p value |
|---|---|---|---|
| Her-2 | 0.50 | 0.31–0.83 | 0.0065 |
| ER | 0.41 | 0.24–0.71 | 0.0015 |
| HK2 | 2.52 | 1.27–4.97 | 0.0080 |
Based on staining results from 113 patients with complete data on the three parameters.
Discussion
Brain metastases are increasing in incidence in metastatic breast cancer patients and often occur when a patient is responding to treatment or has stable disease systemically (3, 25). Several factors suggest that this niche site may be molecularly distinct: We previously documented a prominent neuro-inflammatory response between brain metastatic breast cancer cells and activated host microglia and astrocytes, both in the 231-BR mouse model and resected human brain metastases (26) and others have observed similar trends (27–29). These data suggest the existence of a unique tumor cell-microenvironmental interaction. Second, the brain is known as a sanctuary site (rev in (30)). While the blood-brain barrier (BBB) severely restricts access of drugs and cytokines to the normal brain, it is not well known to what extent the BBB is patent or compromised in developing brain metastases; to the extent that the BBB is patent, this would also contribute to an altered environment. Based on these observations we hypothesized that brain metastases would exhibit distinct gene expression trends. A microarray analysis of gene expression was conducted using flash frozen samples of resected brain metastases of breast cancer. It would be impossible to identify frozen matched primary tissues for these brain metastases, therefore unlinked primary breast tumors matched for hormone receptor and TNM status were used for comparison. While using imperfect matches, and limited in the number of specimens available, the potential insights afforded by actual clinical material as opposed to cell lines is inarguable. Another advantage of the experimental design was that epithelial cells were purified by LCM from each lesion, so that gene expression differences due to contaminating non-tumor cells in the surgical margin were not present. The microarray analysis indicated that brain metastases differentially express several genes by 2–3 fold, including phosphatases, GPI anchoring proteins and those regulating the extracellular matrix and cell adhesion. Differential gene expression was validated using a semi-independent set of brain metastases and primary tumors, the experimental design again dependent on availability of crainiotomy specimens suitable for molecular analysis. Statistically significant differences or trends (p<0.1) in differential gene expression were observed for 6 genes. Given the limited sample size, our conclusion is that all of these trends may be of biological importance. Several of these genes are under further investigation. Recently, Bos, et al. reported 243 genes that were differentially expressed between brain metastatic and primary cell lines. From this data set 2 genes overlap with our findings in the surgical specimens. CASK (calcium/calmodulin-dependent serine protein kinase after up-regulated in the brain metastatic cohorts while TRIM34 (tripartite motif-containing 34) was down-regulated (31).
HK2 was a compelling target for several reasons, including its reported involvement in bioenergetics and apoptosis. Hexokinase expression and activity is up-regulated in breast cancer as compared to normal breast tissue (32), and limited data suggested that it is further up-regulated in metastases (33). In addition, HK2 was one of few genes up-regulated in the brain metastases, so that inhibitors with potential therapeutic benefit could be considered.
Using the 231-BR model system, HK expression was down-regulated ~2-fold using two different shRNAs. Under normal tissue culture conditions, no difference was observed between the control and HK2 shRNA clones in ATP levels/viability or drug-induced apoptosis. However, normal serum glucose levels are 7–10 milligrams per liter, in contrast to a concentration of 4.5 grams per liter in normal cell culture medium. Therefore the experiments were repeated using 10% of the standard glucose concentration, conditions in which we expect that metabolism of glucose by the cells decreases the glucose concentration to physiological levels over the time course of the experiment. Under these conditions the HK2 knockdown cells exhibited altered growth kinetics. Viability, as assessed by both intracellular ATP concentrations and MTT assay, was reduced in the HK2 knockdowns. A reduction in proliferation by down-regulation of HK2 was also reported for Lovo colon cancer cells (34). Circumstantial evidence suggests that conditions of limited nutrient availability may exist in brain metastases. Necrotic centers were observed in at least 8 of 16 resected human brain metastases, indicative of poor vascularization (26). The data therefore suggest a functional role for altered HK2 expression in cell growth under conditions of limited nutrient availability. This difference will be difficult to confirm in vivo. The 231-BR model produces multiple brain metastases after intracardiac injection but mice must be routinely sacrificed one-month post-injection according to ACUC guidelines due to neurological deficits (paralysis) and weight loss, a point too early for the formation of large necrotic metastases (23, 26, 35). To overcome this, new models of breast cancer brain metastasis will be needed that recapitulate necrosis and conditions of nutrient availability.
No difference in apoptosis induced by drugs or radiation was observed between the control and HK2 knockdown clones (data not shown). These data may reflect the recent discovery that glucose metabolism can exert other functions to regulate apoptosis, such as the oxidation of cytochrome c (36). Thus, glucose metabolism pathways downstream of HK2 interaction with the mitochondrion may alter its effects on apoptosis. It is also possible that HK1 could compensate for the reduced expression of HK2 in the apoptotic pathway as HK1 is capable of binding to mitochondria.
The potential significance of HK2 overexpression in brain metastases of breast cancer was confirmed by an immunohistochemical analysis in a cohort of craniotomy specimens. Approximately 77% of the craniotomy specimens exhibited high HK2 expression, and this was associated with reduced survival post-resection (p=0.028). These data may not apply to all brain metastases, as patients with a limited number of brain lesions which are located in accessible regions of the brain are typical surgical candidates. If validated, therapeutic approaches to inhibit HK2 may be considered. While this enzyme is ubiquitous and instrumental for cell function, a reduction in its activity rather than a complete elimination may be an achievable target. Several compounds have been identified against the potential anti-apoptotic role for HK2 (19, 37–39), but would be of limited benefit in the 231-BR model system. Other agents directed at the bioenergetic activity of HK2, if BBB permeable, would be of high interest.
Materials and Methods
Specimens
Frozen brain metastases were collected from Vanderbilt University, the University of Schleswig-Holstein Medical Center (Kiel, Germany) and the National Institutes of Health. Frozen primary invasive breast samples were collected from the University of Schleswig-Holstein Medical Center (Kiel, Germany) or University of Tuebingen (Teubingen, Germany). All samples were anonymized and approved by the National Cancer Institute Office of Human Subjects Research. Diagnosis and histopathological characteristics were confirmed by a single pathologist before use in the study.
Expression profiling of tumor cohorts
All samples were subjected to laser capture microdissection (LCM) and RNA Amplification. Briefly, eight micron frozen tissue sections were thawed 15 seconds at room temperature and stained using Histogene LCM Frozen Section Staining Kit (Arcturus Engineering, Mountain View, CA) following the manufacturer’s instructions with the addition of a 5 minute incubation in 100% ethanol prior to xylene. Approximately 2000 laser captured pulses were collected for each sample using the Pix Cell II Laser Capture Microdissection System (Arcturus Engineering) in a 15 minute period. Captured cells were lysed off the LCM caps in PicoPure RNA Extraction buffer (PicoPure RNA Isolation Kit, Arcturus Engineering) for 30 minutes at 55 °C and frozen at −80 °C until ready to proceed. Total RNA was extracted according to the manufacturer’s instructions for the PicoPure RNA Isolation Kit. Quality and quantity of RNA was assessed using the RNA 6000 Pico Assay for the 2100 Bioanalyzer (Aligent Technologies, Waldbronn, Germany). A minimum of 10 ng of RNA was then subjected to two rounds of a T7 RNA polymerase catalyzed amplification protocol using the RiboAmp RNA Amplification Kit (Arcturus Engineering). Quality and quantity of the amplified aRNA was assessed using the RNA 6000 Nano Assay for the 2100 Bioanalyzer (Agilent Technologies). A reference RNA sample consisting of a pool of six breast cancer cell lines (MCF7, ZR-75, BT-474, BT-549, MDA-MB-231, and T-47-D) was simultaneously amplified.
cDNA microarray analysis: Cy3- or Cy5-dUTP labeled cDNA (GE Healthcare, Chalfont St. Giles, UK) was synthesized from 50–100 μg of amplified aRNA using random primed polymerization with Superscript II reverse transcriptase (Invitrogen, Calsbad, CA). Equal amounts of labeled cDNA for the test sample and the reference sample were hybridized to cDNA array for each tumor. Arrays were constructed on glass slides from PCR products representing the inserts of 28,411 IMAGE consortium cDNA clones in the NHGRI microarray core laboratory. Fluorescent intensities were measured using a Agilent scanner and scanned images were analyzed using DeArray software. Significance of Microarray (SAM) analysis was used for data mining and analysis. Supervised clustering was preformed where each sample was identified as brain metastasis or primary tumor and the ER or Her-2 status identified.
Quantitative real-time PCR (Q-PCR)
Double stranded cDNA was prepared using the second round synthesis protocol from the RiboAmp RNA amplification kit (Arcturus Engineering). Q-PCR reactions consisted of 1X SybrGreen Supermix, (BioRad Laboratories, Hercules, CA) 0.2 mM forward and reverse primers and 10 ng cDNA. Cycling conditions consisted of three-step amplification and melt curve analysis using the iQ5 Real Time PCR Detection System (BioRad Laboratories). For generating a standard curve, amplified cDNA from the reference sample detailed above was used in a 5 fold dilution series. Relative gene expression was calculated by dividing the specific expression value (starting quantity, ng) by the GAPDH expression value.
Cell Culture and Knockdown of HK-2 with shRNA
A brain metastatic derivative of the breast cancer cell line, MDA-MB-231, was isolated and described previously (24). These cells, designated 231-BR, were maintained in High Glucose DMEM medium (Invitrogen) and supplemented with 10% fetal bovine serum (FBS, Invitrogen) and penicillin-streptomycin (Invitrogen) at 37 °C in 5% CO2. Clones of 231-BR cells with stable knockdown of HK2 expression were generated using a commercial lentiviral system for introduction of short hairpin RNAs (shRNAs, Sigma, St. Louis, MI). Sigma Mission non-target shRNA (SHC002V) was used to generate control cell lines, and two shRNAs, TRCN 0000037669 and TRCN 0000037673 were used for knockdown of HK2 expression. Lentiviral particles (2×105) were added to 231-BR cells for 24 hours and cells were subsequently cultured in medium containing 1 μg/ml puromycin. Cells were maintained and expanded in this selective medium to select individual clones with stable knockdown.
Western blots analysis
Western blots were performed using standard protocols. Briefly, 231-BR cell lysates were prepared in RIPA buffer (20mM Tris-HCl, pH8; 100mM NaCl; 10% glycerol, 1% NP40; 0.5% Na deoxycholate, 0.1% SDS) supplemented with a complete, mini protease inhibitor cocktail (Roche, Mannheim, Germany). Protein lysates (50–100 μg) were separated using SDS –PAGE. Proteins were transferred to nitrocellulose and blots were incubated for 2 hours to overnight with the following antibodies: rabbit anti-Hexokinase type I polyclonal antibody (Millipore, Billerica, MA), 1:2000 dilution; rabbit anti-type II Hexokinase polyclonal antibody (Millipore, Billerica, MA), 1:5000 dilution; rabbit anti-HK-2 polyclonal antibody (Abgent, San Diego, CA), 1:500 dilution; rabbit anti-Hexokinase 3 polyclonal antibody (Abgent, San Diego, CA), 1:500 dilution; and rabbit anti-GCK (glucokinase, Hexokinase 4, Santa Cruz Biotechnology, Santa Cruz, CA), 1:200 dilution. Goat-anti-rabbit-HRP (1:5000, Santa Cruz Biotechnology, Santa Cruz, CA) was use as a secondary antibody. Enhanced chemiluminescence was used for detection.
Cell viability assay
Cells were seeded in normal growth medium (High Glucose DMEM, 10% FBS with no additional additives). This medium was diluted 1:10 with glucose-free DMEM for glucose limiting experiments and viability was measured at various time points. The Cell Titer-Glo Luminescent cell viability assay (Promega, Madison, WI) was used to measure cellular ATP levels. This assay was conducted in 96 well opaque-walled luminescence plates (Perkin-Elmer, Waltham, MA) and relative ATP levels measured using a luminometer (Promega GloMax). Replicate 96 well plates were prepared and each sample analyzed in quadruplicate wells.
Immunohistochemistry
Archival paraffin-embedded tumor samples were obtained at MD Anderson Cancer Center (Houston, TX) from patients with a history of breast cancer who presented with metastases to the CNS parenchyma. The median age of the cohort was 51 with a range from 27 to 75 years of age. Thirty-three of 124 patients (26.6%) had ER+ brain metastasis and 45 of 124 (36.2%) had Her-2 amplification. Sections from these samples were confirmed to have metastatic breast cancer present and subjected to immunohistochemistry (IHC) for HK2. Sections were processed and stained as previously described (40) using an anti-HK2 antibody (Millipore) at a 1:500 dilution with microwave antigen retrieval in 10 mM sodium citrate buffer. Scoring was performed using a 3-tiered system (no, low or high) for HK2 protein levels.
Statistical analysis
Survival of patients was determined from date of craniotomy until date of death or date of last follow-up as appropriate. The probability of survival as a function of time was determined by the Kaplan-Meier method, and the statistical significance of the difference between two Kaplan-Meier curves was determined by a two-tailed log-rank test. A Cox proportional hazards model was used to determine the statistical significance of HK2 on survival when evaluated along with ER and Her2 status. A likelihood ratio test was used to determine the statistical significance of HK2 when this parameter was added to a model which had consisted of ER and Her2 status, and provides the additional effect of this parameter when added to the model. A one-way ANOVA was performed on the average results from control clones and 3 HK2 knockdown clones, from n=3 experiments, for the in vitro data in Figure 3. The results were adjusted using Holm’s method. Comparisons of expression levels of genes were performed using an exact Wilcoxon rank sum test. In view of the number of tests performed, only p-values <= 0.01 should be interpreted as being statistically significant, while those for which 0.01 <p <0.05 would be considered strong trends.
Figure 4.
High HK2 expression in brain metastases predicts poor survival post-craniotomy. A. The HK2 expression of 123 resected brain metastases of breast cancer was determined by IHC; representative images of low and high levels of HK2 staining are shown. B. A Kaplan-Meier plot of patient survival post-craniotomy shows reduced survival in the high HK2 subset (P=0.028 by two-tailed log rank test).
Acknowledgments
We would like to thank Dr. Abdel Elkahloun for construction of the cDNA microarrys used herein. This research was supported by the Intramural program of the National Cancer Institute and by grant W81XWH-062-0033 from the Department of Defense Breast Cancer Research Program.
References
- 1.Lin N, Bellon J, Winer E. CNS metastases in breast cancer. J Clin Oncol. 2004;22:3608–17. doi: 10.1200/JCO.2004.01.175. [DOI] [PubMed] [Google Scholar]
- 2.Weil R, Palmieri D, Bronder J, Stark A, Steeg P. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167:913–20. doi: 10.1016/S0002-9440(10)61180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin NU, Winer EP. Brain metastases: the HER2 paradigm. Clin Cancer Res. 2007;13:1648–55. doi: 10.1158/1078-0432.CCR-06-2478. [DOI] [PubMed] [Google Scholar]
- 4.Evans A, James J, Cornford E, et al. Brain metastases from breast cancer: Identification of a high-risk group. Clin Oncol. 2004;16:345–9. doi: 10.1016/j.clon.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–14. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 6.Slimane K, Andre F, Delaloge S, et al. Risk factors for brain relapse in patients with metastatic breast cancer. Ann Oncol. 2004;15:1640–4. doi: 10.1093/annonc/mdh432. [DOI] [PubMed] [Google Scholar]
- 7.Pestalozzi BC, Zahrieh D, Price KN, et al. Identifying breast cancer patients at risk for Central Nervous System (CNS) metastases in trials of the International Breast Cancer Study Group (IBCSG) Ann Oncol. 2006;17:935–44. doi: 10.1093/annonc/mdl064. [DOI] [PubMed] [Google Scholar]
- 8.Carey LA, Ewend MG, Metzger R, et al. Central nervous system metastases in women after multimodality therapy for high risk breast cancer. Breast Cancer Res Treat. 2004;88:273–80. doi: 10.1007/s10549-004-0999-3. [DOI] [PubMed] [Google Scholar]
- 9.Ryberg M, Nielsen D, Osterlind K, Andersen PK, Skovsgaard T, Dombernowsky P. Predictors of central nervous system metastasis in patients with metastatic breast cancer. A competing risk analysis of 579 patients treated with epirubicin-based chemotherapy. Breast Cancer Res Treat. 2005;91:217–25. doi: 10.1007/s10549-005-0323-x. [DOI] [PubMed] [Google Scholar]
- 10.Tham YL, Sexton K, Kramer R, Hilsenbeck S, Elledge R. Primary breast cancer phenotypes associated with propensity for central nervous system metastases. Cancer. 2006;107:696–704. doi: 10.1002/cncr.22041. [DOI] [PubMed] [Google Scholar]
- 11.Hicks D, Short S, Prescott N, et al. Breast cancers with brain metastases are more likely to be estrogen receptor negative, express teh basal cytokeratin CK5/6 and over-express Her-2 or EGFR. Am J Surg Path. 2006;30:1097–104. doi: 10.1097/01.pas.0000213306.05811.b9. [DOI] [PubMed] [Google Scholar]
- 12.Peng SY, Lai PL, Pan HW, Hsiao LP, Hsu HC. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol Rep. 2008;19:1045–53. [PubMed] [Google Scholar]
- 13.Lyshchik A, Higashi T, Hara T, et al. Expression of glucose transporter-1, hexokinase-II, proliferating cell nuclear antigen and survival of patients with pancreatic cancer. Cancer Invest. 2007;25:154–62. doi: 10.1080/07357900701208931. [DOI] [PubMed] [Google Scholar]
- 14.Rho M, Kim J, Jee CD, et al. Expression of type 2 hexokinase and mitochondria-related genes in gastric carcinoma tissues and cell lines. Anticancer Res. 2007;27:251–8. [PubMed] [Google Scholar]
- 15.Smith TA. Mammalian hexokinases and their abnormal expression in cancer. Br J Biomed Sci. 2000;57:170–8. [PubMed] [Google Scholar]
- 16.Pedersen PL. Voltage dependent anion channels (VDACs): a brief introduction with a focus on the outer mitochondrial compartment’s roles together with hexokinase-2 in the “Warburg effect” in cancer. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9165-7. [DOI] [PubMed] [Google Scholar]
- 17.Galluzzi L, Kepp O, Tajeddine N, Kroemer G. Disruption of the hexokinase-VDAC complex for tumor therapy. Oncogene. 2008;27:4633–5. doi: 10.1038/onc.2008.114. [DOI] [PubMed] [Google Scholar]
- 18.Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–96. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 19.Machida K, Ohta Y, Osada H. Suppression of apoptosis by cyclophilin D via stabilization of hexokinase II mitochondrial binding in cancer cells. J Biol Chem. 2006;281:14314–20. doi: 10.1074/jbc.M513297200. [DOI] [PubMed] [Google Scholar]
- 20.Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paudyal B, Oriuchi N, Paudyal P, Higuchi T, Nakajima T, Endo K. Expression of glucose transporters and hexokinase II in cholangiocellular carcinoma compared using [18F]-2-fluro-2-deoxy-D-glucose positron emission tomography. Cancer Sci. 2008;99:260–6. doi: 10.1111/j.1349-7006.2007.00683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada K, Brink I, Bisse E, Epting T, Engelhardt R. Factors influencing [F-18] 2-fluoro-2-deoxy-D-glucose (F-18 FDG) uptake in melanoma cells: the role of proliferation rate, viability, glucose transporter expression and hexokinase activity. J Dermatol. 2005;32:316–34. doi: 10.1111/j.1346-8138.2005.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 23.Palmieri D, Bronder JL, Herring JM, et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;67:4190–8. doi: 10.1158/0008-5472.CAN-06-3316. [DOI] [PubMed] [Google Scholar]
- 24.Yoneda T, Williams P, Hiraga T, Niewolna M, Nishimura R. A bone seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking cloe in vivo and in vitro. J Bone and Mineral Res. 2001;16:1486–95. doi: 10.1359/jbmr.2001.16.8.1486. [DOI] [PubMed] [Google Scholar]
- 25.Bendell J, Domchek S, Burstein H, et al. Central Nervous System Metastases in Women who Receive Trastuzumab-Based Therapy for Metastatic Breast Carcinoma. Cancer. 2003;97:2972–7. doi: 10.1002/cncr.11436. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald D, Palmieri D, Hua E, et al. Reactive glia are recruited by highly proliferative brain metastases of breast cancer and promote tumor cell colonization. Clin Exp Metast. 2008;25:799–810. doi: 10.1007/s10585-008-9193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetti D, Li J, Shen R. Astrocytes contribute to the brain-metastatic specificity of melanoma cells by producing heparanase. Cancer Res. 2000;60:4767–70. [PubMed] [Google Scholar]
- 28.Sierra A, Price J, Garcia-Ramirez M, Mendez O, Lopez L, Fabra A. Astrocyte derived cytokines contribute to the metastatic brain specificity of breast cancer cells. Lab Invest. 1997;77:357–68. [PubMed] [Google Scholar]
- 29.Zhang M, Olsson Y. Reactions of astrocytes and microglial cells around hematogenous metastases of the human brain. Expression of endothelin-like immunoreactivity in reactive astrocytes and action of microglial cells. J Neurol Sci. 1995;134:26–32. doi: 10.1016/0022-510x(95)00227-9. [DOI] [PubMed] [Google Scholar]
- 30.Deeken JF, Loscher W. The blood-brain barrier and cancer: transporters, treatment, and trojan horses. Clin Cancer Res. 2007;13:1663–74. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]
- 31.Bos P, Zhang X, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balinsky D, Platz C, Lewis J. Enzyme activities in normal, dysplastic and cancerous human breast tissues. J Nat’l Cancer Inst. 1984;72:217–24. [PubMed] [Google Scholar]
- 33.Hennipman A, Oirschot Bv, Smits J, Rijksen G, Staal G. Glycolytic enzyme activities in breast cancer metastases. Tumour Biol. 1988;9:241–8. doi: 10.1159/000217568. [DOI] [PubMed] [Google Scholar]
- 34.Peng Q, Zhou Q, Zhong J, Pan F, Liang H. Stable RNA interference of hexokinase II gene inhibits human colon cancer LoVo cell growth in vitro and in vivo. Cancer Biol Ther. 2008;7:1128–35. doi: 10.4161/cbt.7.7.6199. [DOI] [PubMed] [Google Scholar]
- 35.Gril B, Palmieri D, Bronder JL, et al. Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain. J Natl Cancer Inst. 2008;100:1092–103. doi: 10.1093/jnci/djn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughn A, Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nature Cell Biol. 2008;10:1477–85. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penso J, Beitner R. Lithium detaches hexokinase from mitochondria and inhibits proliferation of B16 melanoma cells. Mol Genet Metab. 2003;78:74–8. doi: 10.1016/s1096-7192(02)00203-2. [DOI] [PubMed] [Google Scholar]
- 38.Penso J, Beitner R. Clotrimazole decreases glycolysis and the viability of lung carcinoma and colon adenocarcinoma cells. Eur J Pharmacol. 2002;451:227–35. doi: 10.1016/s0014-2999(02)02103-9. [DOI] [PubMed] [Google Scholar]
- 39.Goldin N, Heyfets LAA, Israelson A, et al. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase. Oncogene. 2008;27:4636–43. doi: 10.1038/onc.2008.108. [DOI] [PubMed] [Google Scholar]
- 40.Simmons ML, Lamborn KR, Takahashi M, et al. Analysis of complex relationships between age, p53, epidermal growth factor receptor, and survival in glioblastoma patients. Cancer Res. 2001;61:1122–8. [PubMed] [Google Scholar]