SUMMARY
Epigenetic mechanisms set apart the active and inactive regions in the genome of multicellular organisms to produce distinct cell fates during embryogenesis. Here we report on the epigenetic and transcriptome genome-wide maps of gastrula-stage Xenopus tropicalis embryos using massive parallel sequencing of cDNA (RNA-seq) and DNA obtained by chromatin immunoprecipitation (ChIP-seq) of histone H3 K4 and K27 trimethylation and RNA Polymerase II (RNAPII). These maps identify promoters and transcribed regions. Strikingly, genomic regions featuring opposing histone modifications are mostly transcribed, reflecting spatially regulated expression rather than bivalency as determined by expression profile analyses, sequential ChIP, and ChIP-seq on dissected embryos. Spatial differences in H3K27me3 deposition are predictive of localized gene expression. Moreover, the appearance of H3K4me3 coincides with zygotic gene activation, whereas H3K27me3 is predominantly deposited upon subsequent spatial restriction or repression of transcriptional regulators. These results reveal a hierarchy in the spatial control of zygotic gene activation.
INTRODUCTION
Changes in the chromatin state play an important role in developmental gene regulation; covalent posttranslational modifications of the N-terminal tails of histone proteins affect chromatin accessibility and serve to recruit effector molecules to mediate this regulation (reviewed in Strahl and Allis, 2000; Bhaumik et al., 2007; Taverna et al., 2007). Deposition of three methyl groups on lysine 4 of histone H3 (H3K4me3) generally occurs at the promoters of transcribed genes (Santos-Rosa et al., 2002). During Drosophila embryogenesis Trithorax group proteins (trxG) deposit this mark and positively regulate the expression of homeotic genes (Breen and Harte, 1993). In embryonic stem (ES) cells H3K4me3 was identified at active promoters including genes without elongation of transcription (Guenther et al., 2007). The H3K4me3 mark can directly interact with for example TAF3, a subunit of the general transcription initiation factor TFIID (Vermeulen et al., 2007), but it also recruits the chromatin-remodeling complex NURF (Wysocka et al., 2006) and CHD1, a protein involved in transcription elongation and pre-mRNA splicing (Sims et al., 2007).
Polycomb group (PcG) proteins, a well conserved group of transcriptional repressors, are required during the development of multicellular organisms (Schuettengruber et al., 2007). The methyl groups of lysine 27 of histone H3 (H3K27me3) are respectively deposited and bound by Polycomb repressor complexes PRC2 and PRC1 (Cao et al., 2002; Schuettengruber et al., 2007). In cultured embryonic stem cells H3K4me3 and H3K27me3 can co-occupy a subset of promoters (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). This ‘bivalent’ configuration results in gene repression but may poise developmental regulator genes for later transcriptional activation. During differentiation of the stem cells one of the two marks is lost at most loci, leading to either activation or stable repression, but the co-occurrence of H3K4me3 and H3K27me3 does not exclusively arise in pluripotent cells (Barski et al., 2007; Mikkelsen et al., 2007; Pan et al., 2007; Cui et al., 2009). Bivalency is infrequent in Drosophila embryos (Schuettengruber et al., 2009). The Drosophila trxG and PcG proteins act as antagonistic regulators at the Drosophila Hox gene Ultrabithorax where the H3K4 methyl transferase Ash1 selectively prevents lysine 27 methylation at the promoter (Papp and Muller, 2006). Antagonism of the two histone H3 modifications in mammalian cells is mediated by Rbp2 (Jarid1a), a histone H3 lysine 4 demethylase that interacts with PRC2, and UTX, a histone H3 lysine 27 demethylase that interacts with MLL H3K4 methyl transferase complexes (Lee et al., 2007; Pasini et al., 2008).
Characterizing epigenetic control in vertebrate embryos could shed more light on early events such as zygotic gene activation and the spatial control of chromatin states underlying germ layer specification, patterning and morphogenesis in vertebrates. In Xenopus, the onset of embryonic transcription takes place at the mid-blastula transition (MBT; Newport and Kirschner, 1982a, b). Global gene repression is relieved by a change in the cytoplasm-to-nucleus ratio during the blastula stages (Newport and Kirschner, 1982a, b) which, at least in part, is mediated by the competing effects of repressive chromatin and the transcription machinery (Prioleau et al., 1994; Almouzni and Wolffe, 1995; Veenstra et al., 1999). Knockdown of DNA methyltransferase 1 (DNMT1) led to precocious transcription, revealing a role in maintaining transcriptional repression before the MBT (Stancheva and Meehan, 2000; Dunican et al., 2008). During gastrulation histone B4 is replaced by histone H1 (Smith et al., 1988; Dimitrov et al., 1993), which causes selective repression of oocyte but not somatic 5S transcription, as well as a loss of mesoderm competence (Wolffe, 1989; Bouvet et al., 1994; Steinbach et al., 1997). Global assessment of histone modifications by western blotting and mass spectrometry in oocytes, sperm, sperm nuclei incubated in egg extract, cell lines and erythrocytes revealed that the differentiation status is correlated with histone modification signatures (Nicklay et al., 2009; Shechter et al., 2009).
These observations suggest that chromatin plays an important role during early development, however chromatin state maps and the dynamics of epigenetic control in early vertebrate embryos are unknown. Here we report on genome-wide profiles of H3K4me3, H3K27me3, RNA polymerase II (RNAPII) and the transcriptome of Xenopus tropicalis gastrula embryos using chromatin immunoprecipitation (ChIP) experiments and massive parallel sequencing. The H3K27me3 mark correlates with spatial regulation of expressed transcription factor genes. The bivalent configuration is not predominant, and chromatin-bound H3K4me3 and H3K27me3 emerge after the MBT, coincidental with zygotic transcription activation and transcriptional repression respectively. The analyses reveal a hierarchy in activating the embryonic genome by activating and repressive histone marks and spatially restricted transcriptional regulators.
RESULTS
Histone methylation profiles and the transcriptome of Xenopus tropicalis gastrula embryos
To generate epigenetic profiles, ChIP was performed using specific antibodies against trimethylated H3K4 and H3K27 in Xenopus gastrula-stage embryos (Nieuwkoop-Faber stage 11–12), followed by deep sequencing (ChIP-seq). In addition, polyA-selected RNA (stages 10-13) was reverse-transcribed and sequenced (RNA-seq). We also carried out a RNAPII ChIP-seq. The reads were mapped to the X. tropicalis genome, Joint Genome Institute version 4.1 (Klein et al., 2002; Klein et al., 2006). For details see experimental procedures and supplementary information (Table S1).
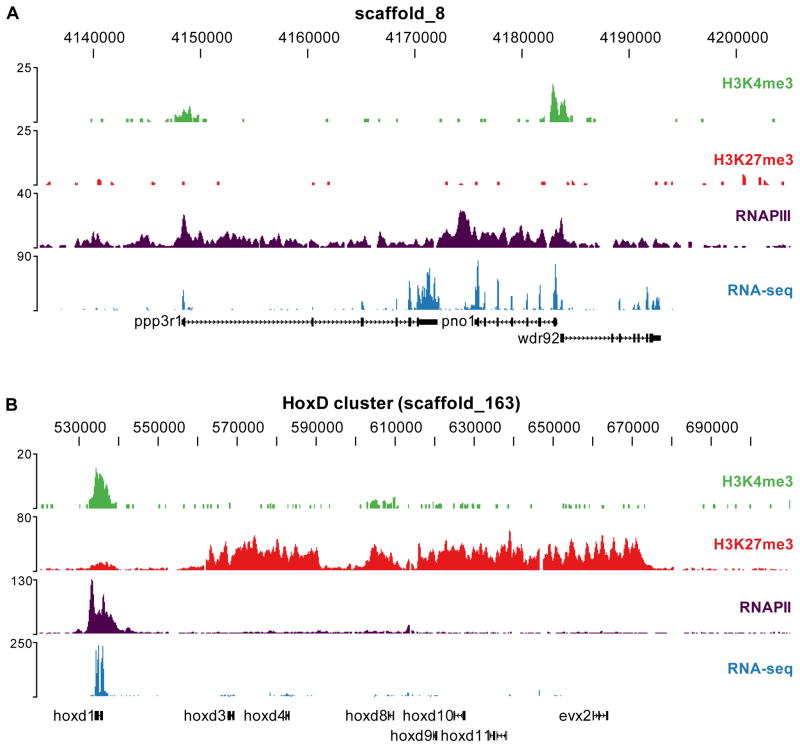
The high resolution profiles show that H3K4me3 is enriched at the 5′ end of genes (shown for a number of representative loci; Figure 1A). The H3K4me3 signals coincide with transcribed loci as is shown by a strong RNAPII signal throughout the gene body as well as RNA-seq evidence. H3K27me3 is enriched in broad domains as is illustrated for example by the HoxD cluster (Figure 1B). The hoxd1 gene is marked with H3K4me3 and shows expression, whereas a widespread presence of repressive H3K27me3 decorates the other HoxD genes at this stage. To determine the extent of genome-wide H3K4me3- and H3K27me3-enrichment, peaks were identified with a sliding window approach (see experimental procedures). A total of 12,281 and 3,599 regions were found enriched for H3K4me3 and H3K27me3 respectively. These numbers are comparable to those previously found (Zhao et al., 2007) for the same modifications in human ES cells (17,167 and 4,392 regions). To validate these sets, randomly chosen regions were experimentally validated and a false discovery rate of <0.06 was found (Figure S1). In addition, EZH2, the catalytic PRC2 subunit responsible for H3K27 methylation, was detected solely at genes that are enriched for H3K27me3 (Figure S2). In conclusion, robust epigenetic profiles and a sensitive transcriptome profile were obtained using deep sequencing of gastrula X. tropicalis embryos.
Figure 1. Histone methylation profiles and the transcriptome of the Xenopus tropicalis gastrula embryo.
Profiles of H3K4me3 (green), H3K27me3 (red), RNAPII (purple) and RNA-seq (blue) are visualized using the UCSC Genome Browser for two genomic regions. (A) The upper panel shows a region on scaffold_8 with three expressed genes (ppp3r1, pno1 and wdr92). (B) The lower panel shows the HoxD cluster on scaffold_163.
Genes with both H3K4me3 and H3K27me3 are expressed in gastrula embryos
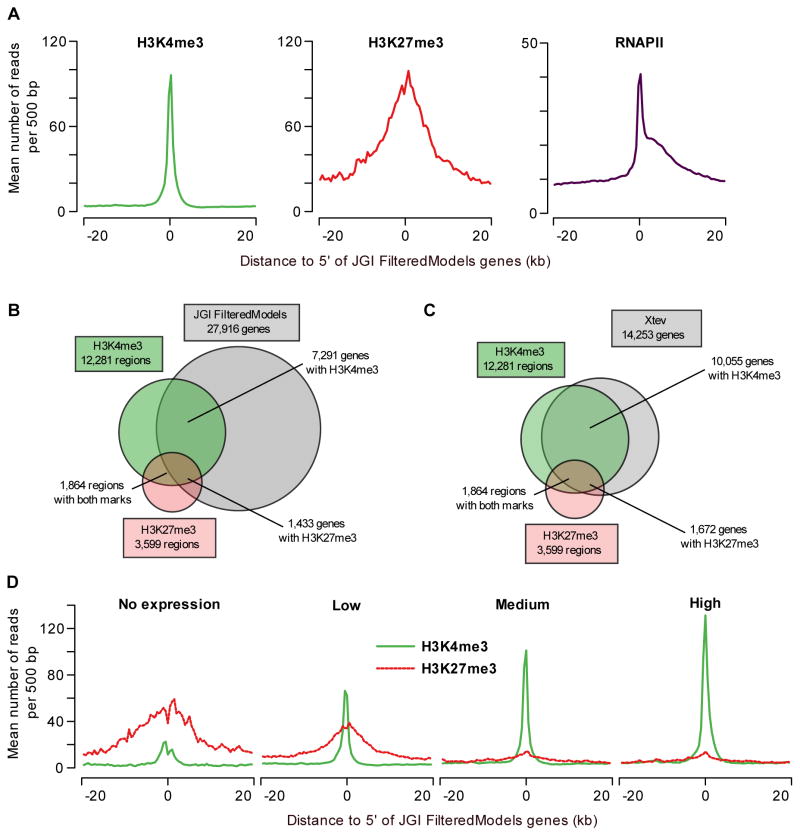
The Joint Genome Institute generated a collection of gene models based on gene prediction algorithms (FilteredModels genes v1, JGI FM). We compared the distribution of both chromatin modifications and RNAPII around the transcription start site (TSS) of JGI FM genes (Figure 2A). H3K4me3- and H3K27me3-enriched regions are strongly associated with the 5′ ends of genes; H3K27me3 however is distributed more broadly compared to H3K4me3. RNAPII enrichment also peaks at the TSS but is also detected in the gene body, as expected. To determine the number of peaks that associate with JGI FM genes, the peaks were correlated to annotation within 1kb of the transcription start site. A considerable number of H3K4me3 peaks cannot be correlated to the present annotation (Figure 2B). These H3K4me3-enriched regions that show no overlap with JGI FM genes likely represent upstream promoters of suboptimal annotated regions and novel transcribed regions. We also determined the overlap with other gene collection databases. The Xenopus model organism database (Xenbase) contains gene annotations based on a subset of JGI FM genes, improved by manual curation (Bowes et al., 2008). In addition, we include Ensembl genes, RefSeq genes and human genes mapped to X. tropicalis. To compare these gene collections the occurrence of H3K4me3 peaks within 1kb of the annotated 5′ ends was determined (Figure S3). The JGI FM genes (of 27,916 gene models), represented the most inclusive Xenopus gene set and overlapped with the most H3K4me3-enriched regions. However, all these databases contained a unique set of genes overlapping with H3K4me3 peaks that were not present in any other collection of genes (Figure S3), showing that the current annotation is not optimal yet. We used the RNA-seq expression and splice junction information in combination with H3K4me3 and RNAPII ChIP-seq data to combine the (unique) models of different gene databases and improve the current gene annotation based on experimental evidence. Gene models from Xenbase, JGI, Ensembl and Refseq were combined and updated with EST (Gilchrist et al., 2004) and RNA-seq data, and H3K4me3 and RNAPII data were used to validate or update the 5′ ends of the gene models (for details see Figures S4 and S5). This annotation pipeline resulted in a collection of 14,253 Xenopus tropicalis experimentally validated gene models (Xtev genes; Tables S2 and S3). A comparison of the overlap between H3K4me3- and H3K27me3-enriched regions at both JGI FM genes and Xtev genes is shown in Figures 2B and 2C. Out of the 14,253 Xtev genes 10,055 (71%) have a H3K4me3-enriched region within 1kb of the 5′ end of the gene, which is a marked improvement over 7,291 JGI FM genes (26%) associated with a H3K4me3-enriched region. For further analyses the Xtev genes have been used, except when a genome-wide coverage including non-expressed genes was appropriate.
Figure 2. Primary data analysis and comparison to gene models.
(A) Profiles of H3K4me3, H3K27me3 and RNAPII at annotated genes. The average H3K4me3 read coverage for genes with a H3K4me3-enriched region within 1kb of the JGI FM genes annotated 5′ end is shown in the left panel (mean number of reads per 500bp, green). The middle panel shows the equivalent for H3K27me3 (red) and the right panel for RNAPII (purple). (B) Overlap of H3K4me3- and H3K27me3-enriched regions with JGI FM genes (within 1kb of the annotated 5′ end). (C) Overlap of H3K4me3- and H3K27me3-enriched regions with Xenopus tropicalis experimentally validated genes (within 1kb of the annotated 5′ end). (D) H3K4me3 and H3K27me3 correlate with gene expression levels. JGI FM genes were divided in equal-sized groups based on normalized RNA-seq expression level (no, low, medium and high expression). For these groups H3K4me3 (green) and H3K27me3 (red-dotted) occupancy profiles (mean number of reads per 500bp) are shown.
H3K4me3- and H3K27me3-enriched regions are associated with 10,055 and 1,672 Xtev genes respectively (Figure 2C). Gene Ontology analysis of genes marked by H3K27me3 showed that this epigenetic modification decorates many genes with a function in development and transcriptional regulation (Figure S6 and Table S4). For example, as many as 46 out of 50 top ranking H3K27me3-enriched genes encode transcription factors (Table S5), many of which are known to play important roles during embryonic development. Genes with H3K27me3 in ES cells are also tied to roles in transcriptional regulation and development (Boyer et al., 2006).
The correlation of H3K4me3 with transcriptional activation and that of H3K27me3 with repression has not yet been studied in vertebrate embryos. Therefore, the presence of H3K4me3 and H3K27me3 at the promoter was compared with expression levels as determined by their RNA-seq levels (Figure 2D). For this purpose the RNA-seq reads were mapped to JGI FM genes and normalized expression levels were calculated. The balance between H3K4me3 and H3K27me3 correlates well with gene expression levels and association of RNAPII (Figures 2D and S7A, R2 of H3K4me3~RNAPII = 0.58). On silent genes H3K27me3 dominates, whereas highly expressed genes show a strong enrichment for H3K4me3. Genes with a low expression level show both marks, and conversely, genes with both marks recruit RNAPII (Figures 2D and S7B, Table S6). In total 72% of the JGI FM genes marked with both H3K4me3 and H3K27me3 are expressed in Xenopus embryos. At face value this is in contrast to studies in ES cells in which bivalent, co-occurring H3K4me3 and H3K27me3 histone modifications repress developmental regulators (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). To explore this issue we compared the double-marked genes in Xenopus to bivalent genes in mouse and human ES cells. Among orthologs of human and mouse bivalent genes, on average 18% is marked by both H3K4me3 and H3K27me3 in Xenopus (p<10−69), a majority of which is expressed (64%; Table 1). It has been reported that genes that are bivalent in mouse ES cells frequently have a paused RNAPII (Stock et al., 2007). In Xenopus gastrula embryos however, the RNAPII pausing index (Welboren et al., 2009) of genes with both histone modifications was not higher than that of genes enriched for H3K4me3 only (Figure S8). All these observations raise the question whether the two marks co-occupy the same nucleosomal DNA or occupy the locus in different cells of the embryo.
Table 1. Overlap double-marked regions in Xenopus compared to mouse and human ES cells.
Orthology mapping was based on Ensembl gene IDs. ~20% of double-marked genes in X. tropicalis are registered as bivalent genes in either mouse or human ES cells. In general, these genes are expressed in Xenopus embryos (~64%), whereas these genes are repressed in the ES cells. 72.0% of all JGI FM genes that have double markings are expressed in Xenopus embryos.
| Dataset | organism | bivalent genes | X. trop. orthologs of bivalent genes (Ensembl) | double-marked X. trop. orthologs (Ensembl) | % | double-marked JGI FM genes | expressed genes | % |
|---|---|---|---|---|---|---|---|---|
| Mikkelsen et al | mouse | 2.632 | 2.090 | 388 | 18.6 | 334 | 213 | 63.8 |
| Pan et al | human | 3.244 | 2.432 | 390 | 16.0 | 334 | 209 | 62.6 |
| Zhao et al | human | 1.766 | 1.361 | 283 | 20.8 | 249 | 160 | 64.3 |
H3K4me3 and H3K27me3 decorate different nucleosomal DNA populations
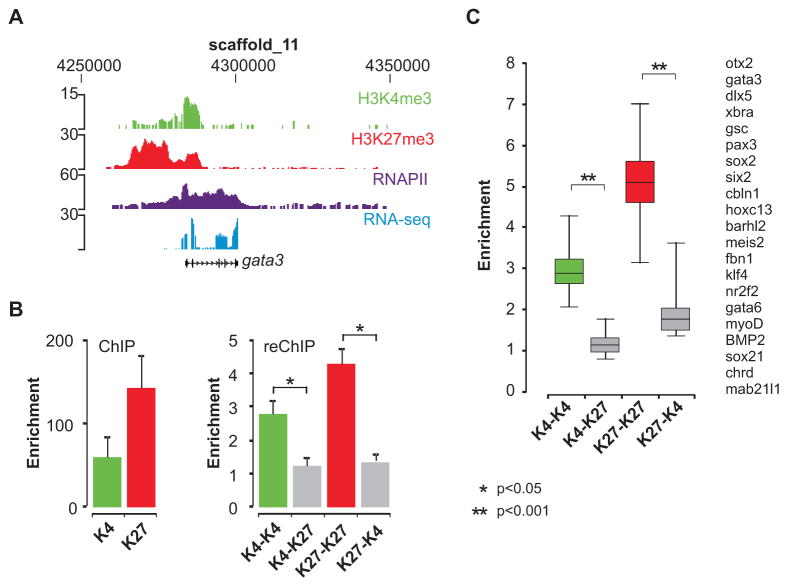
Gata3 is an example of a bivalent gene in ES cells which in embryos displays enrichment for both H3K4me3 and H3K27me3 and is robustly expressed, as indicated by the abundant presence of RNAPII and RNA-seq reads (Figure 3A). To directly examine the bivalent chromatin state of genes in the embryo sequential ChIP was performed. In these experiments the material obtained after the first round of ChIP is subjected to a second immunoprecipitation with a different antibody (reChIP). Positive signals in the reChIP would allow the conclusion that histones with the two different modifications co-occupy the same nucleosomal DNA. Either an H3K4me3 or an H3K27me3 antibody was used in the first ChIP, followed by a reChIP with either the same antibody (positive control) or the antibody for the other histone modification. To account for the signal generated by residual antibody of the first IP, an essential beads-only negative control was also included in the reChIP. The gata3 gene is enriched for H3K4me3 and H3K27me3 in the first round of ChIP (Figure 3B, left panel). In the reChIP with different antibodies (K4–K27 and K27-K4), the signals are comparable to the beads-only control, whereas the positive control reactions (K4-K4 and K27-K27) yielded significantly more chromatin, establishing a functional reChIP (Figure 3B, right panel). In total we tested 21 double-marked genes in the reChIP, including 9 genes identified as bivalent in previous studies (Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007) which are not expressed in Xenopus embryos. This revealed that most of the genes that have both marks show enrichment of less than two-fold when the opposite antibody is used in the reChIP (Figure 3C). The same results are obtained when the order of the antibodies is reversed. We did not observe any difference between expressed and silent double-marked genes. A relatively minor K27-K4 enrichment was observed for 6 out of 21 genes, but this effect is only observed when the sequential ChIP is started with the H3K27me3 antibody and not when the order is reversed (Figure S9). Therefore bivalent marking of genes is not a prevalent configuration in Xenopus embryos. It appears that K4me3- and K27me3-modified histone H3 mark different DNA fragments, presumably derived from different cells in gastrula-stage embryos. This raises the possibility that the two marks decorate loci in a spatially regulated fashion, contributing to the regulation of localized gene expression.
Figure 3. Sequential ChIP experiments for genes enriched for H3K4me3 and H3K27me3.
(A) The gata3 locus is visualized using the UCSC genome browser; H3K4me3 (green), H3K27me3 (red), RNAPII (purple) and RNA-seq (blue). (B) Enrichment of H3K4me3 and H3K27me3 in the first round of ChIP (left panel), calculated as the fold over the background of a negative locus. Sequential ChIP enables to examine the presence of two histone modifications at the same chromatin fragment. The eluted chromatin fraction of the first ChIP was used as the starting material for a second round of ChIP (reChIP). In the reChIP (right panel) enrichment was calculated relative to a beads-only (no antibody) control. This is the most relevant control to determine enrichment due to the relatively high background generated by residual antibody of the first ChIP reaction. Enrichment in the reChIP (K4–K27 and K27-K4, grey bars) was less than two-fold. ReChIP with the same antibody shows strong signals (K4-K4, green bars and K27-K27, red bars). Enrichment values are presented as the mean + SEM of five independent experiments. Asterisks indicate p value <0.05 (t-test). (C) ReChIP results for 21 double-marked genes visualized in a boxplot. Double asterisks indicate p value <0.001 (t-test).
H3K27me3 is associated with spatially regulated genes
To study the spatial epigenetic markup of the embryo, genes with differential spatial expression in the embryo along either the dorso-ventral or the animal-vegetal axis (Zhao et al., 2008) were investigated for the H3K4me3 and H3K27me3 occupancy of their promoters (Figure 4). Genes that are preferentially expressed in the animal or the vegetal pole are both enriched with H3K4me3 at their promoters. However, H3K27me3 is mainly detected at promoters of genes that are preferentially expressed at the vegetal pole. Likewise, genes that show either dorsal- or ventral-specific expression are mostly trimethylated at H3K27. This is in contrast to genes which show no clear differential expression along either axis, where H3K27me3 is hardly observed at the 5′ end (Figure 4). Similarly, genes with preferential expression in anterior mesoderm, posterior mesoderm or notochord (Tanegashima et al., 2008) are also enriched for H3K27me3, in contrast to genes lacking a localized expression (Figure S10). Transcripts that are enriched in the animal pole represent the only group of genes with localized expression that lack H3K27me3; these transcripts are also expressed albeit at lower levels at the vegetal pole. In general however, double-marked loci correspond to spatially regulated genes (Figures 4 and S10).
Figure 4. H3K27me3 is linked to localized repression patterns.
H3K27me3 marks genes spatially regulated along the dorso-ventral and animal-vegetal axis. Xenopus embryo explant microarray data (animal and vegetal cap, dorsal and ventral marginal zone) was compared to H3K4me3 and H3K27me3 occupancy at promoters of differentially regulated genes. H3K27me3 is enriched at 5′ ends of genes which are at least 2-fold higher expressed in the vegetal side of the embryo compared to the animal cap. Genes which are either preferentially expressed (2-fold difference) at the dorsal or at the ventral side of the embryo are also enriched for H3K27me3 at their 5′ end. Number of genes in these groups: animal 211; vegetal 201; dorsal 33; ventral 22; no differential expression 2955.
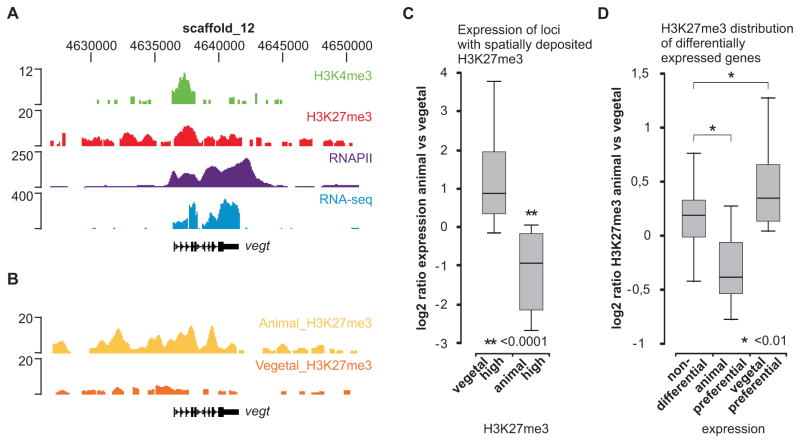
To experimentally validate these findings and test the predictive power of spatially deposited histone marks, gastrula-stage embryos (stage 10–12) were dissected in animal and vegetal halves, and animal and vegetal H3K27me3 ChIP-seq profiles were generated. The profiles observed for the vegt gene, which is decorated with both histone modifications (Figure 5A) and is expressed at the vegetal pole (Zhang and King, 1996), show that H3K27me3 is enriched in the animal hemisphere at this locus (Figure 5B). To assess this relationship in a genome-wide fashion, the reads obtained for the two samples were normalized and we calculated the animal/vegetal H3K27me3 ratio for all genes. Genes with more than two-fold difference in H3K27me3 between animal and vegetal hemispheres (Table S7) displayed localized expression patterns (Figure 5C and S11). Interestingly, the genes with animal hemisphere H3K27me3 include genes that are regulated by vegt like xbra, gsc and sox17a (Engleka et al., 2001; Vonica and Gumbiner, 2002; Messenger et al., 2005). Conversely, genes with higher expression in the animal or vegetal hemisphere according to microarray data (Zhao et al., 2008) have different animal/vegetal H3K27me3 ChIP-seq ratios. Genes that are preferentially expressed in the vegetal half have a significantly higher (p<0.01) animal/vegetal H3K27me3 ratio compared to genes that are not differentially expressed (Figure 5D). Reciprocally, genes with higher expression in the animal hemisphere have a lower animal/vegetal H3K27me3 ratio. In conclusion, these results indicate that the H3K27me3 histone modification is spatially deposited, which is predictive of expression patterns within the embryo.
Figure 5. Spatial deposition of H3K27me3 is predictive of localized expression.
(A) The vegt locus is visualized using the UCSC genome browser; H3K4me3 (green), H3K27me3 (red), RNAPII (purple) and RNA-seq (blue). (B) H3K27me3 ChIP-seq of animal and vegetal halves (stage 10–12). The vegt gene is most enriched for H3K27me3 in the animal hemisphere. (C) qRT-PCR expression ratio (animal/vegetal) of genes with animal-high (n=4) or vegetal-high (n=25) H3K27me3 (normalized H3K27me3 ChIP-seq ratio, 2Log > 1). (D) Normalized H3K27me3 ChIP-seq ratio (2Log animal/vegetal) of genes with expression differences: non-differential (n=195), animal preferential (n=9) and vegetal preferential genes (n=35). Numbers refer to genes that are both enriched for H3K27me3 in whole embryos and included in the microarray data (Zhao et al., 2008).
Temporal delay of H3K27me3 deposition after the mid-blastula transition
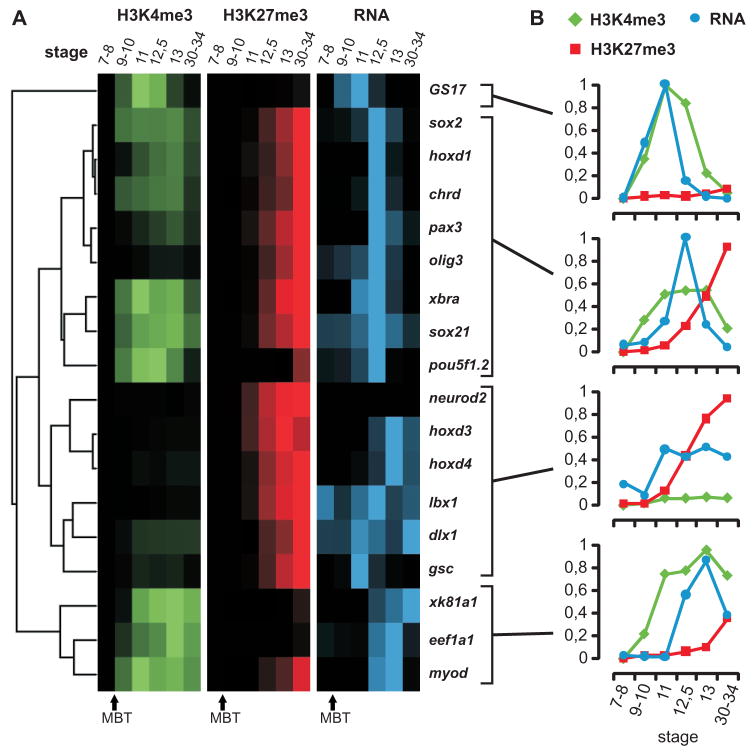
To determine the temporal aspect of epigenetic regulation, the interplay of H3K4me3, H3K27me3 and gene expression levels between blastula and larval stages was explored (st. 7–34). ChIP-qPCR and qRT-PCR was performed with material obtained from different stages of development, including pre-mid-blastula transition stages (pre-MBT; stage 7–8), before the onset of embryonic transcription. Genes known to play important roles in development were selected for this analysis and ChIP values relative to RNA levels were hierarchically clustered and visualized in a heat map (Figures 6 and S12). The clusters found in this analysis differ in both the kinetics and the extent of transcriptional activation during early development. For all tested genes chromatin-bound H3K4me3 and H3K27me3 signals emerge after the MBT, whereas histone H3 is detected before this stage with an H3-core antibody. No substantial trimethylation of H3K27 is observed on genes before the mid-gastrula stage (stage 11), whereas H3K4me3 is observed between the MBT and the start of gastrulation. For most genes transcription activation coincides with or follows the appearance of H3K4me3, and reduced mRNA levels correlate with increasing levels of H3K27me3 and a drop in H3K4me3 (Figure 6 and S12). GS17, one of the early genes that is actively transcribed after the MBT (cluster 1), is an exception showing no deposition of H3K27me3 upon downregulation. Deposition of H3K4me3 precedes accumulation of transcript in the second cluster of genes, with expression peaking toward the end of gastrulation (stage 12.5). These genes are generally robustly expressed during gastrulation, which distinguishes this cluster from the third cluster that is characterized by low H3K4me3; these genes are also expressed at relatively low levels. For example, the neuro2d promoter is only enriched for H3K27me3 in the absence of expression, whereas Lbx1 and dlx1 are low for H3K4me3 and transcription levels fluctuate over time with increasing H3K27me3. Some of these transcripts likely originate from maternal RNA since RNA is detected before the MBT. The fourth cluster consists of genes that have high levels of H3K4me3 and are robustly expressed at the end of gastrulation, whereas H3K27me shows relatively little enrichment, with the exception of myoD which shows an increase in H3K27me3 levels at larval stages. To visualize the ChIP signals appearing at a gene locus, several primers set were developed for the xbra locus and tested in ChIP-qPCR. H3K27me3 appears at the promoter concomitantly with repression and subsequently spreads within the locus (Figure S13). Most of the genes analyzed here show deposition of H3K4me3 prior to or concomitant with transcriptional activation, without significant prior enrichment for H3K27me3 (p<0.001 for difference at stage 9–10).
Figure 6. Dynamic regulation of epigenetic marks and gene expression during development.
(A) Hierarchical clustering results of relative ChIP recoveries for H3K4me3 (green) and H3K27me3 (red) and RT-PCR for stages 7–34 of developmentally regulated genes (blue). Intensities of the signal show high or low ChIP enrichment and expression. (B) The panels show the average relative ChIP recoveries for H3K4me3 (green), H3K27me3 (red) and expression values (qRT-PCR in blue) of the corresponding stages 7–34 for the four clusters. ChIP signals represent the average of a biological duplicate experiment.
DISCUSSION
This study uncovers a key feature of epigenetic regulation in vertebrate embryos. H3K4me3 and H3K27me3 modifications strongly correlate with expression levels but these epigenetic marks do not represent the bivalent configuration as they by and large do not co-occur on the same nucleosomal DNA. Gene loci with both modifications have RNAPII in the gene body and 72% are expressed. Correlation of the epigenetic profiles with spatially regulated genes identified by microarray expression analysis (Tanegashima et al., 2008; Zhao et al., 2008) reveals that the H3K27me3 mark is associated with genes that display spatial regulation of expression. ChIP-sequencing for H3K27me3 in animal and vegetal halves shows that spatial differences in deposition of the Polycomb mark are predictive of spatially regulated expression in the embryo. Concordantly, paused RNAPII was not observed at the double-marked genes in Xenopus embryos, whereas RNAPII elongation is impaired at bivalent genes (Stock et al., 2007). The number of targets of double-marked regions in Xenopus embryos is comparable to the number of regions with bivalent status in mouse and human ES cells (2000–3000 promoters; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). There is a considerable overlap of H3K4me3- and H3K27me3-modified genes between ES cells and Xenopus embryos (~18%). However, two thirds of these double-marked regions in Xenopus embryos correspond to expressed genes, rather than genes that are poised for activation upon differentiation. H3K4me3 and H3K27me3 tend to be separated in embryonic space and time. It is possible that the two marks co-occur on the same nucleosomal DNA with low frequency in Xenopus embryos. We suggest that true bivalency may result from the co-occurring antagonistic forces of activation and repression in a given cell rather than representing a special mechanism of gene activation. It is possible that the key to bivalency is in the dynamics of differentiation. Cells differentiating from a progenitor into different lineages tend to be spatially separated in the embryo but do still express competing activities for alternative transcription programs. Therefore bivalency may particularly be prevalent in situations in which differentiation is inhibited, representing a ‘holding pattern’. The antagonism between the marks is likely mediated by complexes harboring both histone methyltransferase and demethylase activities (Lee et al., 2007; Pasini et al., 2008), a conserved feature of developmental gene regulation, whereas the extent to which bivalency is observed may depend on the dynamics of differentiation, which is variable between systems.
The data reported here reveal a hierarchy in epigenetic and transcriptional regulation that exhibits both spatial and temporal aspects. In most cases H3K4me3 precedes or coincides with transcriptional activation after the mid-blastula stage. Well after the deposition of H3K4me3 and the onset of embryonic transcription, H3K27me3 accumulates and is deposited on many transcription factor genes, coinciding with repression or spatial restriction of gene expression. These spatially regulated transcription factors in turn activate or repress their cognate downstream targets within their expression domains as is seen for the vegt gene regulatory network. The relatively late role for H3K27me3 is concordant with an earlier observation that the RNA of the Xenopus Polycomb homolog, a subunit of the PRC1 complex, is only translated after the MBT (Strouboulis et al., 1999). Consistent with a role well after the onset of transcription, YY1, a homolog of Drosophila Pho, interacts with the PRC2 subunit EED and both proteins are involved in neural induction in Xenopus (Satijn et al., 2001). Experimental validation of the PRC2 subunit EZH2 revealed that this subunit was only detected on gene loci that are enriched for H3K27me3.
The epigenetic hierarchy of activation and repression is likely to extend to other epigenetic marks and to involve other factors, such as maternal activators and induced repressors. For example, activation of mesoderm gene expression is observed in embryos in which protein synthesis is inhibited, however proper spatial restriction of expression requires induced repressors (Latinkic et al., 1997; Lerchner et al., 2000; Kurth et al., 2005). DNA methylation has been implicated in gene activation at the MBT at specific gene loci (Stancheva et al., 2002). Other epigenetic cross-talk likely includes histone acetylation and methylation at different residues, for example H3K9me3, which like H3K4me3 and H3K27me3 is much more abundant in somatic cells when compared to sperm and pronuclei (Shechter et al., 2009). In embryos, stored diacetylated histone H4 becomes incorporated at cleavages stages, but is deacetylated between cleavage stages and gastrulation (Dimitrov et al., 1993). During later development WDR5 is essential for deposition of H3K4me3 and spatial regulation of Hox genes (Wysocka et al., 2005). These and other studies show that epigenetic regulation is highly dynamic and interconnected during development.
Additional genome-wide localization and functional studies will provide important new insights in the role of other epigenetic marks, chromatin modifying enzymes and chromatin remodelers, and how they control spatio-temporal patterns of gene expression during development, which will enhance our current knowledge in transcription regulation and the complex interplay between different cells and tissues in the embryo.
EXPERIMENTAL PROCEDURES
Animal procedures
Xenopus tropicalis embryos were obtained by natural mating, dejellied in 3% cysteine and collected at the indicated stage. Dissections were performed on in vitro fertilized embryos using a Gastromaster microsurgery apparatus (Xenotek Engeneering) equipped with white tips.
Chromatin immunoprecipitation and antibodies
Chromatin harvesting and ChIP was performed as described (Jallow et al., 2004) with minor modifications: 12.5μl of Prot A/G beads (Santa Cruz) were used and during reversal of crosslinking proteinase K was omitted from the buffer. The following antibodies were used: α-H3K4me3 (Abcam, ab8580), α-H3K27me3 (a kind gift from J. Martens and T. Jenuwein; Peters et al., 2003), RNAPII (Diagenode AC-055-100), EZH2 (Active Motif, 39103). For sequential ChIP, chromatin of the first ChIP was eluted from the beads in 100 μl elution buffer (100 mM NaHCO3 pH 8.8; 1% SDS), diluted 10 times in incubation buffer (50 mM Tris pH 8.0; 100 mM NaCl; 2 mM EDTA; 1 mM DTT; 1% NP40; protease inhibitor cocktail, Roche) and a second round of ChIP was performed.
RNA preparation for RNA-Seq
X. tropicalis embryos of stage 10–13 were collected and total RNA was isolated using Trizol and the Qiagen RNeasy Kit. Subsequently, polyadenylated RNA was selected twice with the Oligotex mRNA kit (Qiagen) to remove rRNA. cDNA was prepared with random hexamer primers using Superscript III (Invitrogen) and the second strand was made with DNA polymerase I, DNA ligase and T4 DNA polymerase. The purified double-stranded cDNA was used for Illumina sample preparation.
Quantitative (RT-) PCR
PCR reactions were performed on a MyIQ single color real-time PCR detection system (BioRad) using iQ SYBR Green Supermix (BioRad). Primer sequences are available upon request.
Sample preparation and sequencing
Sequencing samples were prepared according to the manufacturer’s protocol (Illumina). Shortly, adapter sequences were linked to the generated ChIP and cDNA samples, the library was size selected (200–250bp) and amplified by PCR. The subsequent sequencing was carried out on a Genome Analyzer (Illumina).
Alignment of reads
We used ELAND (GAPipeline version 1.0, Illumina) to map sequence reads to the Xenopus tropicalis genome, Joint Genome Institute, assembly version 4.1 (Klein et al., 2002; Klein et al., 2006). All unfiltered (Illumina Chastity Filter) 32-mer reads were aligned allowing up to two mismatches. For the histone modifications all mapped reads were extended to 133 bp (estimated fragment length). See table S1 for statistics.
Data availability
The data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE14025 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14025). Visualization tracks are available at the authors’ web site (http://www.ncmls.nl/gertjanveenstra) and at Xenbase (http://www.xenbase.org).
Detection of enriched H3K4me3 and H3K27me3 regions
Enriched regions were defined using a read count threshold with a sliding window approach (window of 500bp, threshold of 9 and 12 reads for H3K4me3 and H3K27me3 respectively). These thresholds were chosen on basis of a Monte-Carlo simulation where we randomly placed reads on the mappable part of scaffold_1 (ELAND) and called the number of enriched regions in this simulated dataset. This procedure was repeated 1000 times, and a False Discovery Rate (FDR) was calculated for each threshold. We chose the threshold corresponding to the same theoretical FDR (<0.001) for both samples, and validated randomly chosen regions by ChIP-qPCR.
Generation of distribution profiles
The mean number of mapped reads per 500bp was computed for 20kb around the annotated 5′ end of genes with an enriched region for H3K4me3, H3K27me3 and RNAPII within 1kb.
Gene Ontology analysis
Gene Ontology annotations for JGI FilteredModels genes v1 were obtained from Xenbase (http://www.xenbase.org), and the analysis was carried out using Ontologizer (Grossmann et al., 2007). We used the Parent-Child method with Westfall-Young-Single-Step multiple testing correction and set a corrected p-value threshold of 0.01.
Normalized expression level and detection call of expressed genes
Using an approach similar to (Sultan et al., 2008), the normalized expression level was calculated as the number of reads mapped to a gene divided by the total number of unique 32-mers in a gene:
Nunique was determined by generating all possible 32-mers for each gene and mapping these to the Xenopus tropicalis genome and the JGI FM genes using ELAND. All 32-mers that mapped uniquely to the JGI FM genes and did not map to multiple sites in the genome were counted as unique.
Comparison with explant microarray data
Xenopus laevis explant microarray data were downloaded from the Gene Expression Omnibus (accession GSE8990) and ArrayExpress (accession E-MEXP-717). We used RMA normalization (limma package, Bioconductor; Gentleman et al., 2004) on the raw expression values as provided by the submitter. All probe sequences from the microarray design were retrieved from Affymetrix (http://www.affymetrix.com) and were mapped to the JGI FM genes with blat (Kent, 2002). Hits were filtered based on the following criteria: 30% of the probe matches, at least 50% similarity, no more than 4 matches. Using the microarray data we compared the expression in different explants, and selected JGI FM genes with a 2-fold or higher difference in expression. Histone modification profiles were computed as described above.
Supplementary Material
Acknowledgments
We thank Kris Vleminckx for Xenopus tropicalis frogs and Ron Engels for animal care. This work was funded by grants of the US National Institutes of Health (grant 5 R01 HD054356) and the Netherlands Organization of Scientific Research (NWO-ALW VIDI grant 864.03.002) to GJCV.
Footnotes
The supplemental data includes thirteen figures and seven tables and can be found with this article online at http://www.cell.com/developmental-cell/supplemental/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. The EMBO journal. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nature structural & molecular biology. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Dimitrov S, Wolffe AP. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes & development. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic acids research. 2008;36:D761–767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Breen TR, Harte PJ. Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell stem cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Almouzni G, Dasso M, Wolffe AP. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Developmental biology. 1993;160:214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development (Cambridge, England) 2008;135:1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka MJ, Craig EJ, Kessler DS. VegT activation of Sox17 at the midblastula transition alters the response to nodal signals in the vegetal endoderm domain. Developmental biology. 2001;237:159–172. doi: 10.1006/dbio.2001.0366. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist MJ, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Developmental biology. 2004;271:498–516. doi: 10.1016/j.ydbio.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics. 2007;23:3024–3031. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci U S A. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Gerhard DS, Wagner L, Richardson P, Schriml LM, Sater AK, Warren WC, McPherson JD. Resources for genetic and genomic studies of Xenopus. Methods Mol Biol. 2006;322:1–16. doi: 10.1007/978-1-59745-000-3_1. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Kurth T, Meissner S, Schackel S, Steinbeisser H. Establishment of mesodermal gene expression patterns in early Xenopus embryos: the role of repression. Dev Dyn. 2005;233:418–429. doi: 10.1002/dvdy.20342. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Umbhauer M, Neal KA, Lerchner W, Smith JC, Cunliffe V. The Xenopus Brachyury promoter is activated by FGF and low concentrations of activin and suppressed by high concentrations of activin and by paired-type homeodomain proteins. Genes & development. 1997;11:3265–3276. doi: 10.1101/gad.11.23.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science (New York, NY. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Latinkic BV, Remacle JE, Huylebroeck D, Smith JC. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development (Cambridge, England) 2000;127:2729–2739. doi: 10.1242/dev.127.12.2729. [DOI] [PubMed] [Google Scholar]
- Messenger NJ, Kabitschke C, Andrews R, Grimmer D, Nunez Miguel R, Blundell TL, Smith JC, Wardle FC. Functional specificity of the Xenopus T-domain protein Brachyury is conferred by its ability to interact with Smad1. Developmental cell. 2005;8:599–610. doi: 10.1016/j.devcel.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nicklay JJ, Shechter D, Chitta RK, Garcia BA, Shabanowitz J, Allis CD, Hunt DF. Analysis of histones in Xenopus laevis. II. mass spectrometry reveals an index of cell type-specific modifications on H3 and H4. The Journal of biological chemistry. 2009;284:1075–1085. doi: 10.1074/jbc.M807274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell stem cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes & development. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & development. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Mechali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Molecular and cellular biology. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS biology. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Nicklay JJ, Chitta RK, Shabanowitz J, Hunt DF, Allis CD. Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. The Journal of biological chemistry. 2009;284:1064–1074. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Dworkin-Rastl E, Dworkin MB. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes & development. 1988;2:1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Developmental biology. 2002;243:155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & development. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Steinbach OC, Wolffe AP, Rupp RA. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature cell biology. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Damjanovski S, Vermaak D, Meric F, Wolffe AP. Transcriptional repression by XPc1, a new Polycomb homolog in Xenopus laevis embryos, is independent of histone deacetylase. Mol Cell Biol. 1999;19:3958–3968. doi: 10.1128/mcb.19.6.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Tanegashima K, Zhao H, Dawid IB. WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. The EMBO journal. 2008;27:606–617. doi: 10.1038/emboj.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Destree OH, Wolffe AP. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Molecular and cellular biology. 1999;19:7972–7982. doi: 10.1128/mcb.19.12.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vonica A, Gumbiner BM. Zygotic Wnt activity is required for Brachyury expression in the early Xenopus laevis embryo. Developmental biology. 2002;250:112–127. doi: 10.1006/dbio.2002.0786. [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. The EMBO journal. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. The EMBO journal. 1989;8:527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development (Cambridge, England) 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell stem cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE14025 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14025). Visualization tracks are available at the authors’ web site (http://www.ncmls.nl/gertjanveenstra) and at Xenbase (http://www.xenbase.org).