Abstract
It has been proposed that separase-dependent centriole disengagement at anaphase licenses centrosomes for duplication in the next cell cycle. Here we test whether such a mechanism exists in intact human cells. Loss of separase blocked centriole disengagement during mitotic exit and delayed assembly of new centrioles during the following S phase; however, most engagements were eventually dissolved. We identified Polo-like kinase 1 (Plk1) as a parallel activator of centriole disengagement. Timed inhibition of Plk1 mapped its critical period of action to late G2 or early M phase, i.e., prior to securin destruction and separase activation at anaphase onset. Crucially, when cells exited mitosis after downregulation of both separase and Plk1, centriole disengagement failed completely, and subsequent centriole duplication in interphase was also blocked. Our results indicate that Plk1 and separase act at different times during M phase to license centrosome duplication, reminiscent of their roles in removing cohesin from chromosomes.
Introduction
The centrosome is the major microtubule organizing center (MTOC) in most animal cells and strongly influences spindle assembly during mitosis (Luders and Stearns, 2007). As a consequence, centrosome number must be precisely regulated to ensure genome stability. During interphase, actively proliferating cells contain two centrosomes that are juxtaposed to form a single MTOC. Depending on the cell cycle stage, the core of each centrosome consists of either a single centriole, or a pair of orthogonally opposed, or engaged, centrioles, surrounded by pericentriolar material (PCM) that nucleates and organizes microtubule arrays (Azimzadeh and Bornens, 2007; Bettencourt-Dias and Glover, 2007). Because centrioles dictate PCM localization and thus determine the number of centrosomes, from a mechanistic perspective, the problem of centrosome duplication resolves to the question of how centriole duplication is controlled and coordinated with other cell cycle events.
Cells begin G1 phase with two centrosomes that each contain a single centriole. During S phase, a new (daughter) centriole grows from the lateral surface of each pre-existing (mother) centriole, due to the combined influence of Cdk2/cyclin E activity and a conserved set of centriole assembly factors (Azimzadeh and Bornens, 2007; Bettencourt-Dias and Glover, 2007; Nigg, 2007). Importantly, although this event doubles the number of centrioles, each daughter centriole remains engaged with (and shares the same PCM as) its mother. Thus, centriole duplication per se does not cause an immediate change in the total number of centrosomes. Rather, this occurs only upon passage through mitosis and cytokinesis, when each centrosome associates with one of the two spindle poles and is inherited by the corresponding daughter cell. Around the same time, the paired centrioles within each centrosome disengage (Kuriyama and Borisy, 1981), enabling the daughter centriole ultimately to acquire its own PCM and form a new centrosome.
Beyond its temporal restriction to S phase, centriole duplication is also governed by centrosome-intrinsic mechanisms. For example, in normal cells a mother centriole produces only a single daughter centriole, regardless of the length of S phase (Wong and Stearns, 2003). However, this restrictive control does not preclude centriole duplication in G1 centrosomes that are exposed to S or G2 phase cytoplasm via cell fusion (Wong and Stearns, 2003). Conversely, if the daughter centriole within an S phase centrosome is intentionally destroyed, the mother centriole regains its ability to produce a new daughter centriole (Loncarek et al., 2008). Together, these findings suggest that the physical engagement between mother and daughter centrioles creates a cis-acting block to further rounds of centriole assembly that is relieved only as cells pass through M phase, thereby entraining centrosome duplication to the broader cell division cycle.
Despite its fundamental role in centrosome biology, centriole disengagement remains poorly understood at the molecular level. Whereas RNAi screens in nematodes, flies, and mammalian tissue culture cells have uncovered multiple gene products necessary for centriole duplication in S phase (Azimzadeh and Bornens, 2007; Bettencourt-Dias and Glover, 2007; Nigg, 2007), none have thus far been identified which are required for centriole disengagement during M phase exit. Nevertheless, recent in vitro experiments implicate the mitotic protease separase in this process (Tsou and Stearns, 2006b). This enzyme becomes active at anaphase onset and triggers sister chromatid disjunction via endoproteolytic cleavage of cohesin (Nasmyth, 2002), but also controls aspects of M phase exit via nonproteolytic mechanisms (Gorr et al., 2006; Kudo et al., 2006; Stegmeier et al., 2002; Sullivan and Uhlmann, 2003). Specifically, it was observed that purified human centrosomes undergo anaphase-specific disengagement when added to Xenopus egg extracts, unless these extracts are first treated with high levels of nondegradable cyclin B or securin, treatments known to inhibit separase (Tsou and Stearns, 2006b).
While both securin and cyclin B clearly inhibit separase (Gorr et al., 2005), whether separase is in fact their relevant target vis-à-vis centriole disengagement remains unsettled, as flies and mice with hypomorphic or conditional separase alleles lack obvious defects in centrosome duplication, and by implication, centriole disengagement (Kumada et al., 2006; Pandey et al., 2005; Wirth et al., 2006). One possibility is that securin and cyclin B both inhibit an additional component of the egg extract that is distinct from separase. Alternatively, this discrepancy could be explained by technical limitations in the rate or completeness of separase inactivation in vivo, resulting in an intermediate level of function that is inadequate for chromatid disjunction but sufficient for centriole disengagement. A similar situation has recently been described in budding yeast; temperature-sensitive esp1 (separase) alleles that fully block cohesin cleavage delay mitotic exit only slightly (Stegmeier et al., 2002; Sullivan and Uhlmann, 2003), but if these alleles are genetically engineered to enhance their thermolability, then both chromosome segregation and mitotic exit are blocked with high penetrance (Queralt et al., 2006), as also occurs in strains that express nondegradable securin (Cohen-Fix and Koshland, 1999; Tinker-Kulberg and Morgan, 1999). Finally, separase could be essential for centriole disengagement only in meiotic cells or extracts, but partially or completely rescued by a second disengagement pathway in somatic cells. A similar redundancy has been suggested to explain why separase is required for M phase exit in oocytes but not mouse embryonic fibroblasts (MEFs) (Gorr et al., 2006; Kudo et al., 2006).
To elucidate the mechanisms responsible for centriole disengagement and replicative licensing in vivo, we engineered human somatic cells with conditional-null alleles of hESPL1 (the locus encoding hSeparase) so that the endogenous protease could be fully depleted. We also developed a correlated timelapse/immunofluorescence microscopy assay that allowed us to monitor the temporal coupling between centriole disengagement and mitotic exit in individual cells. Using these tools, we report that without separase, engaged centrioles are no longer disengaged in synchrony with M phase exit, but instead persist well into G1 phase. Centriole disengagement requires the proteolytic activity of separase but is not affected by overexpression of noncleavable cohesin, suggesting that separase promotes disengagement through cleavage of an additional non-cohesin substrate. However, longer-term observation revealed that engaged centrioles are not completely stable in cells lacking hSeparase, implying the existence of an additional disengagement-promoting activity. Using highly specific pharmacologic and chemical genetic inhibitors, we identified Polo-like kinase 1 (Plk1) as a positive regulator of centriole disengagement, and further mapped its execution point temporally to late G2 or early mitosis, upstream of securin destruction and separase activation in anaphase. Combined inactivation of both separase and Plk1 eliminated disengagement, and most importantly, prevented cells from assembling new centrioles upon entry into S phase. We conclude that Plk1 and separase act during M phase to license centrioles for duplication in the following cell cycle. Taken together, these results provide a molecular description of the mechanism that limits centrosome duplication to a single round per cell cycle in vivo.
Results
To test the role of separase in centriole disengagement, we wished to analyze cells with null mutations in the gene encoding separase. Although mice with conditional mESPL1 alleles exist, these alleles may permit the synthesis of fragments of mSeparase that, although proteolytically inactive, retain some cell-cycle regulatory functions (Kumada et al., 2006; Wirth et al., 2006). To eliminate this potential caveat we targeted the gene encoding the human enzyme, for which both N-terminal and C-terminal antibodies exist (Chestukhin et al., 2003; Jallepalli et al., 2001; Papi et al., 2005; Waizenegger et al., 2000). Briefly, HCT116 cells (a diploid and karyotypically stable human colorectal cell line) were sequentially infected with two adeno-associated virus (AAV)-based gene-targeting vectors, such that exon 21 of the hESPL1 locus was either flanked by loxP sites or deleted outright (Supplementary Fig. S1A). Three independent hESPL1flox/Δ clones were derived in this manner and verified by Southern blotting (Supplementary Fig. S1B).
To inactivate separase, hESPL1flox/Δ cells were infected with an adenovirus expressing Cre recombinase (AdCre), or with an adenovirus expressing β-galactosidase (Adβgal) as a negative control. Within 48 hours of AdCre infection, both full-length hSeparase (220 kDa) and its autocleavage fragments (160 kDa and 60 kDa) were lost, without the appearance of any new immunoreactive species (Fig. 1A). Cytologically, homozygous deletion of hESPL1 resulted in a severe block to anaphase chromosome segregation, as judged by live imaging of cells expressing GFP-tagged histone H2B (Supplementary Movies 1 and 2) and immunofluorescence microscopy of fixed cells stained with antibodies to centromere-specific autoantigens (CREST) and INCENP, which dissociates from centromeres at anaphase onset (Fig. 1B). Despite failing anaphase, hESPL1Δ/Δ cells exited M phase and progressed through further rounds of DNA replication, resulting in polyploidy (Supplementary Fig. S1C) and the accumulation of so-called “diplochromosomes” (sets of four cohesed sister chromatids) by cytogenetic analysis (Supplementary Fig. S1D). Notably, this chronic retention of chromosome cohesion occurred despite timely loss of the cohesin protector Sgo1 from centromeres (Fig. 1B). In contrast, the Drosophila Sgo1 ortholog MeiS-332 remains stably bound to anaphase centromeres in the absence of separase (Lee et al., 2004). Based on these results, we conclude that our conditional gene-targeting strategy eliminates expression of all hSeparase-derived polypeptides and blocks sister chromatid separation but not overall cell cycle progression, similar to mESPL1-deficient MEFs (Kumada et al., 2006; Wirth et al., 2006).
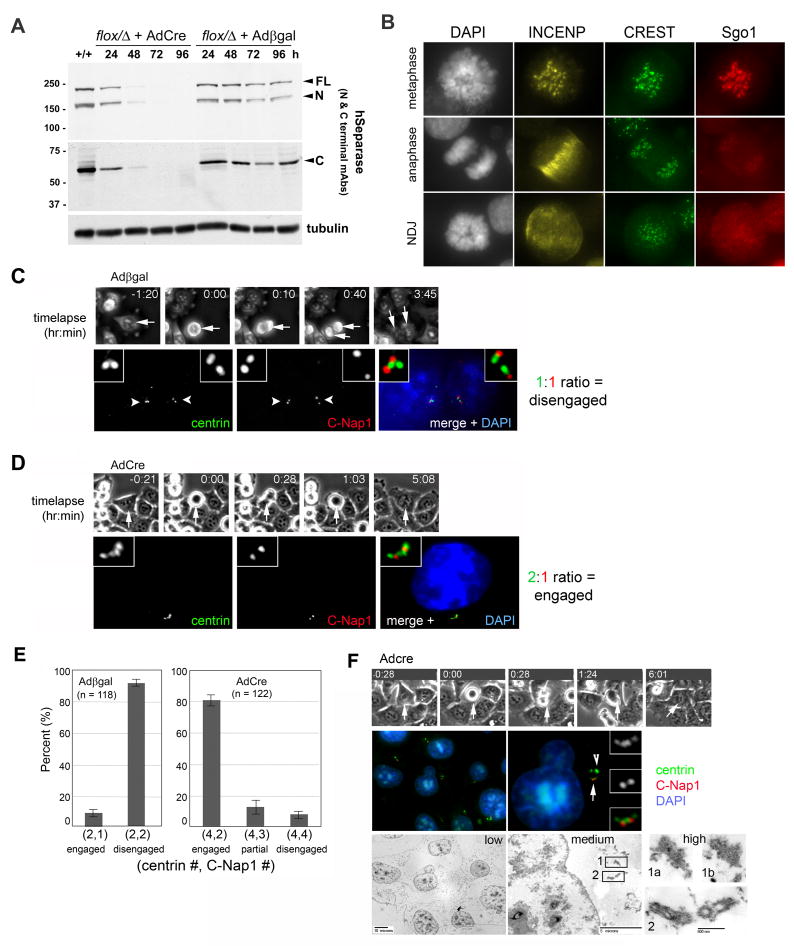
Figure 1. hSeparase is essential for centriole disengagement during M phase exit.
(A) Homozygous deletion of hESPL1 generates cells devoid of hSeparase-derived polypeptides. hESPL1flox/Δ cells were generated by homologous recombination (Supplementary Fig. S1) and infected with adenoviruses expressing Cre recombinase (AdCre) or β-galactosidase (Adβgal) as a negative control. Cells were harvested at the indicated times post-infection and analyzed by immunoblotting. Bands corresponding to full-length hSeparase (FL) and auto-cleaved N- and C-terminal fragments (N and C) are highlighted with arrowheads. (B) hESPL1flox/Δ cells in metaphase and anaphase (top two rows) and hESPL1Δ/Δ cells (48 hours after AdCre infection) undergoing nondisjunction in anaphase (NDJ, bottom row) were stained with antibodies to INCENP (yellow), CREST antiserum (green), and Sgo1 (red). (C and D) hESPL1flox/Δ cells infected with Adβgal (C) or AdCre (D) were traced by timelapse microscopy. After fixation and staining, cells that had exited mitosis 2 to 6 hours earlier were relocated and scored for centriole engagement with anti-centrin (green) and anti-C-Nap1 (red) antibodies. Nuclei were visualized with DAPI (blue). (E) Quantification of C and D. Error bars indicate standard deviations from three independent experiments. (F) A hESPL1Δ/Δ cell was traced through M phase exit by timelapse microscopy, permeabilized, and stained with anti-centrin and anti-C-Nap1 antibodies. Arrow and arrowhead indicate centrioles and centrin aggregates respectively. After acquiring fluorescent images, the same cell was fixed and processed for electron microscopy. Electron micrographs are shown at three different magnifications to facilitate correlation between images and visualization of centriolar structures. Box 1 highlights two consecutive sections of an electron-dense centrin aggregate; box 2, two pairs of engaged centrioles.
We next used this system to address hSeparase's role in centriole disengagement. We developed a correlated timelapse/immunofluorescence microscopy assay for analyzing both centriole configuration and cell cycle position in individual cells (Fig. 1C-E). Briefly, hESPL1flox/Δ cells were plated onto gridded coverslips and infected with AdCre or Adβgal as above. Beginning 40 hours after infection, coverslips were imaged by phase-contrast microscopy for 12 hours to identify cells entering and exiting mitosis, then fixed and stained with antibodies to centrin and C-Nap1, which differentially mark engaged versus disengaged centrioles (Tsou and Stearns, 2006b). From the timelapse recordings, cells that had exited mitosis 2.5 to 6 hours earlier, and thus were in G1 phase at the time of fixation, were identified. These cells were relocated on the coverslip and scored for their pattern of centrin and C-Nap1 staining. 93% of Adβgal-infected hESPL1flox/Δ cells exhibited a 1:1 ratio of centrin and C-Nap1 foci in each G1-phase daughter cell (Fig. 1C,E), indicating successful centriole disengagement. In contrast, 81% of AdCre-infected cells that reached G1 after anaphase failure exhibited a G2-like centriole pattern, with four centrin foci and two C-Nap1 foci (a 2:1 ratio; Fig. 1D,E). To confirm that these centrioles were engaged, we performed correlated timelapse/immunofluorescence/electron microscopy (Fig. 1F). hESPL1Δ/Δ cells that had exited mitosis 4 to 6 hours earlier were located and immunostained with antibodies to centrin and C-Nap1. After acquisition of fluorescent signals, the same cell was further fixed and processed for transmission electron microscopy (n = 4). Both pairs of centrioles in each of these cells were engaged (i.e., in an orthogonal configuration; Fig. 1F), in agreement with the localization of centrin and C-Nap1 by light microscopy. We also occasionally observed additional centrin-containing aggregates of variable size in the cytoplasm (Fig. S2). The nature of these aggregates is not known, but they lack typical microtubule structures that define centrioles (Fig. S2 and Fig. 1F, box 1a,b). In addition, although centrin is enriched in these structures, the centriolar proteins pericentrin, HsSas-6, and C-Nap1 are absent (data not shown). These data demonstrate that hSeparase is needed for centriole pairs to disengage in vivo after M phase exit.
In principle, the lack of disengagement in hESPL1Δ/Δ cells could reflect a direct positive requirement for separase, or an indirect negative effect of anaphase failure. To distinguish between these two alternatives, we analyzed the centrioles of cells that express a noncleavable form of the cohesin subunit Scc1 (Scc1NC) under the control of a tetracycline-regulated promoter (Hauf et al., 2001). We again used correlative timelapse microscopy and focused our analysis on the most severely affected population of Scc1NC cells (∼ 5% of the total) in which anaphase chromatid disjunction failed completely, giving rise to a cell with a single undivided nucleus (Fig. 2A). All cells examined (n = 9) exhibited 1:1 ratios of centrin and C-Nap1 staining, indicating that centriole disengagement does not depend on successful anaphase chromosome segregation. Thus, the absence of chromatid disjunction cannot explain the failure of centriole disengagement in hESPL1Δ/Δ cells. Rather, it appears that hSeparase triggers both events in parallel during M phase exit.
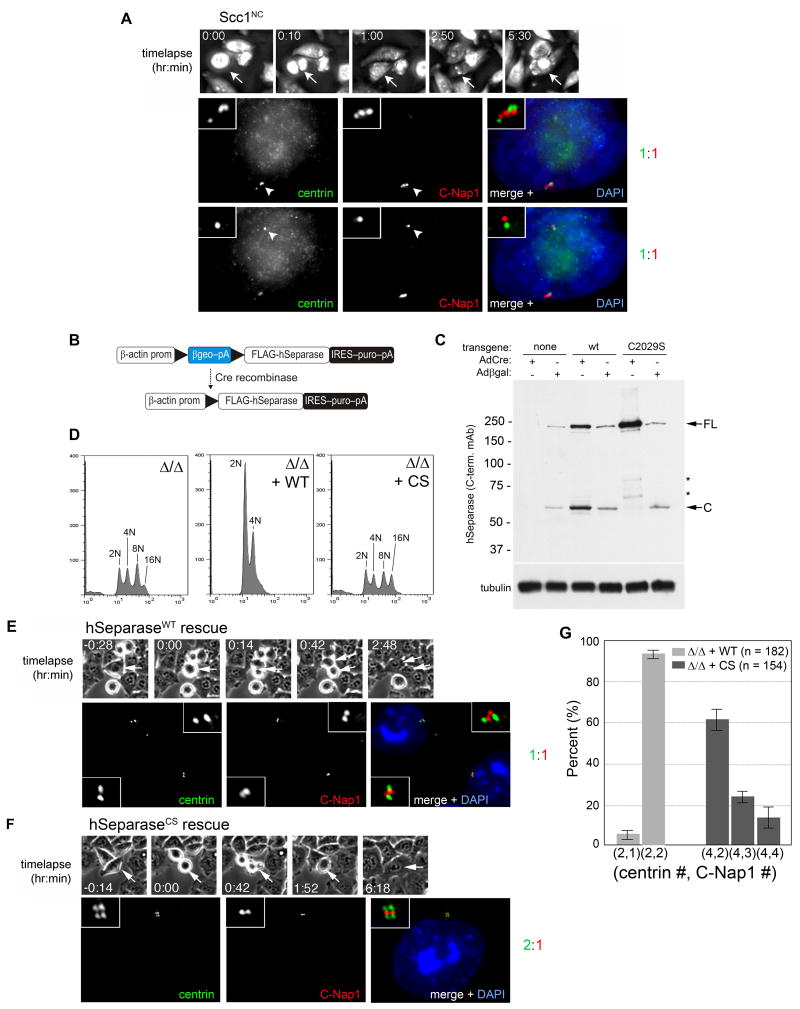
Figure 2. Centriole disengagement requires hSeparase-mediated proteolysis but not sister chromatid disjunction.
(A) HeLa cells expressing noncleavable Scc1 (Scc1NC) from a tetracycline-regulated promoter were analyzed by correlative timelapse-immunofluorescence microscopy as in Fig. 1. Top and bottom rows display disengaged centrioles in two different focal planes. (B) Strategy for Cre recombinase-dependent expression of hSeparase transgenes. (C) hESPL1flox/Δ cells harboring the indicated transgenes were infected with AdCre or Adβgal and analyzed by immunoblotting with a monoclonal antibody specific for the C-terminus of hSeparase. Asterisks denote breakdown products. Tubulin was used to confirm equal loading. (D) Cells in C were analyzed by flow cytometry 96 hours after AdCre infection. Peaks corresponding to diploid (2N), tetraploid (4N), octoploid (8N), and hexadecaploid (16N) DNA content are indicated. (E, F) hESPL1Δ/Δ cells expressing wildtype (WT) or protease-dead (CS) separase were examined by correlated timelapse-immunofluorescence microscopy as in A. (G) Quantification of E and F. Error bars indicate standard deviations from three independent experiments.
In both yeast and vertebrate oocytes, separase functions other than cohesin destruction do not depend on its cysteine protease activity, but are nonetheless sensitive to inhibition by securin (Gorr et al., 2005; Herbert et al., 2003; Kudo et al., 2006; Stegmeier et al., 2002; Sullivan and Uhlmann, 2003). To address whether centriole disengagement requires separase's protease activity, hESPL1flox/Δ cells were stably transfected with constructs that direct Cre-dependent expression of FLAG epitope-tagged wildtype (WT) or protease-dead (C2029S) hSeparase (Fig. 2B). In these cells, endogenous hSeparase disappeared upon AdCre infection, and the transgene-encoded proteins were induced (Fig. 2C). As expected, protease-dead hSeparaseCS failed to rescue the chromosome segregation defect of hESPL1Δ/Δ cells, in contrast to hSeparaseWT (Fig. 2D). hSeparaseCS was also unable to rescue the centriole disengagement defect of hESPL1Δ/Δ cells (Fig. 2E-G). These data indicate that the cysteine protease activity of hSeparase is required to separate sister chromatids and disengage centriole pairs at mitotic exit.
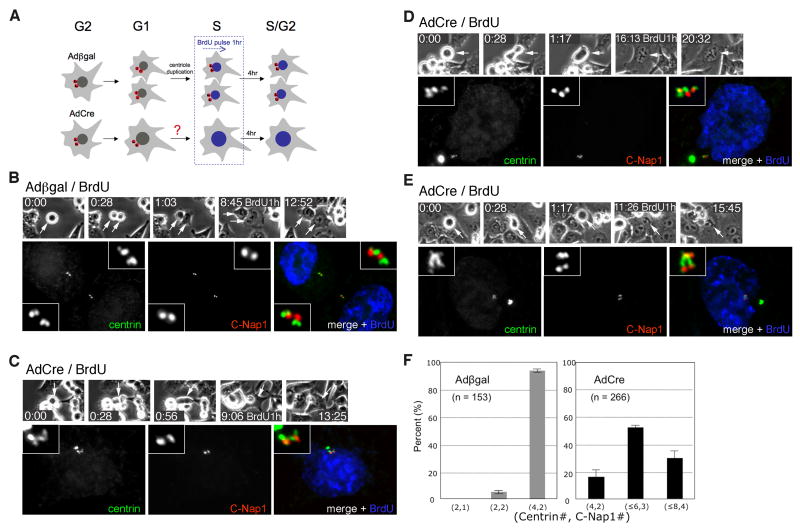
Having established that hSeparase-mediated proteolysis is important for centriole disengagement in vivo, we next investigated the fate of centriole pairs that remain engaged in its absence. We filmed hESPL1Δ/Δ cells for longer periods, during which we included a 1 hour pulse-labeling with 5-bromo-2-deoxyuridine (BrdU) and a 4 to 5 hour BrdU-free chase to mark cells progressing through S phase (Fig. 3A). Both hESPL1flox/Δ and hESPL1Δ/Δ cells entered S phase about 7 hours after mitosis (data not shown). As expected, more than 90% of the previously disengaged centrioles in control hESPL1flox/Δ cells underwent duplication, resulting in two pairs of engaged centrioles (i.e., 4 centrin foci and 2 C-Nap1 foci per cell; Fig. 3B). In contrast, hESPL1Δ/Δ cells displayed a wide range of aberrant centriole configurations that reflected a significant but variable block to duplication. 17% of cells exhibited the strongest phenotype, wherein only the two pairs of engaged centrioles inherited from the previous mitosis and failed cell division were present (4 centrin foci and 2 C-Nap1 foci per cell; Fig. 3C, F). However, we noticed that the C-Nap1 focus in some centriole pairs was elongated (Fig. 3C), rather than a compact dot as seen in G1 phase (Fig. 1D & F), suggesting that these juxtaposed centrioles were actually in an early stage of disengagement. Consistent with this interpretation, the majority (53%) of cells displayed 5 to 6 centrin foci and 3 C-Nap1 foci (Fig. 3D and F), indicating that one of the two centriole pairs had overtly disengaged and duplicated. Such asynchronous duplication was never observed in control cells. In the remaining 30%, full centriole disengagement and duplication were evident (Fig 3E, F). These results indicate that hSeparase is essential for the telophase-specific dissolution of centriole engagement, and probably as a consequence, for the timely and synchronous assembly of new centrioles in S phase. However, it also appears that mammalian cells possess a second activity that can disengage centrioles, albeit inefficiently, when hSeparase is absent or limiting.
Figure 3. Many centrioles eventually disengage and duplicate in hESPL1-null cells, revealing the existence of a second licensing pathway.
(A) Experimental scheme. Asynchronous hESPL1flox/Δ cells were infected with Adβgal or AdCre and traced by timelapse microscopy. Cells that had exited mitosis and progressed through the G1/S transition were labeled by a 1 hour BrdU pulse, followed by a 4 hour chase into BrdU-free medium. Cells were then fixed and stained with antibodies to centrin, C-Nap1, and BrdU, and then examined for evidence of centriole disengagement and/or duplication in S phase. (B) A pair of hESPL1flox/Δ cells exhibiting complete centriole disengagement (2 C-Nap1 foci) and duplication (4 centrin foci). (C-E) Centriole configurations in hESPL1Δ/Δ cells. (C) Absent or incipient centriole disengagement (2 C-Nap1 foci) without duplication (4 centrin foci). (D) Asynchronous disengagement (3 C-Nap1 foci) and duplication (5 or 6 centrin foci). (E) Complete disengagement (4 C-Nap1 foci) and duplication (8 centrin foci). (F) Quantification of S/G2 phase centriole configurations. Error bars indicate standard deviations from three independent experiments.
Separase does not dissolve sister chromatid cohesion in isolation, but rather is assisted by Polo-like kinase 1 (Plk1)-dependent phosphorylation. For example, Plk1-generated modifications on the SA2 subunit of cohesin promote the complex's nonproteolytic removal from chromosome arms in prophase (Hauf et al., 2005; Sumara et al., 2002), while similar modifications on Scc1 or its meiotic counterpart Rec8 drive anaphase-specific cohesin destruction, apparently by making Scc1 and Rec8 better substrates for separase (Alexandru et al., 2001; Brar et al., 2006; Hauf et al., 2005; Hornig and Uhlmann, 2004). Plk1 also localizes to centrosomes and is a key regulator of their structure and MTOC activity during spindle assembly (Barr et al., 2004; Petronczki et al., 2008). We therefore considered Plk1 a candidate for the second disengagement-promoting activity in mammalian cells.
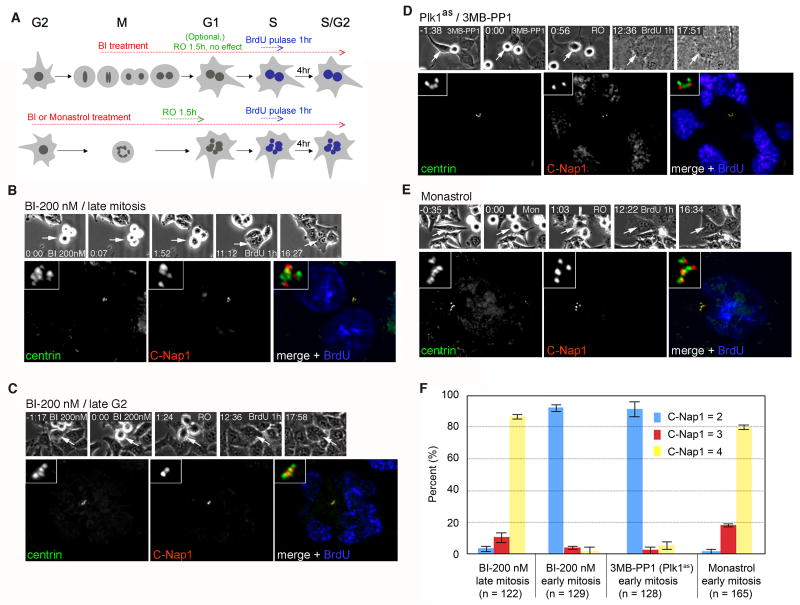
To test this hypothesis, we rapidly inactivated Plk1 using specific small-molecule inhibitors. First, we treated hESPL1flox/Δ cells with BI 2536 (hereafter BI) specifically during late mitosis (Fig. 4A, top panel). In agreement with recent reports (Brennan et al., 2007; Burkard et al., 2007; Petronczki et al., 2007; Santamaria et al., 2007), inhibiting Plk1 in this manner allowed mitotic exit but prevented cytokinesis, resulting in binucleated interphase cells. Nevertheless, centriole disengagement and duplication proceeded on schedule in the next cell cycle, indicating that the late mitotic activity of Plk1 is not needed for these aspects of centriole metabolism (Fig. 4B, G). In comparison, we observed a quite different result when Plk1 was inhibited earlier, in late G2 or early M phase. Briefly, we filmed an asynchronous culture of hESPL1flox/Δ cells treated with BI for 3 hours, during which time some cells entered mitosis but arrested in prometaphase with monopolar spindles, as expected (Lenart et al., 2007). These cells were then induced to exit mitosis by treatment with the Cdk1-specific inhibitor RO-3306 ((Vassilev et al., 2006); hereafter RO) and pulse-labeled with BrdU as above (Fig. 4A, bottom panel). Inhibiting Cdk1 in this manner allows cells to exit mitosis and carry out the subsequent round of DNA replication. In contrast to late mitotic Plk1 inhibition, early M phase Plk1 inhibition resulted in a dose-dependent block to centriole disengagement and duplication in S phase (Fig. 4C, F). For example, about one-third (37%) of cells treated with 25 nM BI failed to disengage their centrioles, whereas an 8-fold higher dose (200 nM) blocked disengagement almost completely (Fig. 4G). To control for the possibility that monopolar spindle geometry indirectly perturbs centriole disengagement, we analyzed centrioles in cells sequentially treated with the Eg5 inhibitor monastrol and RO. Over 90% of all centrioles disengaged and duplicated under these conditions (Fig. 4E, F), indicating that neither spindle bipolarity nor spontaneous relief of the spindle assembly checkpoint are required for centrosome duplication.
Figure 4. Plk1 acts during early mitosis to promote centriole disengagement.
(A) Experimental scheme. Asynchronously proliferating cells were filmed during a 3 hour treatment with the Plk1 inhibitor BI-2536 (BI) or the Eg5 inhibitor monastrol as a control. We note that Plk1 inactivation in late G2 or prophase activates the spindle assembly checkpoint and arrests cells in prometaphase. In contrast, late mitotic Plk1 inactivation in does not block anaphase onset, but instead inhibits cytokinesis. To allow analysis of centriole duplication potential under both treatment regimens, cells in the former population were induced to exit mitosis using the Cdk1-selective inhibitor RO-3306 (RO). Cells transiting through S phase were marked by BrdU pulse-labeling and analyzed as in Fig. 3. (B) A hESPL1flox/Δ cell treated with BI during late M phase (200 nM) exhibits complete centriole disengagement (4 C-Nap1 foci) and duplication (8 centrin foci). (C) A hESPL1flox/Δ cell treated with BI during late G2 (200 nM), showing no disengagement (2 C-Nap1 foci) and no duplication (4 centrin foci). (D) A Plk1as cell (RPE1, retinal pigment epithelial human cells) treated with 3MB-PP1 (10 mM) during late G2, showing no disengagement (2 C-Nap1 foci) and no duplication (4 centrin foci). (E) A hESPL1flox/Δ cell treated with monastrol (50 μM) during late G2 exhibits complete disengagement (4 C-Nap1 foci) and duplication (8 centrin foci, one of which lies in a different focal plane (not shown)). (F) Quantification of results in B-E. Error bars indicate standard deviations from three independent experiments.
Although BI has been developed as a Plk1-specific inhibitor, this compound inhibits multiple Plks and could have additional off-target effects. We therefore sought an independent and gene-specific test of Plk1's role in centriole disengagement. To this end, we repeated the above experiments with a recently described “chemical genetic” system for inhibiting Plk1 in human retinal pigment epithelial cells (Burkard et al., 2007). In this system, cells whose endogenous PLK1 gene has been deleted via homologous recombination are complemented by a genetically modified allele (Plk1as) whose ATP-binding pocket can accommodate (and be inhibited by) bulky purine analogs. Importantly, these analogs are inactive towards isogenic cells lacking the Plk1as allele, providing evidence of their in vivo specificity (Burkard et al., 2007). Sequential treatment of Plk1as cells with one such analog (3-MB-PP1) and RO dramatically inhibited centriole disengagement and duplication (Fig. 4D, F). Together, these findings indicate that the activation of Plk1 at centrosomes in late G2 or early M phase primes the dissolution of centriole engagement in late M phase, and thus plays a crucial role in licensing centriole duplication in S phase.
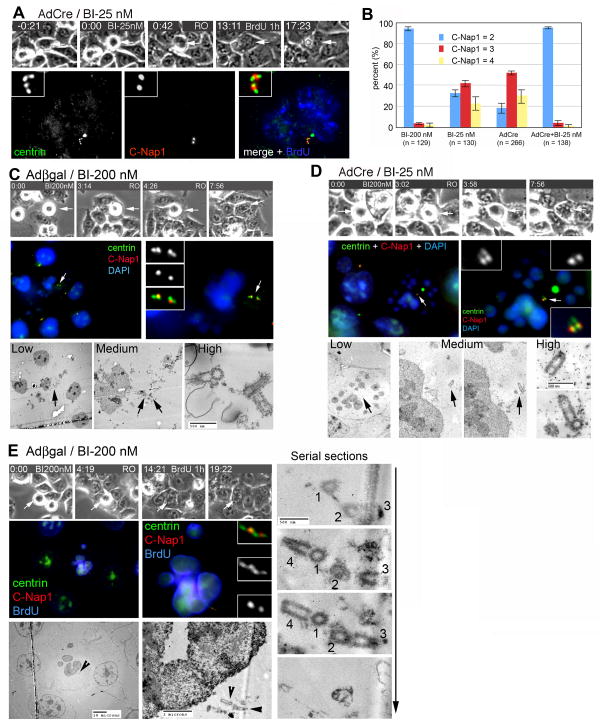
We returned to the issue of whether Plk1 potentiates the eventual disengagement and duplication of centrioles in hESPL1Δ/Δ cells. To address this question, we treated hESPL1Δ/Δ cells with a low dose of BI (25 nM) during late G2 and early mitosis, and then induced mitotic exit with RO as above. Strikingly, combined downregulation of Plk1 and hSeparase exhibited a synergistic effect as compared with either manipulation alone, as more than 95% of all centrioles failed to disengage and duplicate under these conditions (Fig. 5A,B). These results strongly suggest that both Plk1 and separase play important roles in licensing centrioles for duplication.
Figure 5. Separase and Plk1 regulate the mitotic licensing of centriole duplication.
(A) G2 phase hESPL1Δ/Δ cells were treated with 25 nM BI, induced to exit mitosis with RO, and labeled during S phase transit with BrdU. Centrioles remained engaged (2 C-Nap1 foci) and unable to duplicate (4 centrin foci). (B) Quantification of centriole duplication after downregulation of hSeparase, Plk1, or both regulators. Error bars indicate standard deviations from three independent experiments. (C, & D) G2 phase hESPL1Δ/Δ (C) and hESPL1flox/Δ (D) cells were treated with BI 200 nM or 25 nM respectively, induced to exit mitosis with RO, and stained with antibodies to centrin and C-Nap1. Each cell was then processed for serial sectioning and electron microscopy. (E) A late S/G2 phase hESPL1flox/Δ cell treated with BI 200 nM in the previous G2/M phase was stained with antibodies to centrin, C-Nap1, and BrdU, and then serially sectioned. Electron micrographs are shown at three different magnifications to facilitate correlation between images and details of centriole structure (arrowheads).
To strengthen this conclusion, we performed correlated timelapse/IF/EM studies to confirm the defects of centriole disengagement and duplication observed in these cells. G1 phase hESPL1flox/Δ or hESPL1Δ/Δ cells (obtained by sequential treatment with high or low doses of BI (200 or 25 nM) and RO in the previous cell cycle) were stained with centrin and C-Nap1 (Fig 5C & D). After acquiring fluorescent images, the same cell was fixed, embedded, and serially sectioned (Fig. 5C & D). Consistent with the IF pattern, centrioles in these cells were confirmed to be engaged at the ultrastructural level (Fig. 5C & D; n = 3 for each case). Thus, centrin and C-Nap1 are reliable markers of centriole configuration under these conditions. To confirm and extend these results, S phase hESPL1flox/Δ cells (treated with 200 nM BI in the previous G2/M phase) were also subjected to correlative EM analysis (n = 5). One representative is shown in Fig. 5E. As anticipated from the IF pattern, only four engaged centrioles were recovered from the serial sections (Fig. 5E), indicating that both centriole disengagement and duplication were blocked. Taken together, our findings reveal that Plk1 and separase act during M phase to disengage and license centrioles for duplication in the following cell cycle.
Discussion
Like chromosomes, centrosomes duplicate exactly once per cell cycle, but how such regulation is achieved in molecular terms has long been elusive. One attractive hypothesis is that the orthogonal configuration of duplicated centrioles (termed centriole engagement) plays a central role in preventing centriole overduplication (Tsou and Stearns, 2006a). According to this model, centriole engagement, which is first established during procentriole formation in S phase, suppresses further assembly by sequestering or inhibiting one or more activities that are rate-limiting for centriole duplication. Because centriole engagement persists throughout S, G2, and early M phase, this block would be relieved (or equivalently, a new “license” to duplicate granted) only at M phase exit, when centrioles disengage.
This hypothesis gained support from studies in Xenopus egg extracts treated with nondegradable inhibitors of separase, a cysteine protease that becomes active at anaphase onset and known to cleave chromosome-bound cohesin rings (Tsou and Stearns, 2006b). However, given the indirect nature of these experiments, it could not be excluded that these inhibitors actually worked by targeting other components of the extract, especially as Drosophila embryos and MEFs deficient in separase reportedly duplicate their centrosomes correctly. Furthermore, it was unknown if centrioles that have passed through mitosis but never been exposed to separase in fact fail to duplicate.
Here, we have performed experiments that seek to validate and extend this hypothesis by directly examining the fate of centrioles in cells devoid of all detectable hSeparase. To do this, we mutated both alleles of the gene encoding hSeparase in human tissue culture cells and confirmed that these mutations in fact eliminated expression of this large (220 kDa) protease, rather than allowing continued synthesis of a truncated and potentially bioactive N-terminal fragment. Homozygous inactivation of hSeparase strongly inhibited disengagement in the short term (over a period of 2 to 6 hours after mitosis), after which approximately 50% of all centriole pairs disengaged and duplicated during S phase. A similar dose-dependent inhibition of disengagement was observed in cells treated with the Plk1 inhibitor BI 2536 before (but not after) anaphase onset, indicating that Plk1 promotes disengagement at a step prior to securin destruction and separase activation. However, combined downregulation of both Plk1 and hSeparase resulted in a tight block to both centriole disengagement in telophase, and probably as a consequence, complete suppression of centriole assembly in the next S phase. Taken together, these results provide in vivo evidence for the mitotic licensing of centriole duplication, and moreover link this licensing to the action of Plk1 and hSeparase during early and late M phase respectively.
A key question is how Plk1 and hSeparase act to coordinate centriole disengagement with mitotic exit. By analogy to sister chromatid cohesion, Plk1 could promote hSeparase-independent removal of a centriolar “glue” protein in prophase and/or facilitate anaphase-specific cleavage of this “glue” protein by trace amounts of hSeparase that had not yet been cleared from the cell after hESPL1 deletion. It was recently proposed that the telophase/G1-specific destruction of HsSAS-6 (an essential centriole assembly factor present at the mother-daughter interface) also regulates the licensing of centriole duplication (Strnad et al., 2007). While Plk1 inhibition stabilized HsSAS-6 on engaged centrioles, expressing nondegradable HsSAS-6 did not interfere with centriole disengagement (data not shown), arguing that HsSAS-6 destruction does not regulate this decision. Clearly, further progress in this area will require identification of Plk1's and hSeparase's disengagement-specific substrates and detailed analysis of their regulation in the presence or absence of these enzymes. Although the proteomic analysis of the centrosome has been quite difficult due to its small size and low copy number (Andersen et al., 2003), recent advances in the selective labeling of cleaved protease substrates (Mahrus et al., 2008) and kinase substrates (Allen et al., 2007; Blethrow et al., 2008) should be quite helpful in this regard.
In summary, we have described a mechanism for the once-and-only control of centrosome duplication in human cells, whereby the growth of new centrioles in S phase crucially depends on the Plk1- and hSeparase-dependent disengagement of centriole pairs during the preceding M phase. Intriguingly, many aneuploid cancer cells exhibit both centrosome amplification and marked overexpression of Plk1 and hSeparase (Carter et al., 2006; Strebhardt and Ullrich, 2006), raising the question of whether such amplification could in some instances be a consequence of precocious centriole disengagement, rather than cytokinesis failure and subsequent tetraploidization, or simultaneous assembly of multiple daughter centrioles in a single S phase.
Experimental Procedures
Gene targeting and conditional transgenesis
A BAC clone containing the human ESPL1 locus (RP11-680A11) was obtained from BACPAC Resources (Children's Hospital of Oakland Research Institute) and used to amplify 5′ and 3′ homology arms via PCR. A loxP site and BglII restriction-site polymorphism were introduced into the 3′ element via QuikChange mutagenesis (Stratagene). Both homology arms were then cloned into pNX, a pBluescript derivative that contains a central loxP-neo-loxP cassette. The final targeting vector was sequenced in its entirety and then transferred as a NotI fragment into pAAV, yielding pAAV-hESPL1flox. Production of rAAV particles, infection of HCT116 cells, and genomic PCR screening was carried out as described (Papi et al., 2005). hESPL1flox/+ cells were targeted with a second rAAV vector in which exon 21 of hESPL1 was deleted, yielding hESPL1flox/Δ cells. CsCl gradient-purified adenoviruses expressing β-galactosidase (Adβgal) and Cre recombinase (AdCre) were purchased from the Baylor University Vector Development Laboratory and used at a multiplicity of infection (MOI) of 25 plaque-forming units (pfu) per cell. To express hSeparase in trans, FLAG-epitope tagged versions of hSeparase were cloned into pCLIP (George et al., 2007) and stably transfected into hESPL1flox/Δ cells. Transgene expression was activated by AdCre infection as described above.
Cell culture, drug treatments, and timelapse microscopy
HCT116 cells were grown in McCoy's 5A containing 10% fetal bovine serum and 1% penicillin–streptomycin. Human telomerase-immortalized retinal pigment epithelial (hTERT-RPE) cells were cultured in DMEM:F-12 (1:1) medium supplemented with 10% FBS and 1% penicillin-streptomycin. HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Where indicated, BI 2536 (25 or 200 nM), monastrol (50 μM), and RO-3306 (10 μM) were used. For correlative timelapse experiments, cells were imaged on a Zeiss Axiovert microscope configured with a 10 × phase objective, motorized temperature-controlled stage, environmental chamber, and CO2 enrichment system (Zeiss, Germany). Image acquisition and processing were performed using Axiovision software (Zeiss, Germany). 60 fields of cells were filmed with 2 × 2 binning during each experiment. For timelapse fluorescence microscopy, hESPL1flox/Δ cells were stably transfected with a histone H2B-GFP expression plasmid (Kanda et al., 1998) and viewed on a Nikon TE2000 microscope outfitted with 10 ×, 20 ×, and 40 × objectives, Hamamatsu ORCA ER camera, and temperature-controlled stage enclosure and CO2 enrichment system (Solent Scientific). Image acquisition and processing were performed using Metamorph software (Molecular Devices).
Antibodies
A rabbit polyclonal antibody against human C-Nap1 was produced as previously described (Mayor et al., 2000) and used at 1:500 dilution. Other antibodies used in this study include mouse anti-centrin (20H5; a gift from J. Salisbury, Mayo Clinic, Rochester, MN; 1:1000); mouse anti-INCENP (Upstate; 1:500), mouse anti-Sgo1 (Novus; 1:500), rat anti-α-tubulin (Chemicon; 1:500), rat anti-BrdU (Novus; 1:500), mouse anti-hSeparase N-terminus (18H1; MSKCC Monoclonal Antibody Facility; 1:1000) and C-terminus (XJ11-1B12; Dana-Farber Cancer Institute Monoclonal Antibody Facility; 1:1000), and human CREST antiserum (Immunovision; 1:1000). We note that some commercial preparations of XJ11-1B12 predominantly contained antibodies to proteins other than hSeparase, necessitating re-isolation of the hybridoma by the originating facility (Chestukhin et al., 2003).
Immunofluorescence microscopy
Cells were fixed with methanol at −20°C, then blocked with 3% bovine serum albumin (w/v) and 0.1% Triton X-100 in PBS for 30 min. DNA was visualized using 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). For visualizing replicated DNA, BrdU (20 μM) was added to the cells as a 1 hour pulse. After staining for centrosomal antigens, cells were fixed again with -20°C methanol for 10 min and treated with 2 N HCl for 30 min at room temperature. Detection of BrdU-positive cells was performed as above.
Electron microscopy
Cells were traced by phase-contrast microscopy on gridded coverslips made of ACLAR film (EM Sciences), permeabilized in PIPES buffer (pH 6.8) containing 0.1% Triton X-100, stained with anti-centrin and C-Nap1 antibodies as described above, and then fixed in modified Karnovsky's fixative (Murphy et al., 2000) consisting of 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer. For BrdU visualization, fixed cells were treated with 2 N HCl for 30 min at room temperature before staining for BrdU. After acquisition of fluorescence images, cells were maintained on coverslips and further processed for electron microscopy. Cells were first dehydrated in graded series of ethanol, infiltrated with EMbed812 resin and embedded beween two ACLAR films. An area of coverslip containing the cells of interest was selected based on the timelapse movies, excised, and glued to the top of an empty resin block. Mounted samples were sectioned (80-90 nm thickness) on an Ultracut UC6 microtome. Sections were stained with 2% uranyl acetate followed by 1% lead citrate and observed on a FEI TECNAI Spirit G2 microscope. Electron micrographs were captured with the Gatan digital imaging system.
Supplementary Material
Figure S1. Generation and characterization of hSeparase conditional-null cells. (A) Adeno-associated virus (AAV) vectors were used to mutate the hESPL1 locus in HCT116 cells as described in the Experimental Procedures. For the first round of gene replacement, the targeting vector was designed to introduce loxP sites on either side of exon 21 (i.e., a ‘floxed’ allele). After excision of the G418 resistance (neo) cassette with Cre recombinase, the resulting hESPL1flox/+ cells were infected with a second targeting vector in which exon 21 was deleted outright, yielding hESPL1flox/Δ cells. Solid bar marked with an asterisk (*) indicates the probe used for Southern blotting. (B) Verification of gene replacement. Genomic DNAs of the indicated genotypes were digested with Bgl II and Xho I, electrophoretically resolved and transferred to nylon membranes, and probed with the [32P]-labeled fragment indicated in (A). Bands corresponding to the wildtype hESPL1 allele (4.7 kb), the hESPL1flox allele (3 kb), and the hESPL1Δ allele (4.3 kb) are marked with arrowheads. (C) hESPL1flox/Δ cells were infected with AdCre, Adβgal, or left untreated (no tx) as a control. Samples were collected at the indicated timepoints, stained with propidium iodide, and analyzed by flow cytometry. (D) In parallel, metaphase chromosome spreads were prepared and labeled with fluorescence in situ hybridization (FISH) probes specific for chromosome 4 (red) and chromosome 15 (green). The incidence of diplochromosomes was determined from at least 100 metaphase spreads per sample.
Figure S2. Centrin aggregates in HCT116 cells can be significantly larger than centrioles and are comprised of electron-dense material. The image shows a G1 phase hESPL1Δ/Δ cell that had been treated with BI (25 nM) in G2/M, induced to exit mitosis with RO, and stained with antibodies to centrin and C-Nap1. The cell was further processed for serial sectioning and electron microscopy. For comparison, the centrin aggregate and a pair of engaged centrioles (indicated by arrows and arrowhead respectively) are shown separately at high magnification.
Supplementary Movie 1. hESPL1flox/Δ cells stably expressing histone H2B-GFP were infected with Adβgal and filmed 48 hours later.
Supplementary Movie 2. hESPL1flox/Δ cells cells stably expressing histone H2B-GFP were infected with AdCre and filmed 48 hours later.
Acknowledgments
We thank M. Leversha for assistance with FISH and O. Ouerfelli and P. Lin for organic synthesis. This work was supported by National Institutes of Health grants CA107342 (P.V.J.) and GM52022 (T.S.), a Damon Runyon Cancer Foundation postdoctoral fellowship (M-F.B.T.), and a Pew Scholar in the Biomedical Sciences award (P.V.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Brennan IM, Peters U, Kapoor TM, Straight AF. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS ONE. 2007;2:e409. doi: 10.1371/journal.pone.0000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. Processing, localization, and requirement of human separase for normal anaphase progression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4574–4579. doi: 10.1073/pnas.0730733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes & development. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SH, Gertsenstein M, Vintersten K, Korets-Smith E, Murphy J, Stevens ME, Haigh JJ, Nagy A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and cdk1 by two-step complex formation. Mol Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Gorr IH, Reis A, Boos D, Wuhr M, Madgwick S, Jones KT, Stemmann O. Essential CDK1-inhibitory role for separase during meiosis I in vertebrate oocytes. Nat Cell Biol. 2006;8:1035–1037. doi: 10.1038/ncb1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Herbert M, Levasseur M, Homer H, Yallop K, Murdoch A, McDougall A. Homologue disjunction in mouse oocytes requires proteolysis of securin and cyclin B1. Nat Cell Biol. 2003;5:1023–1025. doi: 10.1038/ncb1062. [DOI] [PubMed] [Google Scholar]
- Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. Embo J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Kumada K, Yao R, Kawaguchi T, Karasawa M, Hoshikawa Y, Ichikawa K, Sugitani Y, Imoto I, Inazawa J, Sugawara M, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. The Journal of cell biology. 2006;172:835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. The Journal of cell biology. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Dej KJ, Lopez JM, Orr-Weaver TL. Control of centromere localization of the MEI-S332 cohesion protection protein. Curr Biol. 2004;14:1277–1283. doi: 10.1016/j.cub.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. The Journal of cell biology. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington's disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Pandey R, Heidmann S, Lehner CF. Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. J Cell Sci. 2005;118:733–742. doi: 10.1242/jcs.01663. [DOI] [PubMed] [Google Scholar]
- Papi M, Berdougo E, Randall CL, Ganguly S, Jallepalli PV. Multiple roles for separase auto-cleavage during the G2/M transition. Nat Cell Biol. 2005;7:1029–1035. doi: 10.1038/ncb1303. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Developmental Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Santamaria A, Neef R, Eberspacher U, Eis K, Husemann M, Mumberg D, Prechtl S, Schulze V, Siemeister G, Wortmann L, et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Molecular biology of the cell. 2007;18:4024–4036. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes & development. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006a;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006b;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, Seznec J, Ducos GM, Ricci R, Firnberg N, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. The Journal of cell biology. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Generation and characterization of hSeparase conditional-null cells. (A) Adeno-associated virus (AAV) vectors were used to mutate the hESPL1 locus in HCT116 cells as described in the Experimental Procedures. For the first round of gene replacement, the targeting vector was designed to introduce loxP sites on either side of exon 21 (i.e., a ‘floxed’ allele). After excision of the G418 resistance (neo) cassette with Cre recombinase, the resulting hESPL1flox/+ cells were infected with a second targeting vector in which exon 21 was deleted outright, yielding hESPL1flox/Δ cells. Solid bar marked with an asterisk (*) indicates the probe used for Southern blotting. (B) Verification of gene replacement. Genomic DNAs of the indicated genotypes were digested with Bgl II and Xho I, electrophoretically resolved and transferred to nylon membranes, and probed with the [32P]-labeled fragment indicated in (A). Bands corresponding to the wildtype hESPL1 allele (4.7 kb), the hESPL1flox allele (3 kb), and the hESPL1Δ allele (4.3 kb) are marked with arrowheads. (C) hESPL1flox/Δ cells were infected with AdCre, Adβgal, or left untreated (no tx) as a control. Samples were collected at the indicated timepoints, stained with propidium iodide, and analyzed by flow cytometry. (D) In parallel, metaphase chromosome spreads were prepared and labeled with fluorescence in situ hybridization (FISH) probes specific for chromosome 4 (red) and chromosome 15 (green). The incidence of diplochromosomes was determined from at least 100 metaphase spreads per sample.
Figure S2. Centrin aggregates in HCT116 cells can be significantly larger than centrioles and are comprised of electron-dense material. The image shows a G1 phase hESPL1Δ/Δ cell that had been treated with BI (25 nM) in G2/M, induced to exit mitosis with RO, and stained with antibodies to centrin and C-Nap1. The cell was further processed for serial sectioning and electron microscopy. For comparison, the centrin aggregate and a pair of engaged centrioles (indicated by arrows and arrowhead respectively) are shown separately at high magnification.
Supplementary Movie 1. hESPL1flox/Δ cells stably expressing histone H2B-GFP were infected with Adβgal and filmed 48 hours later.
Supplementary Movie 2. hESPL1flox/Δ cells cells stably expressing histone H2B-GFP were infected with AdCre and filmed 48 hours later.