Abstract
Aims
The ‘rhythmonome’ is the term we have adopted to describe the set of genes that determine the normal coordinated electrical activity in the heart. Elements of this set include pore-forming ion channels, function-modifying proteins and intracellular calcium control elements. Rare mutations in many of these genes are known to cause unusual congenital monogenic arrhythmia syndromes, and single common variants have been reported to modify arrhythmia phenotypes. Here, we report an evaluation of the variation and haplotype structure in six key components of the rhythmonome.
Materials & methods
SNPs were typed using DNA extracted from Coriell cell lines to survey allele frequencies and haplotype structure in six genes (ANK2, SCN5A, KCNE1 and 2 gene cluster, KCNQ1, KCNH2 and RYR2) across four human populations (African—American, European American, Han Chinese and Mexican American).
Results
A total of 307 SNPs were analyzed across the six genes, revealing significant allele-frequency differences between populations and clear differences in haplotype structure.
Conclusions
The pattern of variation we report is an important step towards incorporating common variation across the rhythmonome in studies of arrhythmia susceptibility.
Keywords: arrhythmia, diversity, gene, haplotype, structure
Normal electrophysiological activity of the heart is determined by ordered propagation of excitatory stimuli, resulting in rapid depolarization and slow repolarization and generating action potentials in individual cardiomyocytes. Physiological function of a large set of genes — to which we apply the term ‘rhythmonome’ — and their products is required for such normal activity [1]. Rare mutations in many components of the rhythmonome cause syndromes with the high potential for sudden death due to arrhythmia in the young; these include the long QT syndromes (LQTS) and catecholaminergic polymorphic ventricular tachycardia (CPVT) [2,3].
Ion currents flowing across cell membranes through ion channel proteins determine the shape and duration of cardiac action potentials. The most common disease genes involved in LQTS encode the potassium and sodium channels, KCNQ1, KCNH2 and SCN5A. Other LQTS disease genes encode ion channel ancillary subunits (KCNE1 and KCNE2, located in a single gene cluster KCNE1 and 2) and ankyrin-B (ANK2), which modulates ion channel targeting to appropriate subcellular domains. The disease gene in autosomal dominant CPVT (RYR2) also encodes an ion channel — the intracellular ryanodine release channel, which is responsible for the increase in intracellular calcium that initiates contractile activity in the heart. Importantly, not only do mutations in these genes cause rare congenital arrhythmia syndromes, but polymorphisms — some are ethnicity specific — have also been implicated as a cause of variable susceptibility to common arrhythmias, including those triggered by drug administration [4-9]. However, the haplotype structure of these genes has not been determined and compared across ethnicities. Here, we report an analysis of common variation in these six genes in African—American (AA), European American (EA), Han Chinese of Los Angeles (HC), and Mexican American (MA) populations.
Materials & methods
Study samples
Study subjects were obtained from a large panel of anonymous, unrelated DNA samples from the Human Variation Collection of the National Institute of General Medical Sciences (NIGMS) Repository held by the Coriell Institute (NJ, USA). We specifically used sets of DNA samples obtained from four distinct ethnic groups residing in the USA, including 88 EAs, 88 AAs, 88 HC and 89 MAs.
SNP selection
We selected six genes from the rhythmonome to explore patterns of normal genetic variation: ANK2, SCN5A, KCNE1 and 2, KCNQ1, KCNH2 and RYR2. Using the Ensembl web interface [10], we defined regions using each gene of interest within EnsMart [11]. Regions we examined included exons (coding regions, 5′ and 3′-UTR), introns and 10-kb regions flanking both sides of the gene. Once a region of interest was identified, we then searched the database for SNPs with a minor allele frequency (MAF) of over 10%, searching for validated, nonsynonymous, coding SNPs in particular. In cases where there was a dearth of SNPs using this criteria, we dropped the MAF, but it was never less than 2%. In rare instances, we also accepted nonvalidated, nonsynonymous SNPs. We did this in order to obtain a distribution of one SNP every 300–500 bp in the region of interest.
This list was verified to contain, or was supplemented with, common SNPs in the literature that are related to arrhythmia susceptibility. A final list of 753 SNPs was generated and entered into the SNPlex™ Design Pipeline (Applied Biosystems [CA, USA]). The SNPlex Design Pipeline segregates the selected SNPs into various predetermined design probe pools. These probe pool sets are then used to interrogate the samples on the SNPlex platform.
Statistical analysis
SNPs with a genotyping efficiency below 80% in one or more populations were dropped from all analyses. Hardy—Weinberg equilibrium was assessed for each SNP using Tools For Population Genetic Analyses (TFPGA) software, which is a program for the analysis of allozyme and molecular population genetic data, by conducting exact tests on all genotypes with a Monte Carlo approach (10 batches, 1000 permutations per batch). SNPs that significantly deviated from Hardy—Weinberg equilibrium (p < 0.01) in one or more populations were excluded from further analyses as this was a sign of poor genotyping quality. A total of 307 SNPs out of 753 (40.77%) were used in the remaining analyses.
Pairwise linkage disequilibrium (LD) was estimated for the SNPs in each gene using standardized summary statistics [12] D’ and r2, which were calculated using HaploView software [13]. Haplotype blocks were assigned using the D’ confidence interval algorithm from Gabriel et al., which is implemented in HaploView [14]. Haplotype frequencies for each block were estimated using two methods — the expectation-maximization (EM) algorithm using HaploView software, and the Bayesian haplotype reconstruction program PHASE version 2.1.1 [15,16]. Since KCNE1 and KCNE2 are only 74.5 kb apart, they were considered as one gene cluster (KCNE1 and 2) in this study.
Exact tests for allele-frequency differences in SNPs were conducted in the TFPGA software that employs an algorithm by Raymond and Roussett [17]. For each SNP, we calculated Wright’s fixation index (Fst) — a measure of allelic variation that is owing to variation within a population compared with between-population variation. Fst values were calculated using the algorithm of Weir and Cockerham [18,19].
TagSNPs were identified for each gene within each ethnicity using the Tagger feature of HaploView with an r2 threshold of 0.8. Genotyping platforms were evaluated using probe lists obtained from vendors’ websites. Individual SNPs were matched to probe lists using reference sequence (RS) IDs, and only probes with vendor-specified RS IDs were used for matching.
To determine the presence of copy-number variants (CNVs) in the six genes of interest, we obtained the Affymetrix (CA, USA) SNP 6.0 GeneChip® genotyping data from the Coriell Human Genetic Cell Repository. The data consisted of 100 samples from each of the four different ethnic groups (EA, AA, MA and HC). The analysis was conducted using the Affymetrix Genotyping Console platform [20,21]. The analysis was also conducted using Partek® (MO, USA) software version 6.4.
Results
Differences in allele frequency & haplotype structure
The general haplotype structure of each gene is shown in FIGURE 1 by ethnicity. Detailed results of all statistical analyses for each gene and ethnicity are shown in the context of the haplotype structure in LD-plus [BUSH W, DUDEK S, RITCHIE M ET AL. SUBMITTED] composite figures (FIGURE 2 & 3 for KCNH2 and for other genes, see [101]). A total of five haplotype blocks were common to all ethnicities, and their haplotype frequencies are shown in FIGURE 4. In addition, there were eight blocks common to three of the four populations, and 29 blocks were common to only two of the four populations.
Figure 1. Haplotype structure overview of all rhythmonome genes by population.
Columns indicate the gene region and rows indicate ethnicity.
AA: African—American; EA: European American; HC: Han Chinese; MA: Mexican American.
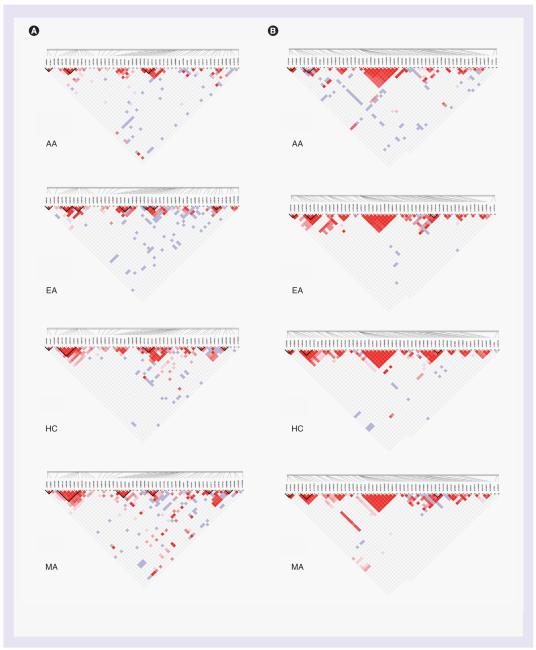
Figure 2. Allele frequencies, haplotype structure and GWAS platform representation of KCNH2 in African—American (A) and European American (B) samples.
Relative physical positions of SNPs are shown at the top of the figure. TagSNPs and SNPs represented on genome-wide association study platforms are indicated below SNP IDs. SNP statistics, such as Fst, -log of the exact test p-value, minor allele frequency, genotyping efficiency and -log of the Hardy—Weinberg equilibrium test are shown as line graphs. Haplotype structures (with Haploview-style D’ and r2 color schemes) are shown at the bottom of the figure. Identified haplotype blocks are indicated with grey shading through the figure, with individual haplotypes and their relative frequencies shown as graphs at the top of the figure.
Figure 3. Allele frequencies, haplotype structure and GWAS platform representation of KCNH2 in Han Chinese (A) and Mexican American (B) samples.
Relative physical positions of SNPs are shown at the top of the figure. TagSNPs and SNPs represented on genome-wide association study platforms are indicated below SNP IDs. SNP statistics, such as Fst, -log of the exact test p-value, minor allele frequency, genotyping efficiency and -log of the Hardy—Weinberg equilibrium test are shown as line graphs. Haplotype structures (with Haploview-style D’ and r2 color schemes) are shown at the bottom of the figure. Identified haplotype blocks are indicated with grey shading through the figure, with individual haplotypes and their relative frequencies shown as graphs at the top of the figure.
GWAS: Genome-wide association study.
Figure 4. Population-specific haplotype frequencies within haplotype blocks common to all populations.
Rs numbers identify the beginning and end of the haplotype block, with the relevant gene label shown above. Haplotype frequencies are displayed as bar graphs.
Allele-frequency analysis revealed significant differences in frequency distributions among the four different ethnic populations. In order to assess the degree of similarity in genetic structure among the different populations, we calculated Fst for all SNPs. Fst values range from 0 to 1 and increase as the allele frequency difference between the populations increases; a value of 0.05 or more is typically considered a significant difference in allele frequencies between the populations.
ANK2
The haplotype structure of ANK2 is most clear in the HC population, with well defined haplotype blocks (FIGURE 1). EA and MA blocks maintain a similar structure with less well-defined edges. In the AA population, the large block from rs2285711—rs29311, which is characteristic of other ethnicities, is much more diffuse. There are two haplotype blocks common to all ethnicities. The first block in ANK2 includes two markers, rs1026089 and rs13108692 (7290 bp), and has two common haplotypes with approximately equal frequency in four populations (FIGURE 4). The second block in ANK2 includes two markers, rs13101312 and rs1351997 (9500 bp), and has two common haplotypes. For this block, the MA (0.70) and AA (0.73) populations have a different haplotype frequency distribution to the HC (0.59) or EA (0.56) populations (FIGURE 4).
African—Americans demonstrated the most statistically significant differences in allele frequency compared with the other ethnicities. There are two notable stretches of allele-frequency difference that constitute haplotypes more prominent in the AA population (rs961294—rs2135351 and rs17045935—rs2272230). Allele frequencies for six SNPs (rs13108692, rs1026089, rs689117, rs6533668, rs13131206 and rs7697312) in ANK2 were remarkably similar across the four populations (exact test: p > 0.05 for all comparisons).
Across all populations, Fst values for 45 of 81 ANK2 SNPs were greater than 0.05. Notable peaks in Fst values (> 0.15) occurred at rs2121928, rs6533660, rs313962, rs29307 and rs17045935. Population differences at these SNPs are mostly due to divergent allele or haplotype frequencies in AA populations.
KCNE1 & 2 gene cluster
The haplotype structure of the KCNE cluster is most distinctive in the AA samples, with fewer and smaller blocks than the other ethnicities. The EA samples have a more well-defined block structure from rs1012945 forward, whereas this region has more diffuse LD in the HC, AA and MA ethnicities. There are two notable stretches of unique allele frequencies in AAs, from rs2834465—rs2834467 and from rs2834477—rs2834480.
For KCNE1 and 2, the fewest statistically significant differences are between AAs and EAs with only 22 of the 60 markers differing in frequency. MA and HC allele frequencies are also similar, with only 26 of the 60 markers showing significant difference.
A total of 25 of 60 markers in the KCNE1 and 2 gene cluster had Fst values greater than 0.05, making this gene cluster the least diverse of all the six surveyed. One SNP, rs2834502, has an Fst of more than 0.15, owing to allele frequency divergence in the EA population. This SNP also occurs frequently in an EA haplotype.
KCNH2
With regard to KCNH2, EAs have a very discrete haplotype structure, and all SNPs had significantly different allele frequencies compared with the HC population. Three markers (rs1805120, rs740952 and rs2269001) remain linked in the AA population and form a frequent CGG haplotype that is part of an infrequent haplotype for a larger block in the HC population (rs2968855—rs2269001, CGCGG) (FIGURE 3). Conversely, the relatively rare TAA haplotype in AAs is part of the dominant CGTAA haplotype of the larger HC block (FIGURE 4). There is also a notable frequent allele sequence in the clear EA block (rs2373885—rs10277237, ATCG) that is less common in the MA and HC populations. The MA and HC allele frequencies for nearly all markers in KCNH2 are significantly different from each other and from the other two populations, so while the haplotype block structure between EA, HC and MA is similar, the haplotype frequencies are quite different.
Nearly all the SNPs in KCNH2 (16 of the 19) had Fst values greater than 0.05, making this an extremely diverse gene. Fst values plateau above 0.15 through the single AA haplotype block, driven by a frequent allele sequence of CGG in AA and EA populations that is of lower frequency in HC and MAs, where the TAA sequence is more prominent.
KCNQ1
The EA and HC populations have very similar haplotype structures as well as allele and haplotype frequencies from rs800338—rs1459825. The AA population has a stretch of significantly different allele frequencies from the other populations between rs6578273 and rs2299618. The general haplotype structure is fairly similar across all ethnicities, with a slightly more developed block from rs2283205 to rs231879 in the EA population (FIGURE 5).
Figure 5. Haplotype structure comparison of KCNQ1 (A) and RYR2 (B) across AA, EA, HC and MA populations.
Haplotype structures are shown with Haploview-style plots, where dark red regions indicate high D’ — collections of markers in the gene that are inherited together through the population — and lighter regions indicate markers that are inherited independently in the population.
AA: African—American; EA: European American; HC: Han Chinese; MA: Mexican American.
There was one common haplotype block in the KCNQ1 gene involving two markers, rs739677 and rs800338 (524 bp). Three frequent haplotypes were observed for these two markers, and each population had a unique haplotype distribution (FIGURE 4).
A total of 37 of 53 SNPs in KCNQ1 had Fst values greater than 0.05, highlighting the diversity of this gene. Notable Fst peaks (>0.15) occurred at rs2283172, rs163183, rs2237896 and rs231356. The high Fst for rs2283172 is owing to strong allele-frequency differences in the AA population. SNPs rs163183 and rs2237896 are part of haplotype blocks that have very different haplotype frequencies between AA, EA and HC populations. The Fst for rs231356 is driven mostly by divergence in the EA population.
SCN5A
European Americans, HC and MA populations all have very similar haplotype structures, but the HC population has a distinct allele frequency (FIGURE 1). The EA and MA populations show very little significant difference in allele frequency, with only five markers differing. Several SNPs (rs7374138, rs7372251, rs6599223 and rs7427574) demonstrate highly significant allele-frequency differences from the other populations.
There is one two-marker haplotype block common to all populations, involving rs6599224 and rs6599223 (2490 bp). The HC population appears to have a different haplotype frequency (0.64) to the other three populations for this haplotype block (0.83–0.92) (FIGURE 4). We observed a completely mismatched pattern in the CG and TC haplotypes of SCN5A, with major (CG) haplotype frequencies of 64–92% and minor (TC) frequencies of 8–36% among the ethnicities.
Only 17 of the 39 SNPs in SCN5A have Fst values greater than 0.05, which is an effect of the similarity between the EA and MA populations, and they punctuate differences in allele frequency in the HC and AA populations. SNP rs7372251 peaks in Fst value owing to highly significant allele-frequency differences in the HC population compared with all others and in the AA population compared with all other population.
RYR2
The general haplotype structure of RYR2 is quite similar across the four populations, with the exception of a stretch of SNPs (rs2805422—rs12116442) that form a diffuse block of LD in the EA and MA populations, and a subset of these (rs1382581—rs12115442) that form a well-defined haplotype block in the HC population (FIGURE 5). These structures are almost completely absent in the AA population.
The RYR2 gene exhibited one three-marker haplotype block involving rs7526245, rs7526759 and rs7537963 (606 bp). There are two frequent haplotypes within this haplotype block, and all haplotypes have different frequencies in each population (FIGURE 4). The ‘yin-yang’ haplotypes are defined as two high-frequency haplotypes composed of completely mismatching SNP alleles; for example, nucleotides differ at every SNP in the haplotype pair [22]. A pair of ‘yin-yang’ haplotypes, GTG and ACT, were observed in the first haplotype block (rs7537963—rs7526245) with frequencies between 37–81% and 19–63%, respectively, among the four populations (FIGURE 4).
For RYR2, there were a large number of statistically significant differences in allele frequency in comparisons involving AAs compared with other groups. A couple of stretches of markers demonstrated significant differences (rs2805422—rs2618661 and rs2819757—rs10925501) between AAs and all other populations. EAs and MAs were the most similar in allele frequency distribution, with only 13 of the 55 markers being significantly different.
Measured by Fst, RYR2 had substantial differences among ethnic groups, with 40 of the 55 SNPs in RYR2 having Fst values greater than 0.05 and nearly half (26 of the 55) had Fst values greater than 0.10. One SNP, rs12410114, had an Fst of nearly 0.30, residing at the tail of a diffuse block in EA and MA ethnicities, and a well-defined HC block. This Fst effect is driven largely by dramatic differences in haplotype frequencies, with the TA haplotype being dominant in HC and the AG haplotype being very frequent in the EA and MA populations.
Identified TagSNPs for the selected genes
Tagger analysis revealed that the HC population required the fewest number of TagSNPs (194) to capture all 307 variants (TABLE 1). The AA population, being the most genetically ancestral population, required the most TagSNPs (263). The large haplotype blocks found in ANK2, KCNE1 and 2 and KCNH2 can be exploited by using TagSNPs to dramatically reduce the amount of genotyping required, in some cases reducing the number of genotyped SNPs by half.
Table 1.
TagSNPs identified for each gene and ethnicity.
| Gene | AA | EA | HC | MA | Total SNPs captured |
|---|---|---|---|---|---|
| ANK2 | 66 | 52 | 42 | 54 | 81 |
| KCNE1 and 2 | 56 | 41 | 36 | 36 | 60 |
| KCNH2 | 14 | 8 | 10 | 8 | 19 |
| KCNQ1 | 51 | 47 | 47 | 45 | 53 |
| RYR2 | 42 | 34 | 32 | 33 | 55 |
| SCN5A | 34 | 31 | 27 | 33 | 39 |
| Total | 263 | 213 | 194 | 209 | 307 |
AA: African—American; EA: European American; HC: Han Chinese; MA: Mexican American.
Copy-number variant analysis
Based on the Affymetrix Genotyping Console platform, we identified one Coriell sample with a CNV in ANK2. We also observed four Coriell samples with a CNV in RYR2. One of these CNVs was replicated using the Partek software — a 4241-bp deletion in RYR2 from 235,341,289 to 235,345,530 (p = 0.0001).
Genome-wide association genotyping platforms
To estimate the amount of information capture obtained in genome-wide association studies, we assessed how many TagSNPs for these genes are represented on the commonly used genotyping platforms, including Affymetrix Mapping 100K (AFFY100), Affymetrix Mapping 500K (AFFY500), Affymetrix Mapping 6.0 (AFFY1M), Illumina (CA, USA) HumanHap 300 (ILMN300), Illumina HumanHap 550 (ILMN550), Illumina HumanHap 650 (ILMN650) and Illumina HumanHap 1M (ILMN1M) (TABLES 2 & 3). As expected, more dense SNP arrays capture more of the 307 identified variants, both directly and indirectly using TagSNPs.
Table 2.
Percentage of total SNPs represented on common genome-wide association study platforms.
| Gene | AFFY100 | AFFY500 | AFFY1M | ILMN300 | ILMN550 | ILMN650 | ILMN1M | Total QC+ in gene |
|---|---|---|---|---|---|---|---|---|
| ANK2 | 3.70 | 22.22 | 32.10 | 17.28 | 25.93 | 28.40 | 38.27 | 81 |
| KCNE1 and 2 | 5.00 | 11.67 | 25.00 | 28.33 | 30.00 | 33.33 | 46.67 | 60 |
| KCNH2 | 0.00 | 5.26 | 5.26 | 21.05 | 26.32 | 26.32 | 47.37 | 19 |
| KCNQ1 | 1.89 | 13.21 | 22.64 | 32.08 | 43.40 | 45.28 | 50.94 | 53 |
| RYR2 | 1.82 | 18.18 | 36.36 | 10.91 | 21.82 | 23.64 | 29.09 | 55 |
| SCN5A | 0.00 | 20.51 | 41.03 | 25.64 | 30.77 | 30.77 | 43.59 | 39 |
AFFY: Affymetrix; ILMN: Illumina; QC+: Quality control positive.
Table 3.
Percentage of TagSNPs represented on common genome-wide association study platforms.
| Population | Gene | AFFY100 | AFFY500 | AFFY1M | ILMN300 | ILMN550 | ILMN650 | ILMN1M | TagSNPs in gene |
|---|---|---|---|---|---|---|---|---|---|
| AA | ANK2 | 4.55 | 24.24 | 33.33 | 19.70 | 27.27 | 30.30 | 42.42 | 66 |
| KCNE1 and 2 | 1.89 | 9.43 | 24.53 | 32.08 | 33.96 | 35.85 | 49.06 | 53 | |
| KCNH2 | 0.00 | 7.14 | 7.14 | 28.57 | 28.57 | 28.57 | 42.86 | 14 | |
| KCNQ1 | 1.96 | 13.73 | 23.53 | 33.33 | 45.10 | 47.06 | 52.94 | 51 | |
| RYR2 | 2.38 | 19.05 | 38.10 | 9.52 | 21.43 | 23.81 | 30.95 | 42 | |
| SCN5A | 0.00 | 23.53 | 41.18 | 26.47 | 32.35 | 32.35 | 41.18 | 34 | |
| EA | ANK2 | 3.85 | 26.92 | 38.46 | 21.15 | 30.77 | 30.77 | 38.46 | 52 |
| KCNE1 and 2 | 4.88 | 14.63 | 26.83 | 29.27 | 31.71 | 36.59 | 48.78 | 41 | |
| KCNH2 | 0.00 | 0.00 | 0.00 | 12.50 | 25.00 | 25.00 | 37.50 | 8 | |
| KCNQ1 | 2.13 | 10.64 | 0.00 | 36.17 | 46.81 | 48.94 | 55.32 | 47 | |
| RYR2 | 2.94 | 17.65 | 41.18 | 8.82 | 17.65 | 17.65 | 23.53 | 34 | |
| SCN5A | 0.00 | 25.81 | 41.94 | 29.03 | 35.48 | 35.48 | 48.39 | 31 | |
| HC | ANK2 | 4.76 | 14.29 | 33.33 | 21.43 | 33.33 | 33.33 | 47.62 | 42 |
| KCNE1 and 2 | 2.78 | 11.11 | 25.00 | 36.11 | 38.89 | 41.67 | 47.22 | 36 | |
| KCNH2 | 0.00 | 10.00 | 10.00 | 30.00 | 30.00 | 30.00 | 40.00 | 10 | |
| KCNQ1 | 2.13 | 12.77 | 23.40 | 31.91 | 42.55 | 44.68 | 51.06 | 47 | |
| RYR2 | 3.13 | 18.75 | 40.63 | 3.13 | 18.75 | 18.75 | 28.13 | 32 | |
| SCN5A | 0.00 | 25.93 | 37.04 | 29.63 | 29.63 | 29.63 | 44.44 | 27 | |
| MA | ANK2 | 1.85 | 20.37 | 35.19 | 22.22 | 33.33 | 33.33 | 40.74 | 54 |
| KCNE1 and 2 | 0.00 | 11.11 | 19.44 | 36.11 | 38.89 | 41.67 | 50.00 | 36 | |
| KCNH2 | 0.00 | 0.00 | 0.00 | 25.00 | 25.00 | 25.00 | 50.00 | 8 | |
| KCNQ1 | 2.22 | 8.89 | 20.00 | 35.56 | 44.44 | 46.67 | 53.33 | 45 | |
| RYR2 | 0.00 | 18.18 | 42.42 | 6.06 | 12.12 | 12.12 | 15.15 | 33 | |
| SCN5A | 0.00 | 24.24 | 45.45 | 27.27 | 30.30 | 30.30 | 45.45 | 33 | |
AA: African—American; AFFY: Affymetrix; EA: European American; HC: Han Chinese; ILMN: Illumina; MA: Mexican American.
Discussion
The rhythmonome is the term we have adopted to describe the set of genes that determine the coordinated, normal electrical activity in the heart. The discovery of rare mutations that induce unusual arrhythmia syndromes and common variants that modify more frequent arrhythmia phenotypes invited a more thorough characterization of ethnic differences in allele frequency, LD structure and haplotype frequencies across six key genes of the rhythmonome (ANK2, SCN5A, the KCNE1 and 2 gene cluster, KCNQ1, KCNH2 and RYR2). A complete characterization of all haplotypes would require resequencing each gene region in many individuals [23,24], but the results of this study provide a basic, working understanding of population differences in these six genes that is necessary for conducting association studies. We also provide an assessment of the representation of the selected SNPs on various large-scale genotyping platforms. These large-scale genotyping platforms are extremely popular for genome-wide association studies; however, as demonstrated in this manuscript, not all genes are covered equally, thus, supplemental genotyping may be advantageous. In addition, most of these platforms are catered for populations of European descent (with some additional SNPs for African descent populations) only. Because of the known ethnic variation in polymorphisms across the genome, there is clearly a need to be careful in selection of SNPs for association studies at the genome-wide or candidate-gene level. A variant of the SCN5A sodium channel (Y1102, rs7626962 — between rs7428882 and rs7431011) has previously been demonstrated to influence the risk of cardiac arrhythmia [8]. This variant is common among individuals of African descent, and the AA population surveyed in this study shows a large degree of allele-frequency differences from the other populations near the location of this variant. There is a relatively high Fst value in the genetic region surrounding the variant, and punctuate stretches of allele-frequency difference suggest that the AA population, on average, harbors significantly different alleles in this gene to the other populations sampled. In addition, the scant patterns of LD in this area make using TagSNPs in this region problematic, and may indicate that common genome-wide association study platforms may not sample the variation of this gene well for AAs.
Studies have also reported rare polymorphisms in the KCNE1 and 2 gene cluster that are associated with increased torsades risk (D85N, rs1805128 — between rs2070357 and rs2070358) [25,26]. Should there also be common variants in this functionally critical region of these potassium channel genes, they are likely to lie on a haplotype background in both AA and MA populations. In EA and HC populations, this region lies on the edge of well-defined haplotype blocks. The region also harbors low genetic diversity, with few differences in allele frequency among the populations surveyed and low Fst values; therefore, it is possible that common variants with similar functional consequences to the rare polymorphisms of this gene would influence risk in multiple populations.
Ion channels are the primary regulators of normal cardiac rhythm, and abundant evidence is now emerging that common and rare variation in ion-channel genes modulate the risk of common arrhythmias, such as atrial fibrillation or sudden cardiac death [27-30]. Susceptibility to arrhythmias also appears to include an ancestry-dependent component [8]; for example, while AA subjects have a high incidence of traditional risk factors for atrial fibrillation, the incidence of the arrhythmia is actually lower than in other ethnic groups [31]. The effect of drugs to treat or prevent arrhythmias is highly variable, and reasons for this include a genetic component factor [32]. Thus, the present data point to future work in unraveling both arrhythmia susceptibility and variability in response to drug therapy.
Executive summary.
Genes of the rhythmonome
The ‘rhythmonome’ is the term we have adopted to describe the set of genes that determine the coordinated normal electrical activity in the heart.
In this study, the allele and haplotype diversity of six genes (ANK2, SCN5A, KCNE1 and 2 gene cluster, KCNQ1, KCNH2 and RYR2) were investigated in four human populations (African—American, European American, Han Chinese and Mexican American).
Genetic diversity of the rhythmonome
There are population-specific patterns of linkage disequilibrium and differences in allele frequency between African—American, European American, Mexican American and Han Chinese ethnicities for the six gene regions of the rhythmonome surveyed in this study.
These variations may have regulatory or other functional consequences that could account for a portion of the ethnic disparity seen in common arrhythmias.
Furthermore, these variations may not be well captured by popular large-scale genotyping technologies.
Acknowledgments
This work was supported by National Institutes of Health grants HL65962. The authors would like to acknowledge Robert Woodhall and Nila Gillani for their contributions to the genotyping.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Rudy Y, Silva JR. Computational biology in the study of cardiac ion channels and cell electrophysiology. Q Rev. Biophys. 2006;39(1):57–116. doi: 10.1017/S0033583506004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2 ■.Knollmann BC, Roden DM. A genetic framework for improving arrhythmia therapy. Nature. 2008;451(7181):929–936. doi: 10.1038/nature06799.Important information for using genetic information to understand treatment of arrhythmias.
- 3.Priori SG, Napolitano C. Role of genetic analyses in cardiology: part I: mendelian diseases: cardiac channelopathies. Circulation. 2006;113(8):1130–1135. doi: 10.1161/CIRCULATIONAHA.105.563205. [DOI] [PubMed] [Google Scholar]
- 4.Bezzina CR, Shimizu W, Yang P, et al. Common sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113(3):338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 5.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin. Pharmacol. Ther. 2007;81(1):35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotti L, Lundquist AL, Insolia R, et al. KCNH2-K897T is a genetic modifier of latent congenital long-QT syndrome. Circulation. 2005;112(9):1251–1258. doi: 10.1161/CIRCULATIONAHA.105.549071. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich JR, Zicha S, Coutu P, Hebert TE, Nattel S. Atrial fibrillation-associated minK38G/S polymorphism modulates delayed rectifier current and membrane localization. Cardiovasc. Res. 2005;67(3):520–528. doi: 10.1016/j.cardiores.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Splawski I, Timothy KW, Tateyama M, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297(5585):1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, Koopmann TT, Pfeufer A, et al. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur. J. Hum. Genet. 2008;16(3):350–357. doi: 10.1038/sj.ejhg.5201952. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard TJ, Aken BL, Beal K, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasprzyk A, Keefe D, Smedley D, et al. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14(1):160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29(2):311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 13 ■■.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457.Haploview is the most commonly used software for exploring linkage disequilibrium patterns in human genetics data.
- 14 ■.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424.Haplotype structure is explored in the human genome through the use of the authors own method.
- 15.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 18.Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 19.Weir B. Population Substructure. In: Sunderland M, editor. Genetic data analysis II. Sinauer Associates; MA, USA: 1996. [Google Scholar]
- 20.Fiegler H, Redon R, Andrews D, et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16(12):1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komura D, Shen F, Ishikawa S, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16(12):1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Rowe WL, Clark AG, Buetow KH. Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am. J. Hum. Genet. 2003;73(5):1073–1081. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429(6990):446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 24.Crawford DC, Carlson CS, Rieder MJ, et al. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 2004;74(4):610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulussen AD, Gilissen RA, Armstrong M, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1 and KCNE2 in drug-induced long QT syndrome patients. J. Mol. Med. 2004;82(3):182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 26.Yang P, Kanki H, Drolet B, et al. Allelic variants in long-QT disease genes in patients with drug-associated torsades de pointes. Circulation. 2002;105(16):1943–1948. doi: 10.1161/01.cir.0000014448.19052.4c. [DOI] [PubMed] [Google Scholar]
- 27.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448(7151):353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 28.Kaab S, Darbar D, van NC, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur. Heart J. 2009;30(7):813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao WH, Arking DE, Post W, et al. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119(7):940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 31.Borzecki AM, Bridgers DK, Liebschutz JM, Kader B, Kazis LE, Berlowitz DR. Racial differences in the prevalence of atrial fibrillation among males. J. Natl Med. Assoc. 2008;100(2):237–245. doi: 10.1016/s0027-9684(15)31212-8. [DOI] [PubMed] [Google Scholar]
- 32.Darbar D, Motsinger AA, Ritchie MD, Gainer JV, Roden DM. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm. 2007;4(6):743–749. doi: 10.1016/j.hrthm.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Supplementary figures 1–30 http://chgr.mc.vanderbilt.edu/ritchielab/rhythmonome.