Abstract
Herein we report the de novo design and synthesis of a geometrically flexible bis-indole carboxamide and a constrained derivative, as a novel class of small molecule scaffold that exhibits high stabilization potential for DNA G-quadruplex sequences associated with the promoters of c-kit2 and c-myc.
Targeting G-quadruplex nucleic acids with small molecules is emerging as a potential strategy for anti-cancer drug design.1,2 G-quadruplex sequence motifs are widespread in genomic regions such as telomeres,3 and in gene promoters.4,5 Small molecules that selectively bind and stabilize the telomeric quadruplex can inhibit telomerase, an enzyme up-regulated in cancer cells.2,3 There is evidence that the binding of small molecules to the G-quadruplex associated with proto-oncogenes including c-myc,5a KRAS, 5b PDGF-A5c and c-kit,6,7 can modulate transcription. Recent data suggest that RNA G-quadruplex in the 5′ untranslated regions may also offer another target for small molecule intervention.8
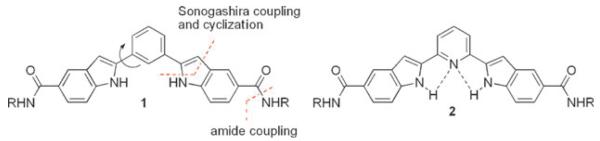
Significant progress has been made during the past few years in the design of small molecules that target G-quadruplex DNA, and a number of classes of ligands have been reported.9 Most of them comprise a planar, aromatic core presumed to stack on the terminal tetrads of G-quadruplex. The scope and utility of G-quadruplex selective molecules were further broadened by a very recent report on a nucleic acid catalyst based on small molecule–G-quadruplex recognition.10 Herein we report the design, synthesis and biophysical evaluation of a conformationally flexible bis-indole carboxamide and a constrained analogue as a new class of G-quadruplex binding ligands (Fig. 1).
Fig. 1.
Design of cationic bis-indole derivatives 1 and 2.
Indole derivatives are privileged scaffolds for medicinal chemistry,11 thus we envisioned that designing G-quadruplex ligands based on such a structure might lead to novel nontoxic compounds suitable for cell biology. Moreover, compounds 1 and 2 exhibit a planar central core,12 having comparable molecular size to the G-quartet (Supporting Information, S.I.†).13 Another relevant feature in our design is the potential for multiple H-bonding sites in the flat bis-indole core of 1 and 2. Ligand 2 may contain internally organized H-bonds between the NH of the indole ring and the pyridine lone pair electrons, whereas ligand 1 can adopt different conformations due to the geometrical freedom (Fig. 1, S.I.†).
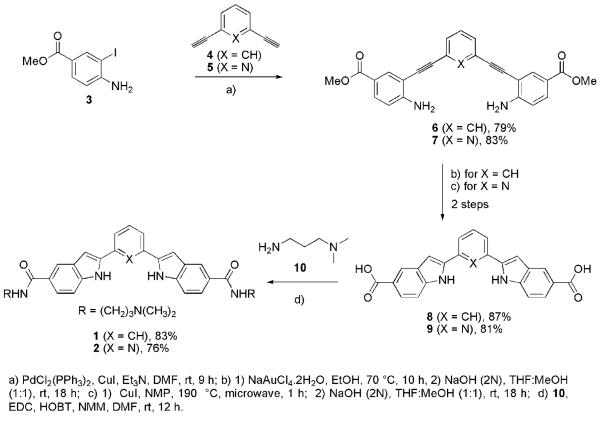
The 2,6-bis(2′-indolyl)pyridine core of ligand 2 has been used as a tridentate ligand12,14 and as synthetic receptors.14 While several methodologies exist for the construction of indole ring systems,11a only a few approaches12,14,15 have been reported for the synthesis of the 2,6-bis(2′-indolyl)pyridine core of ligand 2. Our synthesis was based on the application of Sonogashira coupling,16 and 5-endo-dig cyclization17 (Fig. 1, Scheme 1), and would be amenable to the preparation of diverse functionalized indole derivatives. The Sonogashira coupling reaction of iodoaniline derivative 3 with 1,3-diethynyl-benzene (4) and 2,6-diethynyl-pyridine (5) proceeds with excellent yield to afford the corresponding diester derivatives 6 and 7. The diacids 8 and 9 were acquired in high yields over two steps involving a 5-endo-dig type cyclization18 and base mediated hydrolysis of the corresponding indole diester intermediates. Treatment of diacids 8 and 9 with amine 10 gave the corresponding bis-amides 1 and 2. The syntheses of 1 and 2 were efficiently achieved using 4-step protocols in 57% and 51% overall yield respectively starting from easily accessible starting materials 3–5 (Scheme 1).
Scheme 1.
Synthesis of water soluble bis-indole derivatives 1 and 2.
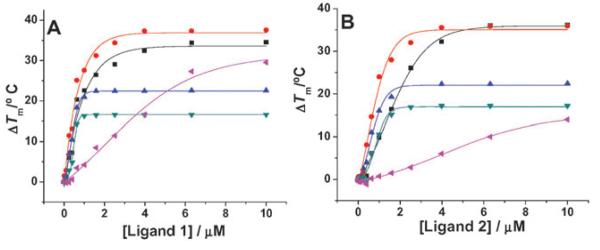
We evaluated the ability of ligands 1 and 2 to interact with four different G-quadruplex forming oligonucleotides based on human genomic sequences using fluorescence resonance energy transfer (FRET) melting.19 FRET melting analysis determines the ligand-induced stabilization of a folded quadruplex by measurement of the ligand induced increase in the melting temperature (ΔTm). The G-quadruplex forming sequences comprised the human telomeric G-quadruplex DNA (h-telo, FAM-d(G3[T2AG3]3)-TAMRA), two G-quadruplexes found in the promoter of c-kit (c-kit1,6b FAM-d(G3AG3CGCTG3AG2AG3)-TAMRA; and c-kit2,6a FAM-d(G3CG3CGCGAG3AG4)-TAMRA) and the G-quadruplex found in the promoter of c-myc5a (c-myc, FAM-d(TGAG3TG3TAG3TG3TA2)-TAMRA). We have also evaluated the melting of a duplex DNA (dup, FAM-d(TATAGCTATA-HEG-TATAGCTATA)-TAMRA) as a non-quadruplex control. The quadruplex melting data are summarized in Table 1 and Fig. 2 (S.I.†).
Table 1.
DNA G-quadruplexes and duplex stabilization by 1 and 2 determined by FRET melting experiments
| Ligand | ΔTm at 1 μM ligand concentration (°C) |
||||
|---|---|---|---|---|---|
| h-telo (59 ± 1)a | c-kit1 (57 ± 1)a | c-kit2b (71 ± 1)a | c-mycb (77 ± 1)a | dup (58 ± 1)a | |
| 1 | 21.5 | 27.7 | 21.7b | 16.5b | 4.2 |
| 2 | 9.8 | 22.1 | 16.0 | 10.5 | 0.5 |
Tm in 60 mM potassium cacodylate buffer, pH 7.4 without ligand.
Maximum measurable ΔTm that can be measured for c-kit2 is 24 °C (i.e. Tm = 95 °C) and for c-myc is 18 °C (i.e. Tm = 95 °C).
Fig. 2.
FRET stabilization curves for ligands (A) 1 and (B) 2 upon binding to h-telo ( ), c-kit1 (
), c-kit1 ( ), c-kit2 (
), c-kit2 ( ), c-myc (
), c-myc ( ) and double stranded (
) and double stranded ( ) DNA.
) DNA.
Ligand 1 exhibits a high stabilization potential for the c-kit2 quadruplex (ΔTm = 21.7 °C at 1 μM, i.e. a Tm of 93 °C) comparable in magnitude to data obtained for the natural product telomestatin (ΔTm= 20.4 °C at 1 μM) and for a quinoline macrocycle (ΔTm = 21.4 °C at 1 μM), which are two of the best reported quadruplex ligands.20 The stabilization potential of 1 for c-myc also saturates at 1 μM concentration (ΔTm = 16.5 °C, i.e.Tm = 93.5 °C). Only a few ligands are reported to interact and modulate c-myc G-quadruplex.9 The ΔTm of ligand 1 for the c-kit1 quadruplex (27.7 °C) was found to be comparable with that of one of the best ligands (27.1 °C) from our recently reported isoalloxazine series, that also shows biological activity against c-kit.7 It is interesting to note the comparative ΔTm values for ligands 1 and 2 (phenylene vs. pyridyl). While ligand 1 shows detectable duplex stabilization (ΔTm = 4.2 °C), diminished duplex stabilization (ΔTm = 0.5 °C) was observed for the more geometrically constrained ligand 2. Similarly there is a large variation of ΔTm of ligand 1 and 2 for h-telo quadruplex. The observed ΔTm of ligand 1 for h-telo was 21.5 °C. Ligand 2 shows stronger stabilization for c-kit1 (ΔTm = 22.1 °C) and c-kit2 (ΔTm = 16.0 °C) as compared to h-telo (ΔTm = 9.8 °C) G-quadruplex DNA at 1 μM concentration.
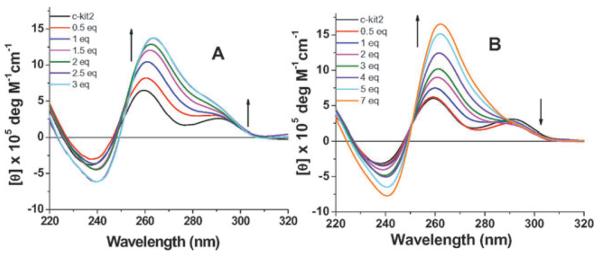
Circular dichroism (CD) spectroscopy on DNA sequences known to fold into G-quadruplexes can be used to monitor the formation of G-quadruplex structure.21 The real time CD titrations of 1 and 2 were conducted for h-telo, c-kit1, c-kit2 and c-myc quadruplex forming DNA sequences at room temperature in the absence of added stabilizing salts22 (S.I.Δ) to find out any selectivity for a particular G-quadruplex conformation. The c-kit2 quadruplex exists as a predominantly parallel structure with a small proportion of an antiparallel conformation.6a The CD spectrum of c-kit2 quadruplex in the absence of any added salt shows a major positive peak at 260 nm and a minor peak at 293 nm, suggestive of a mixture of parallel and antiparallel structures.6a,20 Upon titration with ligand 2 in real time (Fig. 3B), it was noteworthy in the CD spectroscopic analysis that the ligand dependent increase in ellipticity at 260 nm is not accompanied by an associated increase in the peak at 293 nm, supportive of selective induction of a parallel G-quadruplex structure.§ The effect was more prominent at high ligand concentration with the disappearance of the peak at 293 nm. When the titration experiments were carried out with ligand 1, the peaks at both 260 nm and 293 nm were increased, suggesting that ligand 1 does not show any preference to induce folding of a particular quadruplex structure (Fig. 3A). In contrast, for the cases of h-telo, c-kit1 and c-myc an incremental ligand dependent induction in G-quadruplex was not significant and an apparent quadruplex unfolding was observed at high ligand concentration (S.I.†).
Fig. 3.
CD spectra of a 12.5 μM solution of c-kit2 d[G3CG2GCGCGAG3AG4] in Tris buffer (pH 7.4), 0–7 equivalent of ligands (A) 1 and (B) 2.
We have developed an expedient synthetic route to water soluble bis-indole amides, that are a novel class of de novo G-quadruplex binding small-molecule ligands. These ligands have shown a level of G-quadruplex stabilization that compares favourably with the best reported G-quadruplex ligands. CD spectroscopic analysis further reveals strong interaction of the ligands with c-kit2 G-quadruplex DNA. The properties of these ligands make them attractive probes to explore hypotheses for the biological function of G-quadruplexes and such investigations are currently underway.
Acknowledgments
We thank the BBSRC for project funding, Cancer Research UK for programme funding and the European Commission for a Marie Curie Incoming Postdoctoral Fellowship to JD.
Footnotes
Electronic supplementary information (ESI) available: Experimental procedure for FRET, CD and NMR data of indole amides. See DOI: 10.1039/b806042h
CD spectra of ligand 2 annealed with c-kit2 quadruplex and CD titration of ligand 2 with c-kit2 in the presence of K+ are presented in S.I.†
The UV/Vis spectrum for ligand 1 shows absorption at 257, 272, 323 nm, and that of ligand 2 shows absorption at 264, 341 nm. Upon titration with G-quadruplex DNA, both the ligands show peaks at 260 nm. We cannot rule out the possibility that ICD bands belonging to 1 and 2 (at 272 and 264 nm) might overlap in this region.
Notes and references
- 1.Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Royal Society of Chemistry; Cambridge, UK: 2006. [Google Scholar]
- 2.(a) Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJB, Double JA, Neidle S. Cancer Res. 2005;65:1489. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]; (b) Mandine E, Riou J-F, Mergny J-L, Mailliet P, Boussin F. Oncogene. 2005;24:2917. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]; (c) Han H, Hurley LH. TiPS. 2000;21:136. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 3.Sun DY, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. J. Med. Chem. 1997;40:2113. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 4.(a) Huppert JL, Balasubramanian S. Nucleic Acids Res. 2007;35:406. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huppert JL, Balasubramanian S. Nucleic Acids Res. 2005;33:2908. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Todd AK, Johnston M, Neidle S. Nucleic Acids Res. 2005;33:2901. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Nucleic Acids Res. 2007;35:1. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Fernando H, Reszka AP, Huppert JL, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. Biochemistry. 2006;45:7854. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2005;127:10584. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. J. Am. Chem. Soc. 2007;129:12926. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. Nat. Chem. Biol. 2007;3:218. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.For reviews, see: Monchaud D, Teulade-Fichou M-P. Org. Biomol. Chem. 2008;6:627. doi: 10.1039/b714772b. Cuesta J, Read MA, Neidle S. Mini-Rev. Med. Chem. 2003;3:11. doi: 10.2174/1389557033405502. Waller ZAE, Shirude PS, Rodriguez R, Balasubramanian S. Chem. Commun. 2008:1467. doi: 10.1039/b718854d. For specific examples see. Reed JE, Neidle S, Vilar R. Chem. Commun. 2007:4366. doi: 10.1039/b709898g. Gonçalves DPN, Rodriguez R, Balasubramanian S, Sanders JKM. Chem. Commun. 2006:4685. doi: 10.1039/b611731g. Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. J. Am. Chem. Soc. 2006;128:13662. doi: 10.1021/ja064713e. Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-ya K, White E, Wilson WD, Hurley LH. J. Am. Chem. Soc. 2005;127:2944. doi: 10.1021/ja0444482.
- 10.Tang Z, Gonçalves DPN, Wieland M, Marx A, Hartig JS. ChemBioChem. 2008;9:1061. doi: 10.1002/cbic.200800024. [DOI] [PubMed] [Google Scholar]
- 11.For reviews, see: Humphrey GR, Kuethe JT. Chem. Rev. 2006;106:2875. doi: 10.1021/cr0505270. Horton DA, Bourne GT, Smythe ML. Chem. Rev. 2003;103:893. doi: 10.1021/cr020033s. Knoölker H-J, Reddy KR. Chem. Rev. 2002;102:4303. doi: 10.1021/cr020059j.
- 12.For the X-ray crystal structure of the 2,6-bis(2′-indolyl)pyridine core, see: Jia W-L, Liu Q-D, Wang R, Wang S. Organometallics. 2003;22:4070.
- 13.Molecular dimensions of indole derivative 1 are determined from an energy minimized chem 3D diagram and compared with the X-ray crystal structure of G-tetrad. Cian AD, DeLemos E, Mergny J-L, Teulade-Fichou M-P, Monchaud D. J. Am. Chem. Soc. 2007;129:1856. doi: 10.1021/ja067352b. Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876. doi: 10.1038/nature755.
- 14.(a) Liu Q, Thorne L, Kozin I, Song D, Seward C, D'Iorio M, Tao Y, Wang S. J. Chem. Soc., Dalton Trans. 2002:3234. [Google Scholar]; (b) Thummel RP, Hegde V. J. Org. Chem. 1989;54:1720. [Google Scholar]; (c) Hegde V, Hung C-Y, Madhukar P, Cunningham R, Hopfner T, Thummel RP. J. Am. Chem. Soc. 1993;115:812. [Google Scholar]
- 15.(a) Tollari S, Cenini S, Rossi A, Palmisano G. J. Mol. Catal. A: Chem. 1998;135:241. [Google Scholar]; (b) Palmisano G, Santagostino M. Helv. Chim. Acta. 1993;76:2356. [Google Scholar]
- 16.For reviews, see: Sonogashira K. In: Metal-Catalyzed Cross-Coupling Reactions. Diederich F, Stang PJ, editors. Wiley-VCH; New York: 1998. ch. 5. Negishi E-I, Anastasia L. Chem. Rev. 2003;103:1979. doi: 10.1021/cr020377i.
- 17.For a review, see: Muüller TE, Beller M. Chem. Rev. 1998;98:675. doi: 10.1021/cr960433d.
- 18.(a) Arcadi A, Bianchi G, Marinelli F. Synthesis. 2004:610. [Google Scholar]; (b) Pearson SE, Nandan S. Synthesis. 2005:2503. [Google Scholar]
- 19.(a) Darby RAJ, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) De Cian A, Guittat L, Kaiser M, Sacca B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou M-P, Lacroix L, Mergny J-L. Methods. 2007;42:183. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Shirude PS, Gilles ER, Ladame S, Godde F, Shin-ya K, Huc I, Balasubramanian S. J. Am. Chem. Soc. 2007;129:11890. doi: 10.1021/ja073775h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Balagurumoorthy P, Brahmachari SK. J. Biol. Chem. 1994;269:21858. [PubMed] [Google Scholar]; (b) Jin R, Gaffney BL, Wang C, Jones RA, Breslauer KJ. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8832. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Rezler EM, Seenisamy J, Bashyam S, Kim M-Y, White E, Wilson WD, Hurley LH. J. Am. Chem. Soc. 2005;127:9439. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]; (b) Rodriguez R, Pantos GD, Gonçalves DPN, Sanders JKM, Balasubramanian S. Angew. Chem., Int. Ed. 2007;46:5405. doi: 10.1002/anie.200605075. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fu B, Huang J, Ren L, Weng X, Zhou Y, Du Y, Wu X, Zhou X, Yang G. Chem. Commun. 2007:3264. doi: 10.1039/b704599a. [DOI] [PubMed] [Google Scholar]