Summary
A new Cre-reporter strain of mouse has been developed that expresses a fusion protein derived from the lacZ gene fused to GFP with a nuclear localization signal. This construct is expressed from the ROSA26 locus upon Cre-mediated recombination that removes a loxP-flanked PGK-neo cassette, thus allowing for detection of Cre activity in all tissues. This reporter strain, which is similar to prior R26R and R26EGFP strains, has certain advantages related to the nuclear expression and the combined expression of both β-galactosidase and GFP activities. We show that the use of this newly developed reporter line allows for enhanced resolution, detection and co-localization. Thus, we report a previously unrecognized subset of venous endothelial cells derived from Pax3 expressing precursors. genesis 46:200–204, 2008.
Keywords: Pax3, reporter, cardinal vein, GFP, β-gal-actosidase, endothelial cell
Cre recombinase has been used extensively to mediate recombination in the mouse genome, thus allowing for tissue-specific deletion of target genes or genomic sequences. In order to detect Cre activity in vivo, and to identify the derivatives of Cre-expressing cells, a number of Cre-reporter mouse strains have been developed (Chai et al., 2000; Kishigami et al., 2006; Luche et al., 2007; Mao et al., 1999, 2001; Novak et al., 2000; Soriano, 1999; Srinivas et al., 2001; Yamauchi et al., 1999). These Cre-reporter mice share in common the use of a widely expressed promoter that directs expression of a reporter gene (such as lacZ or GFP), and reporter gene expression is prevented by inclusion of a loxP-flanked cassette between the translation initiation site and the reporter gene itself. Reporter strains have been developed as transgenic mice (using the chicken α actin promoter, for example, that expresses widely in mouse tissues) or as knock in mice using homologous recombination in embryonic stem cells. The knock in strategy has the advantage of relative stability and reproducibility, and the lack of potential insertional mutagenesis that can complicate the analysis of new lines of transgenic mice.
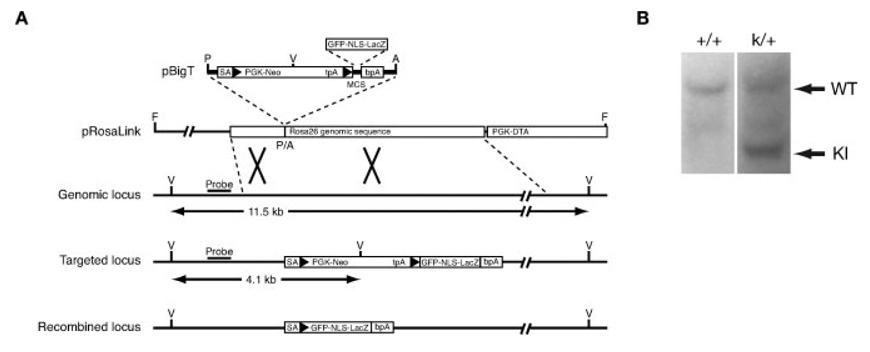
The ROSA26 locus has been used successfully to generate Cre reporter mice, called R26R, that express β-galactosidase upon Cre-mediated recombination (Soriano, 1999). Subsequently, similar reporter mice targeted to the ROSA26 locus that express EGFP (Mao et al., 2001) or EYFP (Srinivas et al., 2001) have been reported. In each case, Cre activity deletes a loxP-flanked PGK-neo cassette that includes stop codons in all three reading frames, and reporter gene expression is produced. One potential disadvantage of some Cre reporter mouse lines is that analysis at the single cell level can be difficult or imprecise due to diffuse signals or “bleeding” after β-galactosidase staining. To address this problem, we have generated a reporter line expressing a 151 kDa nuclear localized GFP/βgal fusion protein which allows for single cell resolution. We targeted the ROSA26 locus using previously described methods to produce mice in which a fusion of GFP and lacZ, including the SV40 nuclear localization signal (NLS), was inserted downstream of the loxP-flanked PGK-neo cassette (Fig. 1a). Correctly targeted ES cells were identified by Southern blotting (Fig. 1b) and used to generate chimeric mice, which produced germline transmission. The resulting line of mice harboring the GFP-NLS-lacZ knock-in is referred to hereafter as GNZ.
FIG. 1. Generation of ROSA26 GNZ knock-in mice.
A: Targeting strategy of GNZ. GFP-NLS-lacZ was cloned into the pBigT vector and subsequently into the targeting vector pRosaLink. The expected sizes of the EcoRV (V) digests recognized by the indicated probe are shown. Arrowheads indicate loxP sites. A, AscI; P, PacI; F, FseI. B: Genomic Southern blot of wild-type (+/+) and targeted GNZ heterozygous ES cells (k/+). Wild-type (WT; 11.5 kb) and knock-in (KI; 4.1 kb) bands are shown.
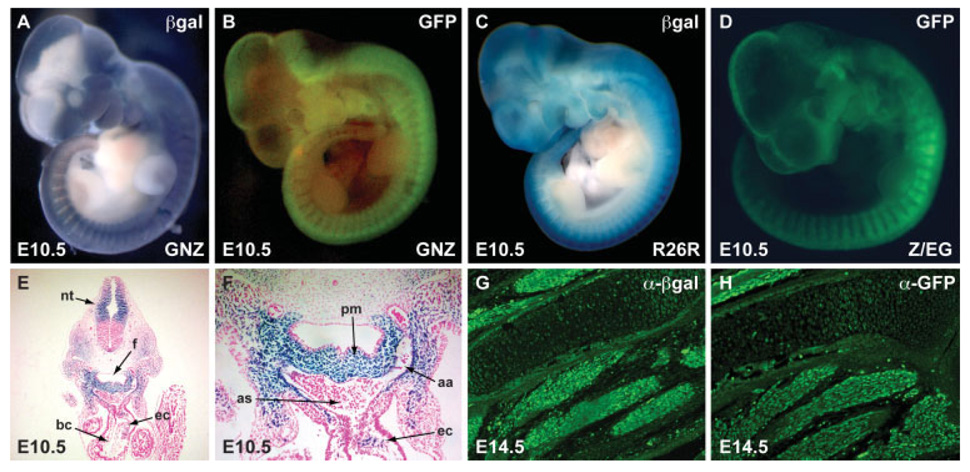
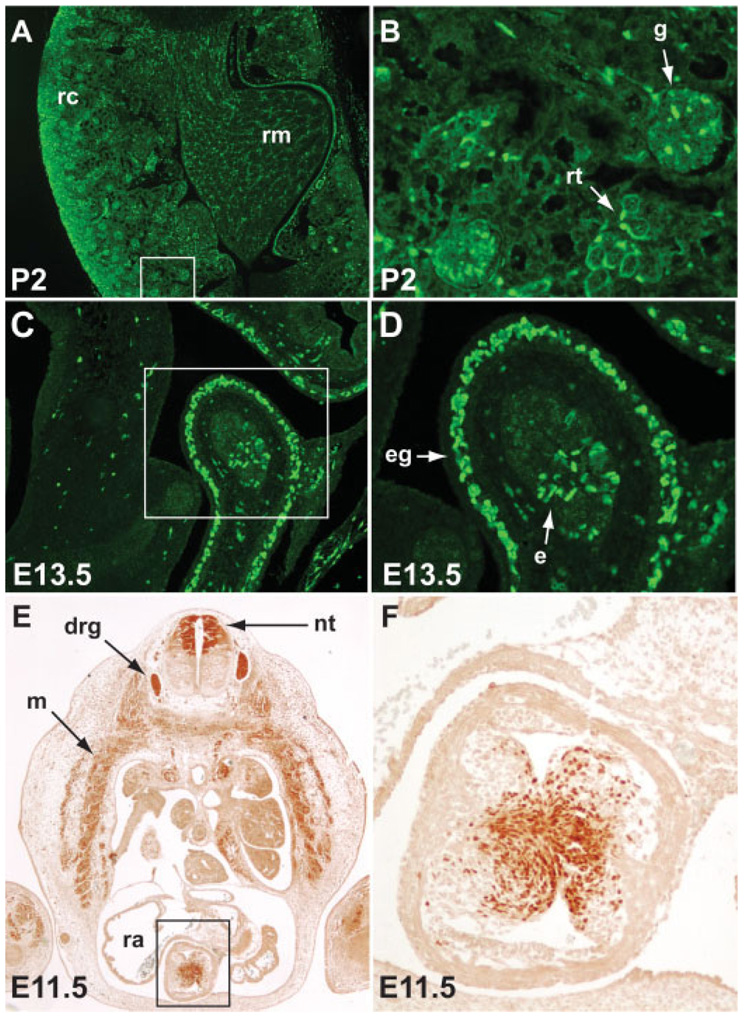
We crossed GNZ mice with Pax3Cre/+ mice in which Cre recombinase has been knocked into the Pax3 locus (Engleka et al., 2005), and analyzed the progeny by immunohistochemistry, X-gal staining, and fluorescence microscopy. We compared the resulting embryos to those derived from crosses of Pax3Cre/+ with R26R or Z/EG. GNZ embryos (E9.5–18.5) never expressed GFP or β-galactosidase activity in the absence of Cre (not shown). However, Pax3Cre/+; GNZ embryos expressed both β-galactosidase (Fig. 2a) and GFP (Fig. 2b) in the dorsal neural tube and somites in a pattern identical to that previously described for Pax3 and its derivatives (Engleka et al., 2005; Milewski et al., 2004). β-galactosidase stained embryos appeared similar to Pax3Cre/+; R26R (Fig. 2c) and to Pax3Cre/+; Z/EG (Fig. 2d) stained embryos, except that punctate nuclear staining was evident upon high power examination (Fig. 2f). While Pax3 derivatives were easily visualized by direct GFP fluorescence, the high resolution nuclear staining was best visualized by immunostaining for either GFP or β-galactosidase. A subset of Pax3 expressing somitic cells give rise to the hypaxial dermomyotome, which subsequently differentiates into skeletal muscle including limb muscle (Esner et al., 2006). These limb muscle Pax3 derivatives were confirmed in E14.5 Pax3Cre/+; GNZ embryos by GFP and β-galactosidase immunostaining (Fig. 2g,h). In addition to the dorsal neural tube, somites, and limb muscle, Pax3 derivatives were also observed in other organs such as the kidney and bowel, consistent with prior reports (Engleka et al., 2005). In the neonatal kidney, Pax3 derivatives were identified in Pax3Cre/+; GNZ pups by immunohistochemistry with an anti-GFP antibody at P2. Labeled cells were present in multiple areas of the renal cortex and medulla including the glomeruli and a subset of renal tubules (Fig. 3a,b). This is in agreement with our prior report (Engleka et al., 2005) although Pax3 derivatives in the kidney had not been reported previously and were unexpected. In the intestine, Pax3 derivatives were visualized in the bowel wall (Fig. 3c,d) in a pattern consistent with that expected for enteric ganglia, which are known to arise from Pax3- expressing neural crest (Lang et al., 2000). Here, we also confirm the existence of Pax3-derived epithelial cells in the distal bowel that we reported previously (Engleka et al., 2005) (Fig. 3e,f).
FIG. 2. β-galactosidase staining and GFP fluorescence using GNZ reporter mouse.
A,B: Reporter gene expression pattern of E10.5 Pax3Cre/+; GNZ embryos. C: β-galactosidase staining of Pax3Cre/+; R26R E10.5 embryo. D: GFP fluorescence of Pax3Cre/+; Z/EG E10.5 embryo. E: β-galactosidase staining of cross section at the level of the heart of an E10.5 Pax3Cre/+; GNZ embryo shows punctate nuclear staining in the dorsal neural tube (nt) and pharyngeal mesenchyme. F: Higher power of image shown in panel E showing Pax3 derivatives in the pharyngeal mesenchyme (pm), surrounding the aortic arch arteries (aa) and within the endocardial cushions (ec) of the cardiac out-flow tract. G,H: Non-adjacent sections of E14.5 Pax3Cre/+;GNZ embryos examined by immunohistochemistry using anti-β-gal (G) and anti-GFP (H) antibodies show nuclear staining of Pax3 skeletal limb muscle derivatives. as: aortic sac; bc: bulbus cordis; f: foregut.
FIG. 3. Pax3 derivatives are seen in the kidney, bowel and outflow tract endocardial cushions.
A,B: Immunohistochemistry with anti-GFP antibody shows Pax3 derivatives in the renal cortex (rc) and medulla (rm) of Pax3Cre/+; GNZ neonates. Staining is evident in the glomeruli (g) and a subset of renal tubules (rt). Panel B is a higher power image of the area shown in the white box in panel A. C,D: Pax3 derivatives are seen in the bowel wall in the region populated by enteric ganglia (eg), and more centrally co-localizing with epithelial cells (e). E,F: Immunohistochemistry with anti-β-galactosidase antibody of an E11.5 Pax3Cre/+; GNZ embryo at the level of the right atrium (ra) shows nuclear staining in the neural tube (nt), dorsal root ganglia (drg), myotome (m) and cardiac outflow tract (black box). F: Higher power of area indicated by black box in panel E reveals nuclear staining of cells within the endocardial cushions.
In the cardiovascular system, Pax3-derived neural crest cells have been reported to contribute to the outflow tract endocardial cushions (Brown et al., 2001; Engleka et al., 2005) and to the smooth muscle layer of the aortic arch (Brown et al., 2001; Li et al., 2000). Some reports (Poelmann et al., 2004; Stottmann et al., 2004), but not others (Cheng et al., 1999; Engleka et al., 2005), have suggested that neural crest derivatives are also found in the epicardium, myocardium or conduction system of the heart. In Pax3Cre/+; GNZ embryos examined between E10.5 and E14.5, we did not observe any labeled myocardial or epicardial cells, while Pax3 derivatives were easily identified in the outflow tract and aortic arch (Fig. 3e,f, and Fig. 4a) in a pattern identical to that predicted for cardiac neural crest derivatives (Jiang et al., 2000).
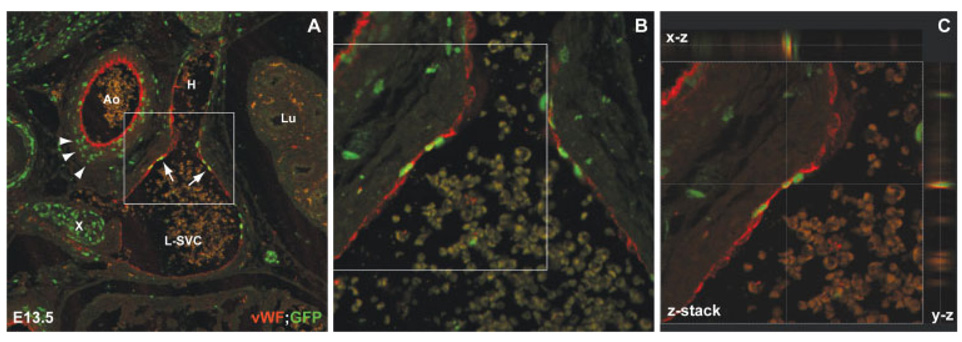
FIG. 4. Pax3 progenitors contribute to venous endothelial cells.
A: Cross section of an E13.5 Pax3Cre/+; GNZ embryo co-stained with antivWF (red) and anti-GFP (green) analyzed by confocal microscopy shows nuclear GFP expression by aortic (Ao) smooth muscle cells (arrowheads) and venous endothelial cells (arrows) of the left superior vena cava (L-SVC) and hemiazygous vein (H). B: Higher power image of area indicated by white box in panel A shows co-localization of GFP and vWF staining. C: An orthogonal view of a confocal z-stack of area indicated by white box in panel B shows cytoplasmic vWF surrounding nuclear GFP in a single endothelial cell. The top and right sides depict x–z and y–z views, respectively, of the stack at the location of the cross hair. Lu: lung; X: left vagal trunk.
The high sensitivity at the single cell level afforded by the GNZ reporter mice allowed us to unequivocally identify a novel Pax3 derivative during development. We identify Pax3 derivatives in the endothelial layer of the cardinal veins, a domain that has not, to our knowledge, been previously reported. The endothelial identity of these Pax3 derivatives was confirmed by co-staining with anti-von Willebrand Factor (vWF) antibodies and confocal microscopy (see Fig. 4). The Pax3 derived cells we observed comprise a portion of the cardinal vein derived left superior vena cava. Interestingly, Delot et al. have proposed that specific defects in anterior cardinal vein development seen in Chordin mutant mice may be a non-cell autonomous defect related to abnormal neural crest cell migration (Delot et al., 2007). Our finding that Pax3 derived cells contribute directly to the endothelium of specific veins may provide an alternative explanation involving a cell autonomous role of Pax3-derived venous endothelial cells to account for these defects.
Our results confirming patterns of Pax3-derived cells in multiple organs, along with our description of a previously unrecognized population of venous endothelial cells derived from Pax3-expressing precursors, highlights the utility of GNZ reporter mice and the single cell resolution that is afforded when GNZ is used to detect Cre activity.
METHODS
Vector Construction and Gene Targeting
The GFP-NLS-lacZ targeting construct was constructed by cloning the 4 kb GFP-NLS-lacZ XhoI/HindIII partial digest fragment of pHM840 (Sorg and Stamminger, 1999) into the XhoIHindIII sites of pEGFP-N1 (Clontech). The 4 kb GFP-NLS-lacZ AccI/XhoI cut fragment of this vector was cloned into the corresponding sites of pBigT (Srinivas et al., 2001) to generate pBigT-GNZ. The KpnI site of pRosa26PA was replaced with an FseI site to generate pRosaLink. The PacI/AscI fragment of pBigT-GNZ was cloned into the corresponding sites of pRosaLink to generate the final targeting vector. This vector was linearized with FseI and used for electroporation. R1 ES cells were targeted using standard techniques and four correctly targeted clones were obtained after screening 192 G418-resistant colonies. Two independent clones were injected into C57BL/6J blastocysts and resulting chimeric mice were bred to produce germline offspring that were used for these studies. These mice were maintained on a mixed C57BL/6J;129 genetic background. Genomic Southern blot hybridization was performed on DNA from ES cells digested with EcoRV. The 5′ probe used detects an 11.5 kb wild type band and a 4.1 kb targeted band. Genotyping of GNZ mice was performed using standard PCR techniques with an annealing temperature of 60°C and the following primers GNZ-1F, 5′-TCCAGTTCAACATCAGCCGCTACA-3′; GNZ-1R 5′-ATCTCCGAGGCGGATACAAGCAAT-3′. The resulting PCR product was 764 bp. The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all animal protocols.
Confocal Microscopy
Confocal images were acquired with the Leica Microsystems DM600 confocal microscope using 5×, 0.15 NA dry and 10×, 0.4 NA dry objectives. Z spacing between slices was 1.3 µm.
X-Gal Staining
Whole mount embryos and paraffin embedded sections were stained for β-galactosidase activity using previously described methods (Brown et al., 2001).
Immunohistochemistry
All sections were deparaffinized and pretreated using heat antigen retrieval with Bull’s Eye Decloaker (BioCare Medical). Endogenous peroxidase was then blocked with 3% hydrogen peroxide in PBS for 10 min. Sections were then washed with 0.1% PBST and blocked with 10% goat serum, 0.5% PBST for 30–60 min at 25°C. For GFP immunostaining, the sections were incubated with rabbit polyclonal anti-GFP (1:250; Invitrogen) in 0.1% PBST overnight at 4°C. After washing with 0.1% PBST, sections were incubated with biotinylated goat anti-rabbit IgG (1:200; Vector Laboratories) for 1 h at 25°C. After washing with 0.1% PBST, sections were then incubated with Vectastain ABC (Vector Laboratories) for 45 min. After washing with 0.1% PBST, sections were incubated with Biotinyl Tyramide Amplification Reagent (TSA Biotin System, PerkinElmer) at 25°C for 4 min. After washing with 0.1% PBST, sections were incubated with Steptavidin-Fluorescein (1:500; PerkinElmer) in 0.1% PBST for 1 h at 25°C. Slides were then washed with 0.1% PBST and mounted with Vectashield mounting medium (Vector Laboratories). For HRP sections, a rabbit polyconal anti-β-galactosidase (1:1000; Cortex Biochem) was used and following the Vectastain ABC incubation, sections were washed with 0.1% PBST followed by 0.1 M Tris (pH 7.5) and 0.3 M NaCl. Peroxidase activity was then detected with DAB (Sigma). For vWF staining, sections were incubated with rabbit anti-von Willebrand Factor (1:200; Sigma) in 0.1% PBST overnight at 4°C. After washing with 0.1% PBST, sections were incubated with Alexa Fluor 568 goat anti-rabbit IgG (1:250; Invitrogen) for 1 h at 25°C. Slides were then washed with 0.1% PBST and mounted with Vectashield mounting medium (Vector Laboratories). Protocols are available at http://www.uphs.upenn.edu/mcrc.
ACKNOWLEDGMENTS
The authors thank Andrea Stout for confocal imaging expertise, Thomas Stamminger and Etienne Weiss for the pHM840 vector.
Contract grant sponsor: NHLBI, Contract grant number: K08-HL086633, Contract grant sponsor: Pediatric Scientist Development Program, Contract grant number: NICHD K12-HD00850, Contract grant sponsor: NHLBI, Contract grant number: 1P01HL075215, Contract grant sponsor: NHLBI, Contract grant number: RO1-HL62974
LITERATURE CITED
- Brown CB, Feiner L, Lu MM, Li J, Ma X, Webber AL, Jia L, Raper JA, Epstein JA. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch D, Soriano P, McMahon A, Sucov H. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Cheng G, Litchenberg WH, Cole GJ, Mikawa T, Thompson RP, Gourdie RG. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126:5041–5049. doi: 10.1242/dev.126.22.5041. [DOI] [PubMed] [Google Scholar]
- Delot EC, Shneyder N, Zhang H, Bachiller D. Abnormal venous and arterial patterning in Chordin mutants. Dev Dyn. 2007;236:2586–2593. doi: 10.1002/dvdy.21287. [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Esner M, Meilhac SM, Relaix F, Nicolas JF, Cossu G, Buckingham ME. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Komatsu Y, Takeda H, Nomura-Kitabayashi A, Yamauchi Y, Abe K, Yamamura K, Mishina Y. Optimized β-galactosidase staining method for simultaneous detection of endogenous gene expression in early mouse embryos. Genesis. 2006;44:57–65. doi: 10.1002/gene.20186. [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling H. Faithful activation of an extra-bright red fluorescent protein in “knock-in”. Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin S. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin S. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci USA. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–837. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe C. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Poelmann RE, Jongbloed MR, Molin DG, Fekkes ML, Wang Z, Fishman GI, Doetschman T, Azhar M, Gittenberger-de Groot AC. The neural crest is contiguous with the cardiac conduction system in the mouse embryo: A role in induction? Anat Embryol (Berl) 2004;208:389–393. doi: 10.1007/s00429-004-0401-6. [DOI] [PubMed] [Google Scholar]
- Sorg G, Stamminger T. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to beta-galactosidase. Biotechniques. 1999;26:858–862. doi: 10.2144/99265bm12. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin C, William C, Tanabe Y, Jessell T, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]