SUMMARY
Ubiquitin-derived peptides are bactericidal in vitro and contribute to the mycobactericidal activity of the lysosome. To further define interactions of ubiquitin-derived peptides with mycobacteria, we screened for mutants with increased resistance to the bactericidal activity of the synthetic ubiquitin-derived peptide Ub2. The four Ub2-resistant M. smegmatis mutants were also resistant to the bactericidal action of other antimicrobial peptides and macrophages. Two mutants were in the mspA gene encoding the main M. smegmatis porin. Using a translocation-deficient MspA point mutant, we showed that susceptibility of M. smegmatis to Ub2 was independent of MspA channel activity. Instead, the M. smegmatis Ub2-resistant mutants shared a common phenotype of decreased cell wall permeability compared to wild type bacteria. Expression of mspA rendered M. tuberculosis CDC1551 more susceptible to both ubiquitin-derived peptides in vitro and to lysosomal killing in macrophages. Finally, biochemical assays designed to assess membrane integrity indicated that Ub2 treatment impairs membrane function of M. smegmatis and M. tuberculosis cells. The M. smegmatis Ub2-resistant mutants were more resistant than wild type M. smegmatis to this damage. We conclude that Ub2 targets mycobacterial membranes and that reduced membrane permeability provides mycobacteria intrinsic resistance against antimicrobial compounds including bactericidal ubiquitin-derived peptides.
Keywords: Mycobacterium tuberculosis, Mycobacterium smegmatis, antimicrobial peptides, ubiquitin, MspA, porin
INTRODUCTION
Tuberculosis is a global health concern due to high rates of disease among HIV patients and a worldwide increase in multi-drug resistant strains. The World Health Organization estimates that Mycobacterium tuberculosis infects one third of the world population and 8 million new cases of tuberculosis are reported annually (WHO, 2007). M. tuberculosis is an obligate pathogen and has no known natural reservoir outside of humans. The ability of these bacteria to survive within the host macrophage is key to its pathogenesis. It does so by arresting phagosome maturation and preventing fusion of the bacteria-containing vacuole with the lysosome. M. tuberculosis resides in a vacuole that resembles an early endosome with a pH of 6.4 and retains markers such as the Rab5 GTPase (Clemens and Horwitz, 1996; Clemens et al., 2000; Sturgill-Koszycki et al., 1994; Via et al., 1997).
The ability of M. tuberculosis to arrest phagosome maturation in resting macrophages is a complex process that depends upon both cell wall lipids and protein effectors to promote bacterial survival (Rohde et al., 2007). While the pathogen is well-adapted to modulate phagosome trafficking in resting host macrophages, activation of macrophages shifts the balance towards mycobacterial clearance (Schaible et al., 1998; Via et al., 1998). Oxidative killing of mycobacteria occurs in activated macrophages, and studies in mice indicate that the reactive oxygen and nitrogen intermediates (ROI and RNI) play a role in control of tuberculosis infection: M. tuberculosis infection of phagosome oxidase (phox)-deficient mice indicates a role for ROI in the lung, and NOS2-/- mice are quickly killed upon infection with M. tuberculosis (Adams et al., 1997; Chan et al., 1992; MacMicking et al., 1997). Activated macrophages also promote bacterial killing by delivering the mycobacteria-containing vacuole to the lysosome.
Studies by Deretic and colleagues showed that IFN-γ activation induces autophagy, a process that promotes delivery of mycobacteria to the lysosome via autophagosomes (Gutierrez et al., 2004). The role of autophagy in the innate and adaptive immune response to infection by intracellular pathogens is increasingly understood (Schmid and Munz, 2007). The induction of autophagy in M. bovis BCG- and M. tuberculosis-infected macrophages by serum-starvation, rapamycin-treatment, or IFN-γ activation leads to killing of intracellular bacteria as a result of trafficking bacteria to the lysosome (Alonso et al., 2007; Gutierrez et al., 2004).
We recently showed that ubiquitin-derived peptides contribute to the mycobactericidal activity of the lysosome (Alonso et al., 2007). Solubilized lysosomes from murine bone-marrow macrophages were bactericidal against both M. tuberculosis and M. smegmatis. Upon fractionation by high performance liquid chromatography (HPLC), the bactericidal activity was restricted to a single fraction that was identified as ubiquitin by mass spectroscopy. Ubiquitin digested by cathepsin proteases or a twelve-amino acid synthetic ubiquitin-derived peptide Ub2 (STLHLVLRLRGG) were bactericidal against mycobacteria in vitro. Ubiquitin was localized to the lysosome by immuno-electron microscopy. The induction of autophagy not only enhanced the association of ubiquitin with the lysosome but also increased bactericidal capacity of the lysosomal extract against mycobacteria. We propose that ubiquitinated proteins are delivered to the lysosomal compartment in a process that is enhanced by autophagy. In the lysosome, cathepsin proteinases release ubiquitin-derived peptides that have antimicrobial activity. The action of these peptides is bactericidal in vitro and likely contributes to mycobacterial clearance in the macrophage. In support of this model, immuno-electron microscopy confirmed that ubiquitin co-localizes with M. tuberculosis in autophagic macrophages (Alonso et al., 2007).
To further define the interactions of ubiquitin-derived peptides with mycobacteria, we screened a transposon mutant library for mycobacterial mutants that exhibited increased resistance to the bactericidal activity of the synthetic ubiquitin-derived peptide Ub2. The Ub2-resistant mutants were more resistant to macrophage killing, consistent with ubiquitin-derived peptides contributing to lysosomal killing. A unifying feature of the Ub2-resistant mutants is that they exhibited reduced outer membrane permeability relative to wild type bacteria.
RESULTS
Isolation of M. smegmatis mutants resistant to killing by the ubiquitin-derived peptide Ub2
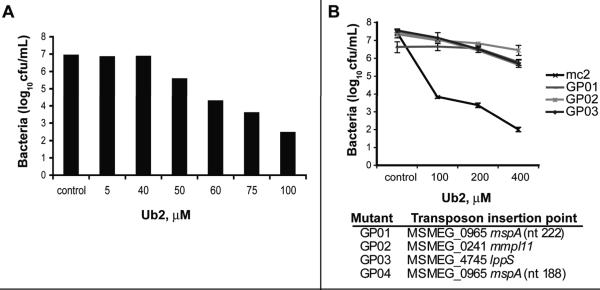
We hypothesized that isolation of mutants with altered resistance to the bactericidal ubiquitin-derived peptide Ub2 would provide insight into its mechanism of action. Ub2 is bactericidal against M. smegmatis at concentrations higher than 50 μM (Fig. 1A). To identify mutants exhibiting higher resistance to the ubiquitin-derived peptide Ub2, pools representing a transposon library of 7680 M. smegmatis mc2 155 mutants were sub-cultured in 7H9 media containing 200 μM Ub2. After 18 h incubation, the surviving bacteria were recovered by plating to 7H11 agar plates. Putative Ub2-resistant colonies were picked and individually grown in 7H9 broth. Southern analysis of forty confirmed hyper-resistant mutants indicated they represented multiple siblings of four independent mutants (data not shown). The susceptibility to Ub2 of representative clones and wild type M. smegmatis mc2 155 was determined in a liquid-culture bactericidal assay (Fig. 1B). The survival of the four mutants was three to four logs greater than wild type upon treatment with Ub2 peptide concentrations greater than 100 μM.
Fig. 1. Ub2 treatment of M. smegmatis hyper-resistant mutants.
A. Killing of M. smegmatis with the Ub2 peptide. M. smegmatis mc2155 was subcultured to 5×105 cfu/mL in 7H9 media containing the indicated concentration of Ub2 peptide. A representative experiment is shown. B. Mutant and wild type bacteria were sub-cultured to 5×105 cfu/mL in 7H9 media or 7H9 containing the indicated concentration of Ub2 and incubated overnight. Bacterial viability was determined by plating serial dilutions. The average +/- standard deviation of three independent experiments is shown. The identities of the transposon mutants GP01-GP04 are shown below the graph.
The transposon insertion junctions were sequenced and two of the four mutants (GP01 and GP04) had independent insertions into the mspA gene (MSMEG_0965) that encodes the M. smegmatis porin MspA (Fig. 1B). Of the two mspA mutants, transposon mutant GP01 was chosen for further characterization. GP02 and GP03 contained transposon insertions in the mmpL11 and the lppS genes of M. smegmatis, respectively.
Presence of the MspA porin corresponds with susceptibility to the Ub2 peptide and lysosomal extracts
MspA is the main porin in the M. smegmatis outer membrane for the uptake of small hydrophilic nutrients (Stahl et al., 2001). MspA mutants are more resistant to several antibiotics than wild-type M. smegmatis indicating that these drugs exploit the MspA porin to enter the cell. Indeed, uptake of some β-lactams, fluoroquinolones and chloramphenicol by M. smegmatis was slower in porin mutants (Danilchanka et al., 2008b; Stephan et al., 2004).
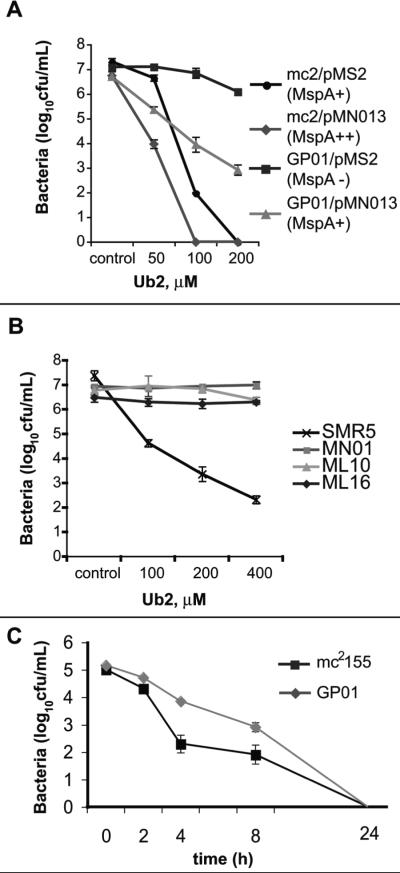
To determine if Ub2 susceptibility correlated with the relative number of MspA porins present in the membrane, we generated a panel of strains that expressed varying levels of MspA. Wild type M. smegmatis mc2155 and the mspA transposon mutant GP01 were transformed with the vector control pMS2 or the mspA expression construct pMN013. In pMN013 mspA expression is controlled by the imyc promoter. In previous studies this plasmid complemented the porin deficiency of the M. smegmatis mspA deletion mutant MN01 (Mailaender et al., 2004). A bactericidal assay was performed to determine the relative susceptibility to the Ub2 peptide. There was a correlation between the levels of MspA expressed and the bactericidal effects of Ub2 (Fig. 2A). M. smegmatis mc2155/pMN013, which overexpresses mspA, was the most susceptible to treatment with 50 μM Ub2, exhibiting a three-log reduction in the number of viable bacteria compared to the untreated control. The mspA mutant transformed with the vector control (GP01/pMS2) was the most resistant, showing little difference in viable bacteria upon treatment with Ub2 compared to the control. GP01 was restored to the Ub2-susceptible phenotype when mspA was expressed from pMN013.
Fig. 2. Bacterial susceptibility to Ub2 and solubilized lysosomes (SF) correlates with the relative levels of MspA.
A. Bactericidal activity of Ub2 was determined against wild type M. smegmatis (mc2155) and the mspA mutant GP01 carrying either the vector control (pMS2) or mspA expression vector (pMN013). Bacteria were incubated overnight in 7H9 media with either a buffer control or the indicated concentration of Ub2. B. Bactericidal activity of Ub2 was determined against wild type M. smegmatis (SMR5) and isogenic porin mutants MN01 (ΔmspA), ML10 (ΔmspA, ΔmspC) and ML16 (ΔmspA, ΔmspC, ΔmspD). Bacteria were incubated overnight in 7H9 media with either a buffer control or the indicated concentration of Ub2. C. Bacterial susceptibility to solubilized lysosomes (SF) was determined against wild type M. smegmatis mc2 155 (black line) and the mspA mutant GP01 (grey line). Bacteria were incubated in the presence of 50 μg/mL SF and bacterial viability determined at the indicated time points.
While MspA is the major porin of M. smegmatis, the genome encodes three additional porins MspB, MspC, and MspD that are highly similar to MspA (Stahl et al., 2001). The role of these porins in ubiquitin-peptide susceptibility was investigated using a panel of previously characterized isogenic porin deletion strains: MN01 (ΔmspA), ML10 (ΔmspA, ΔmspC) and ML16 (ΔmspA, ΔmspC, ΔmspD). There was no significant increase in resistance of the multiple porin mutants beyond that of the mspA mutant MN01 at the concentrations tested (Fig. 2B). Therefore, loss of MspA confers acquisition of Ub2 resistance, and this phenotype is not enhanced upon loss of additional porins. It is unlikely that the remaining porin MspB plays a role in Ub2 resistance. Previous work showed that mspB is not expressed by M. smegmatis grown in standard 7H9 medium (Stephan et al., 2005). Upon deletion of mspA, the authors showed that mspB was expressed and concluded that MspB may compensate for the absence of MspA. Therefore, the absence of mspB expression in wild type M. smegmatis does not confer susceptibility to Ub2. Stephan et al further showed that the expression levels of mspB in the porin mutants ML10 and ML16 were so low that they did not complement their permeability defects.
Solubilized lysosomes (SF) is a complex mixture containing lysosomal enzymes that likely act synergistically. Since porin mutants were resistant to the bactericidal action of Ub2, we examined whether these mutants were also resistant to killing by SF. The mspA mutant GP01 and wild type M. smegmatis mc2 155 were treated with 50 μg/mL SF and survival measured over time. The mspA mutant GP01 was killed at a slower rate than wild type M. smegmatis mc2 155, showing up to 100-fold greater survival several hours post-treatment (Fig. 2C). After 24-hour treatment, no bacteria from either strain were recovered on agar plates. We conclude that the Ub2-resistance phenotype of the mspA mutant GP01 confers a modest increase in resistance to SF.
Ubiquitin contributes to the bactericidal capacity of solubilized lysosomes
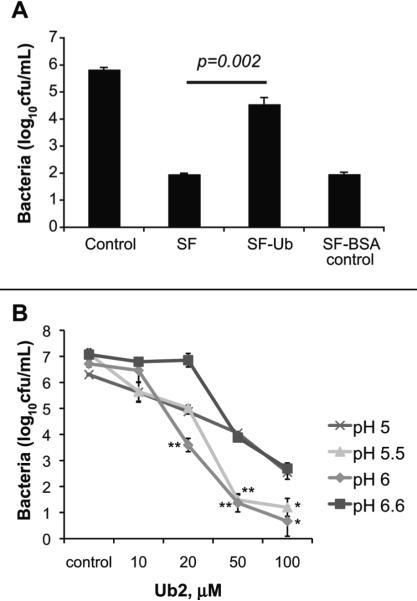
Previously we showed that the bactericidal capacity of the solubilized lysosomes correlated with the relative levels of ubiquitin (Alonso et al., 2007). An additional way to illustrate that ubiquitin was a source of bactericidal activity in the SF is to immuno-deplete ubiquitin and ubiquitin-derived peptides from the SF using polyclonal antibody against ubiquitin. Quantification by immunoblot indicated there was a reduction in the detectable amount of ubiquitin by immunoblot (Fig. S1). A standard bactericidal assay on wild type M. smegmatis using the complete extract (SF) and SF depleted of ubiquitin (SF-Ub) was performed. As a control, SF was incubated with rabbit polyclonal antibody against an unrelated protein, bovine serum albumin, (SF-BSA) (Fig. 3A). Treatment of wild type M. smegmatis mc2 155 with SF or SF-BSA resulted in greater than a three-log reduction in viable bacteria compared to the control-treated bacteria. Treatment with ubiquitin-depleted SF resulted in only a one-log reduction in viable bacteria. These results demonstrate that ubiquitin is a major SF component with bactericidal activity for M. smegmatis.
Fig. 3. Ubiquitin contributes to the bactericidal capacity of lysosomal extracts.
A. Bactericidal activity of SF upon immunodepletion of ubiquitin. M. smegmatis mc2 155 was incubated overnight with either buffer control, 50 μg/mL SF, 50 μg/mL SF pre-depleted of ubiquitin, or 50 μg/mL SF pre-depleted of BSA (negative control). Statistical significance between SF and SF-Ub was determined by Student's t test: p= 0.002. B. The bactericidal activity of Ub2 was determined in 7H9 media at pH 6.6 (standard), pH 6, pH5.5, and pH5. Statistical significance between pH 6.6 and pH 5.5 or pH 6 was determined by Student's t test: *, p<0.05; **, p<0.005. In A and B, the number of viable bacteria was determined by plating serial dilutions, and the average of three independent experiments +/- standard deviation is shown.
Our in vitro experiments with the synthetic ubiquitin peptide Ub2 are performed in standard 7H9 media at pH 6.6. Immature and mature mycobacteria-containing phagosomes are at pH 6.2 and pH 5-5.5, respectively (de Chastellier and Thilo, 2006; Schaible et al., 1998; Sturgill-Koszycki et al., 1994; Via et al., 1998). To determine whether there was an effect of pH on the bactericidal capacity of Ub2, we treated bacteria in 7H9 media at pH 6.6, pH 6, pH 5.5 and pH 5. Ub2 exhibited the greatest bactericidal activity at pH 6.0 and pH 5.5 (Fig. 3B). There was no statistical significance between the bactericidal activity of Ub2 at pH6.0 and pH 5.5. This indicates that ubiquitin-derived peptides are functional at the physiological relevant pH of the phagolysosomal pathway.
Ub2 impairs M. smegmatis membrane integrity
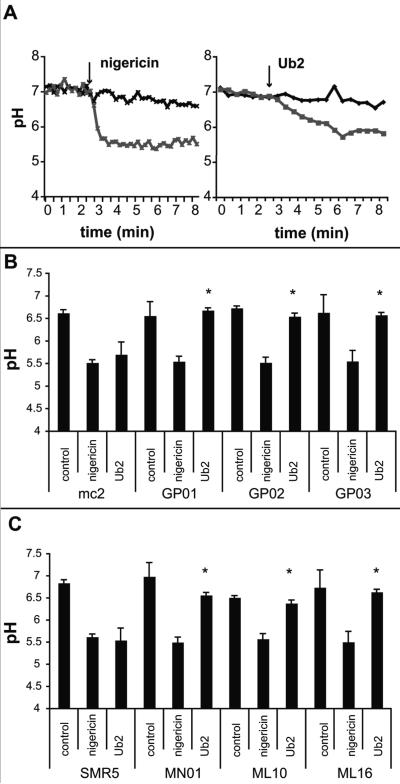
Ub2 is a 12 amino acid positively charged peptide. We hypothesized that Ub2 may behave like a cationic antimicrobial peptide to disrupt the mycobacterial inner membrane. To directly measure membrane integrity, an assay based upon a pH-sensitive fluorophore was performed on intact bacteria. The cytoplasm of M. smegmatis was labeled with the membrane-permeable probe 5-chloromethylfluorescein diacetate (CMFDA), which is processed into a membrane-impermeable form following uptake. The fluorescence emission of fluorescein at 520 nm is pH-sensitive upon excitation at 490 nm, but is pH insensitive when excited at 450 nm. The intracellular pH of the labeled bacteria was neutral, and the bacteria were placed in suspension buffered at pH 5.5. The loss of membrane integrity was therefore measured as a shift in fluorescence upon exposure of the intracellular fluorophore to the extracellular pH. As a positive control, bacteria were treated for five minutes with the ionophore nigericin. This resulted in a fluorescence emission shift indicating that the intracellular pH had dropped from pH 7 to pH 5.5 (Fig. 4A). A five-minute treatment with Ub2 resulted in a drop of intracellular pH to pH 5.8. We noted the kinetics of pH shift upon treatment with Ub2 were slower than those upon treatment with nigericin, which occurred within one minute.
Fig. 4. Action of Ub2 on intact M. smegmatis bacteria.
A. CMFDA-labeled M. smegmatis mc2155 was resuspended in buffer (pH 5.5) and treated with either 10 mM nigericin (left) or 100 μM Ub2 (right) and fluorescence emission at 520 nm measured at excitation wavelengths of 450 nm and 490 nm. The excitation ratio was converted to pH by regression. A representative of five independent experiments is shown. B-C. CMFDA-labeled M. smegmatis strains were resuspended in buffer (pH 5.5) and treated with either nigericin or 100 μM Ub2. Fluorescence emission at 520 nm was measured at excitation wavelengths of 450 nm and 490 nm, and the excitation ratio converted to pH by regression. The average +/- standard deviations from three assays are shown. In panel B, the difference in pH observed between mc2155 and GP01, GP02, and GP03 upon treatment with Ub2 was determined to be statistically significant by a Student's t test, (*, p<0.01). Likewise, in panel C the difference in pH observed between SMR5, MN01, ML10, and ML16 was determined to be statistically significant (*, p<0.01).
These experiments show that treatment with Ub2 results in exposure to external pH. To determine whether the Ub2-resistant mutants had altered susceptibility to Ub2 membrane damage, CMFDA-labeled wild type and mutant bacteria were incubated with Ub2 and fluorescence emission at 520 nm monitored at the excitation wavelengths of 450 nm and 490 nm. After a five-minute Ub2 treatment of mc2155 with 100 μM Ub2, a shift to pH 5.6 occurred (Fig. 4B). Identical treatment of the Ub2-resistant mutants resulted in the cytoplasmic pH shifting to pH 6.5-6.7. The panel of porin mutants was tested and these mutants were also more resistant than wild type to the Ub2-mediated membrane damage (Fig. 4C). These biochemical studies reinforce the observed phenotype of the Ub2-resistant mutants in bactericidal assays.
The susceptibility of M. smegmatis to Ub2 is independent of MspA channel activity
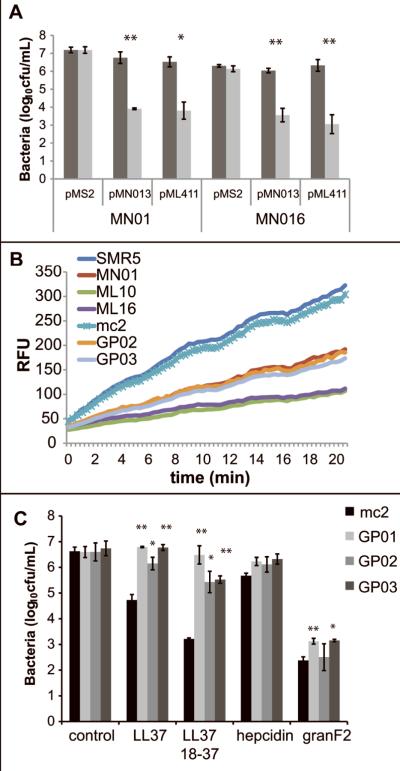
Our observation that M. smegmatis porin mutants are Ub2-resistant raises the interesting possibility that Ub2 may translocate across the outer membrane through the MspA porin. To determine if Ub2 uses the MspA porin in this manner, we assessed the ability of a translocation-deficient porin to complement the Ub2-resistance phenotype of the porin mutants. To this end, we used an MspA mutant with a D90L point mutation in the porin constriction zone. This mutant did not form stable pores in lipid bilayer experiments and showed almost no permeability for larget solutes such as glucose and pnitrophenylphosphate (Huff, Hoffmann and Niederweis, submitted). The M. smegmatis ΔmspA mutant MN10 and the triple porin mutant ML16 were transformed with plasmid pMN013 or pML411 for expression of wild type MspA or MspA D90L porins, respectively. When a bactericidal assay was performed to determine the relative susceptibility to the Ub2 peptide, expression of wild type mspA and the mspA point mutant restored the wild type Ub2-susceptible phenotype (Fig. 5A). There was no significant difference observed in the Ub2-susceptibility of MN10/pMN013 and MN10/pML411 or MN016/pMN013 and MN016/pML411. This data indicates that while loss of the MspA porin reduces Ub2 susceptibility, Ub2 does not utilize the MspA channel directly to reach the inner membrane.
Fig. 5. Reduced mycobacterial cell wall permeability contributes to antimicrobial peptide resistance.
A. Porin mutants MN01 (ΔmspA) and ML16 (ΔmspA, ΔmspC, ΔmspD) carrying pMS2, pMN013 (MspA), or pML411 (MspAD90L) were incubated overnight in buffer control (dark grey) or in 100 μM Ub2 (light grey). Statistical significance between bacterial viability in control and Ub2-treated samples was determined by Student's t test: *, p<0.05; **, p<0.005. B. M. smegmatis strains were incubated in the presence of 20 μM ethidium bromide. Accumulation of ethidium bromide was followed over time by monitoring emission at 590 nm upon excitation at 530 nm and expressed as Relative Fluorescence Units (RFU). C. Mutant and wild type bacteria were sub-cultured to 5×105cfu/mL in 7H9 media or 7H9 containing the indicated antimicrobial peptide and incubated overnight. Statistical significance between wild type and mutant viability in each condition was determined by Student's t test: *, p<0.02; **, p<0.005. In A and C, the number of viable bacteria was determined by plating serial dilutions, and the average of three independent experiments +/- standard deviation is shown.
Ub2-resistant mutants have reduced membrane permeability
Decreased membrane permeability to hydrophobic compounds in porin mutants has been observed previously (Stephan et al., 2004). We speculated that this phenotype might be associated with the Ub2-resistance observed in the M. smegmatis mspA mutant GP01 and by extension the Ub2-resistant mutants GP02 and GP03. To determine whether Ub2-resistant mutants also possessed less permeable membranes, an ethidium bromide uptake assay on intact mycobacteria was performed. This assay relies on the increase in fluorescence of ethidium bromide upon uptake and binding to bacterial nucleic acids (Danilchanka et al., 2008a). The Ub2-resistant mutants had reduced ethidium bromide uptake compared to wild type M. smegmatis (Fig. 5B). When the porin mutants were analyzed, MN01 (ΔmspA) had similar ethidium bromide uptake kinetics as GP01, GP02, and GP03. The porin mutants ML10 (ΔmspA, ΔmspC) and ML16 (ΔmspA, ΔmspC, ΔmspD) had the lowest uptake of ethidium bromide (Fig. 5B).
The affect of membrane permeability on susceptibility to other antimicrobial peptides
Cationic antimicrobial peptides are important components of innate immunity and contribute to human resistance to bacterial infection and colonization (Lehrer and Ganz, 2002; Nizet et al., 2001; Zasloff, 2002). Cationic antimicrobial peptides include cathelicidins, hepcidin and granulysin. To determine whether the reduction in membrane permeability in Ub2-resistant mutants rendered these bacteria more resistant to other antimicrobial peptides, the M. smegmatis Ub2-resistant mutants were exposed to the cathelicidin LL-37, LL-37 (18-37), hepcidin, and granulysin-derived peptide granF2. The cathelicidin fragment LL-37 (18-37) was reported to exhibit enhanced antimicrobial activity relative to full-length LL-37 (Murakami et al., 2004). GranF2 is a synthetic peptide fragment that encompasses a helix-loop-helix region in the central region of the full-length granulysin and is lytic against B. megaterium, E. coli and M. tuberculosis (Andreu et al., 1999). GP01, GP02, and GP03 were over 1-log more resistant than wild type M. smegmatis upon overnight treatment with LL37 and LL37 18-37. GP01 and GP03 were slightly more resistant than wild type to killing by granF2. Consistent with previous work with M. tuberculosis we observed a 15% reduction in cfu/mL following 24h treatment of wild type M. smegmatis with 100 μg/mL hepcidin. The mutants GP01, GP02, and GP03 tended to be more resistant than wild type. These data indicate that reduced membrane permeability confers resistance to some host antimicrobial peptides, but suggests that bactericidal peptides such as granulysin access the membrane in a permeability-independent manner (Fig. 5C).
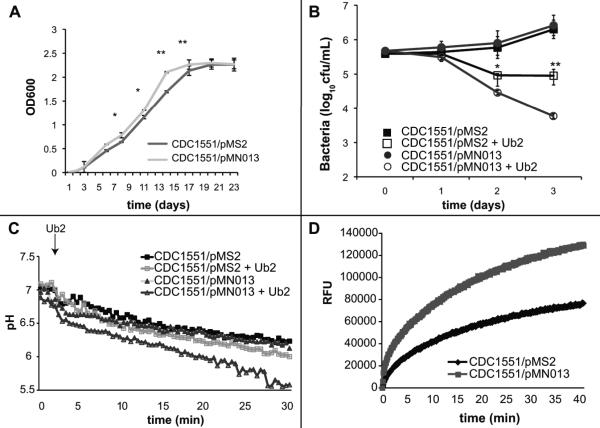
M. tuberculosis expressing mspA grows at a faster rate, but is more susceptible to ubiquitin-derived peptides
There is no readily identifiable homologue of MspA in the slow-growing organisms M. tuberculosis or Mycobacterium bovis BCG. Heterologous expression of mspA increased the growth rate of M. bovis BCG, and expression of mspA in M. tuberculosis results in increased antibiotic susceptibility of the pathogen (Mailaender et al., 2004). To determine the effect of heterologous expression of mspA on growth and susceptibility to Ub2, the M. tuberculosis clinical isolate CDC1551 was transformed with pMS2 and pMN013. Expression of mspA in CDC1551/pMN013 was confirmed by Western analysis (Fig. S2). We observed that M. tuberculosis CDC1551/pMN013 grew faster than CDC1551/pMS2 suggesting that the porin is beneficial to growth in vitro (Fig. 6A). During logarithmic phase, bacteria had a generation time of 1.3 days for wild type CDC1551/pMS2 and every 0.95 days for mspA expressing strain CDC1551/pMN013. This result is consistent with the previous observation that expression of mspA increased the growth rate of M. bovis BCG (Mailaender et al., 2004). When a bactericidal assay was performed, CDC1551/pMN013 was more susceptible to the Ub2 peptide than CDC1551/pMS2 (Fig. 6B). Using the fluorescence-based Ub2 membrane integrity assay described above, we determined that M. tuberculosis CDC1551/pMN013 was more susceptible to the membrane disrupting action of Ub2 than CDC1551/pMS2 (Fig. 6C). There was no significant difference in detergent susceptibility between the two strains to suggest inherent weakness in the CDC1551/pMN013 membrane relative to CDC1551/pMS2 (Fig. S4). We did find increased membrane permeability of CDC1551/pMN013 compared to CDC1551/pMS2 as measured by ethidium bromide accumulation (Fig. 6D). Thus, M. tuberculosis showed the same MspA-dependent in vitro phenotypes of Ub2-resistance and permeability as for M. smegmatis.
Fig. 6. Expression of mspA in M. tuberculosis enhances growth rate, but renders bacteria hyper-susceptible to ubiquitin-derived peptides.
A. Expression of mspA by CDC1551 increases growth rate in broth culture. Growth of M. tuberculosis CDC1551 carrying either pMS2 (black) or pMN013 (grey) was followed over the course of 21 days. Three independent experiments were performed. Statistical significance between CDC1551/pMS2 and CDC1551/pMN13 was determined by Student's t test: *, p<0.05; **, p<0.005. B. Bactericidal susceptibility to Ub2 killing. M. tuberculosis CDC1551 carrying either pMS2 (squares) or pMN013 (circles) was incubated in 7H9 media containing either buffer control (filled) or 100 μM Ub2 (empty). Bacterial viability was determined by plating serial dilutions at the indicated time points. The average +/- standard deviation of three replicates from a representative experiment is shown. Statistical significance between CDC1551/pMS2 and CDC1551/pMN013 upon treatment with Ub2 was determined by Student's t test: *, p<0.05; **, p<0.005. C. Bacterial susceptibility to Ub2 membrane damage. CMFDA-labeled M. tuberculosis CDC1551 carrying either pMS2 (squares) or pMN013 (triangles) was resuspended in buffer (pH 5.5) and treated with either buffer control (filled) or 100 μM Ub2 (empty) and fluorescence emission at 520 nm measured at excitation wavelengths of 450 nm and 490 nm. The excitation ratio was converted to pH by regression. A representative of three independent experiments is shown. C. Ethidium bromide accumulation of M. tuberculosis CDC1551 carrying either pMS2 (black) or pMN013 (grey). Strains were incubated in the presence of 20 μM ethidium bromide and emission at 590 nm upon excitation at 530 nm monitored over time and expressed as Relative Fluorescence Units (RFU). A representative of three independent experiments is shown.
Ub2-resistant mutants are more resistant to killing by macrophages
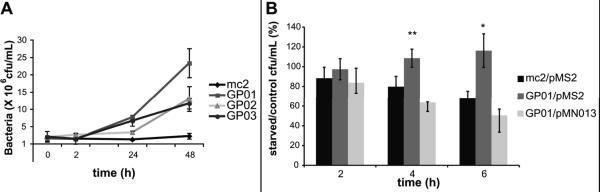
Unlike the pathogen M. tuberculosis, M. smegmatis does not arrest phagosome maturation and the bacteria are ultimately delivered to the bactericidal lysosomal compartment. To determine whether Ub2-resistant mutants were also more resistant to the bactericidal action of macrophages, an infection of RAW 264.7 macrophages was performed. GP01, GP02, and GP03 survived better than wild type M. smegmatis mc2155 in macrophages. The bacterial counts at 24h and 48h post infection indicated that the mutants were multiplying (Fig. 7A). This phenotype for GP01 is consistent with previous data from Niederweis and colleagues that showed absence of mspA enhanced M. smegmatis survival within macrophages, suggesting that macrophages were less able to eliminate porin mutants compared to wild type M. smegmatis (Sharbati-Tehrani et al., 2005).
Fig. 7. Survival in macrophages is enhanced in the M. smegmatis mspA mutant.
A. Bacterial survival of wild type in resting RAW 264.7 macrophages. Macrophages were infected with M. smegmatis mc2 155, the mspA mutant GP01, the mmpL11 mutant GP02, and the lppS mutant GP03. Bacterial survival over a 48 h infection was determined at the indicated time points, by harvesting the infected macrophages and the number of viable bacteria determined by plating serial dilutions. The average and standard deviations of three independent infections are shown. Statistical significance between wild type and mutants at 48 h was determined by students t test: GP01, p=0.02; GP02, p=0.01; GP03, p=0.037. B. Bacterial survival of wild type M. smegmatis mc2155/pMS2, the mspA mutant GP01/pMS2, and the complemented mspA mutant GP01/pMN013 was determined in resting and autophagic RAW264.7 macrophage-like cells. At the indicated time points, the infected macrophages were harvested and the number of viable bacteria determined by plating serial dilutions. The average and standard deviations of three independent experiments are shown. Statistical significance between survival of mc2155/pMS2 and GP01/pMS2 was determined by Students t test: *, p<0.05; **, p<0.01.
Induction of autophagy not only promotes fusion of the bacteria-containing vacuole with lysosomes but also increases the concentration of ubiquitin in the lysosome (Alonso et al., 2007). To determine whether mutant survival was enhanced in ubiquitin peptide-enriched lysosomes, RAW264.7 macrophages were infected and autophagy induced by starvation. Bacterial survival of wild type M. smegmatis, GP01, GP02, and GP03 after 2h was determined in resting and autophagic macrophages. The number of viable bacteria was determined, and relative survival in autophagic macrophages compared to control macrophages was calculated. Autophagy enhanced bacterial killing in macrophages infected with wild type M. smegmatis mc2155. All three mutants had increased survival in autophagic macrophages at this time point relative to wild type (Fig. S3). The mspA mutant GP01 had the strongest phenotype after 2 h in autophagic macrophages. To further assess the ability of GP01 to survive in autophagic macrophages, M. smegmatis mc2155/pMS2, GP01/pMS2 and GP01/pMN013 viability over a six-hour period was observed by harvesting infected resting and autophagic macrophages every two hours. At four and six hours after induction of autophagy, there was a significant increase in viability of the mspA mutant compared to wild type (Fig. 7B). The enhanced survival was abolished upon complementation of GP01 with the MN013 plasmid.
To confirm that increased survival relative to wild type did not result from the ability of the M. smegmatis mspA mutant to prevent phagosome-lysosome fusion, immunolofluorescence was performed on infected resting and autophagic RAW 264.7 macrophages. Macrophages were infected with M. smegmatis mc2155 and GP01 transformed with a GFP expression plasmid. We localized both mc2155 and GP01 to LAMP1-positive compartments indicating that both strains are delivered to the lysosome (Fig. S5).
M. tuberculosis expressing mspA is more susceptible to killing by macrophages
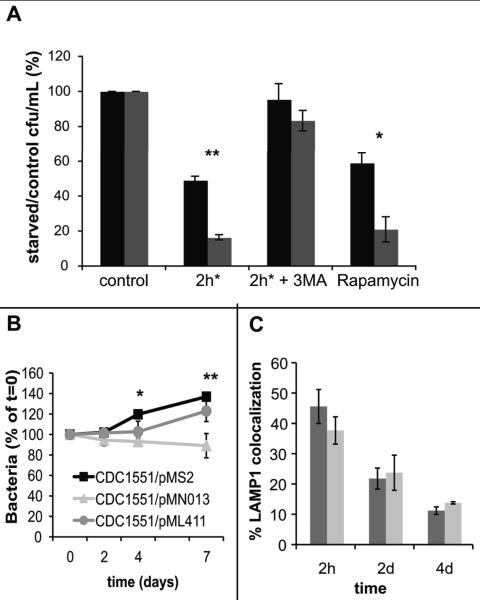
To determine if M. tuberculosis expressing mspA was more susceptible to lysosomal killing in autophagic macrophages, RAW 264.7 macrophages were infected with CDC1551/pMS2 or CDC1551/pMN013. Autophagy was induced by the classical inducer of autophagy, starvation (2h*), or pharmacologically by rapamycin. These treatments were chosen over IFN-γ, a physiologically relevant inducer of autophagy, because starvation and rapamycin promote delivery of the bacterium to the lysosome and are associated with an increase in the concentration of ubiquitin but do not promote the generation of ROI and RNI, which could complicate our interpretation. After two hours, the infected macrophages were harvested and the number of viable bacteria determined in resting (control) and autophagic macrophages. By expressing survival as a ratio of bacteria in starved vs. control, we exclude any potential differences in phagocytosis between experiments. The viability of M. tuberculosis expressing mspA in autophagic macrophages was reduced compared to wild type M. tuberculosis CDC1551/pMS2 (Fig. 8A). The addition of 3MA, an inhibitor of autophagy, reduced the affects of starvation-induced autophagy. From these data, it appears that the presence of MspA in M. tuberculosis reduces the ability of the bacterium to survive within the lysosomal milieu.
Fig. 8. Expression of mspA in M. tuberculosis renders bacteria more susceptible to killing by autophagic macrophages and impairs long-term intracellular survival.
A. RAW 264.7 were infected with CDC1551/pMS2 (black bars) or CDC1551/pMN013 (grey bars). Bacterial survival in resting macrophages and autophagic macrophages was determined and compared to resting control macrophages over a two-hour time course. Statistical significance between CDC1551/pMS2 and CDC1551/pMN13 upon starvation or rapamycin treatment was determined by Student's t test: *, p<0.001; **, p<0.01. B. RAW 264.7 macrophages were infected with CDC1551/pMS2 (black squares), CDC1551/pMN013 (grey triangles) and CDC1551/pML411 (grey circles) and bacterial survival monitored over ten days. The difference between CDC1551/pMS2 and CDC1551/pMN13 was determined to be significant by a Student's t test: *, p=0.012; **, p=0.015. C. RAW 264.7 seeded onto coverslips were infected with CMFDA-stained CDC1551/pMS2 (black bars) or CDC1551/pMN013 (grey bars). At 2 hours, 2 days and 4 days, the coverslips were fixed, pearmeabilized and probed with primary antibody against the lysosomal marker LAMP1 and Texas Red-conjugated secondary antibody. Co-localization of bacteria with LAMP1-positive vacuoles was scored using confocal microscopy. Over 30 bacteria-containing phagosomes were scored for each time point. Each bar represents the average of three experiments, +/- standard deviation.
When M. tuberculosis survival in resting macrophages was followed over a two-week infection, heterologous expression of mspA impaired the ability of CDC1551 to establish and maintain an infection (Fig. 8B). CDC1551/pMS2 survived better than CDC1551/pMN013 bacteria in macrophages. While it is possible that MspA in the membrane may disrupt membrane architecture and render the bacterium less fit, survival of CDC1551/pML411 expressing the MspA porin point mutant was similar to CDC1551/pMS2 in resting macrophages (Fig. 8B). It was possible that the presence of the MspA porin prevented M. tuberculosis from blocking phagosome maturation arrest in resting macrophages. To determine the localization of CDC1551/pMS2 and CDC1551/pMN013 relative to the lysosome, immunofluorescence was performed with antibody against LAMP1. There was no significant difference in LAMP co-localization between CDC1551/pMS2 and CDC/pMN013 at 2 hours, 2 days and 4 days post infection (Fig. 8C, Fig. S6). Therefore, the expression of mspA by M. tuberculosis does not inhibit the establishment of an arrested phagosome. A number of bactericidal compounds are encountered by M. tuberculosis in the macrophage. We infected resting macrophages, but ROI and RNI are generated by activated macrophages and are mycobactericidal. In vitro assays indicated that there was no significant difference between CDC1551/pMS2, CDC1551/pMN013, and CDC1551/pML411 survival upon treatment with 5mM H2O2 (Fig. S7).
Therefore, the presence and porin translocation activity of MspA renders M. tuberculosis unable to establish a successful infection in resting macrophages. Since CDC1551/pMN013 failed to co-localize with LAMP1, we propose that the bacterium expressing mspA arrest phagosome maturation, but may encounter a bacteriostatic or mildly bacteriocidal environment in the arrested phagosome. Wild type M. tuberculosis is more protected due to the intrinsic resistance provided by fewer and/or less efficient, endogenous porins in the outer membrane.
DISCUSSION
We used biochemical and genetic approaches to better understand the interactions of ubiquitin-derived peptides with mycobacteria. The synthetic ubiquitin-derived peptide Ub2 is a 12 amino acid positively charged peptide, and we considered the possibility that it may behave like a cationic antimicrobial peptide. These molecules are thought to multimerize and insert into membranes, although recent data suggests antimicrobial peptides may also directly inhibit intracellular processes such as nucleic acid or protein synthesis (Brogden, 2005). Consistent with the former activity, we showed that Ub2 disrupted the membrane integrity of M. smegmatis and M. tuberculosis cells using fluorimetric assays. While our functional assays indicate that Ub2 impairs membrane function, we can not rule out additional metabolic targets.
Four transposon mutants resistant to the Ub2 peptide were isolated in liquid-culture bactericidal assays. The M. smegmatis Ub2-resistant mutants GP01, GP02 and GP03 were resistant to both the bactericidal and membrane damaging properties of Ub2. Two of the M. smegmatis Ub2-resistant mutants, GP01 and GP04, lacked the major porin MspA. Subsequent experiments indicated that the presence and levels of MspA were associated with susceptibility to the ubiquitin-derived peptide Ub2. From these initial observations, it was tempting to speculate that the Ub2 peptide directly utilized the MspA porin to translocate across the mycobacterial outer membrane. An MspA point mutant that lacks translocation activity restored Ub2 susceptibility as well as wild type MspA, suggesting that the Ub2 peptide does not directly utilize the MspA channel. The absence of MspA is known to decrease permeability of the M. smegmatis outer membrane (Stephan et al., 2004). Thus, the Ub2-resistance phenotype of the mspA mutant likely results from decreased outer membrane permeability.
Biochemical and genetic evidence indicates that M. tuberculosis has porins different from those of M. smegmatis (Niederweis, 2003; Niederweis, 2008). No homologue of MspA exists in M. tuberculosis, and BLAST analysis of available genomes only identifies MspA homologues in fast-growing mycobacterial species such as M. fortuitum, M. peregrinum, and M. abcessus (Niederweis, 2008). The presence of MspA in M. smegmatis and heterologous expression of mspA in M. tuberculosis is associated not only with increased susceptibility to ubiquitin-derived peptides, but also with increased lysosomal killing during infection of autophagic macrophages. When resting macrophages were infected with M. tuberculosis for a standard ten-day survival assay, CDC1551/pMN013 expressing mspA was less able than wild type CDC1551/pMS2 to establish an infection. This observation is in contrast to the results obtained by Sharbati-Tehrani and colleagues that indicated expression of mspA by M. bovis BCG resulted in greater bacterial multiplication in the J774 macrophage cell line and in A549 epithelial cells (Sharbati-Tehrani et al., 2004). While we used an expression plasmid where expression of mspA is driven by an intermediate promoter, Sharbati-Tehrani et al. transformed M. bovis BCG with an integrative plasmid carrying a mspA under the control of its own promoter. They reported that chromosomal expression of mspA was lower than episomal expression of mspA by pMN013 (Sharbati-Tehrani et al., 2004). Therefore it is likely that more MspA porins were expressed in the outer membrane of M. tuberculosis in our experiments.
M. tuberculosis containing an mspA expression vector grew at a faster rate than wild type in broth cultures, but was compromised for survival inside macrophages. CDC1551/pMN013 expressing mspA was not only more susceptible than CDC1551/pMS2 to lysosomal killing in autophagic macrophages, but also exhibited reduced survival in resting macrophages. By immunofluorescence microscopy, we showed that expression of mspA did not prevent M. tuberculosis from arresting phagosome maturation and establishing an infection in resting macrophages. A potential explanation for the survival defect in resting macrophages is that the bacterium is exposed to bacteriostatic and/or bactericidal compounds in the arrested phagosome. The presence of a functional MspA renders M. tuberculosis more susceptible than wild type bacteria to these compounds. Indeed, it has been observed that porins mediate NO diffusion across the M. smegmatis outer membrane during macrophage infections (Fabrino et al., 2009). Macrophage infection is a dynamic process, and mathematical modeling suggests that a small proportion of M. tuberculosis is killed in “early” phagosomes (Jordao et al., 2008). While wild type M. tuberculosis is able to arrest phagosome maturation, it would be an oversimplification to say that all phagosomes arrest at a distinct point in the spectrum of the endosomal pathway. It is likely that some mycobacteria-containing phagosomes mature further along the endosomal continuum, exposing bacteria to an increasingly antimicrobial environment, and this process is reversible (de Chastellier and Thilo, 2006).
An alternative explanation of the attenuation of M. tuberculosis expressing mspA in resting macrophages is that the normal organization of the outer membrane is disrupted, resulting in an organism with compromised fitness. M. tuberculosis expressing mspA grows at a faster rate in vitro, so this growth defect is only apparent in the intracellular environment. In assessing susceptibility to detergent we did not see significant differences from wild type. In fact, M. tuberculosis expressing the mspA D90L porin point mutant survived like wild type in resting macrophages, which suggests that the reduced fitness of the mspA expressing M. tuberculosis strain is due to the channel function. Our findings emphasize that changes in membrane permeability impact the ability of macrophages to kill mycobacteria. We have shown this using both in vitro assays with ubiquitin-derived peptides and by infecting resting and autophagic macrophages. Porins and other membrane proteins that affect membrane permeability are important contributors to the intrinsic resistance of mycobacteria. In addition, it is clear that the relative paucity of porins in M. tuberculosis is an added barrier to small bacteriostatic or bactericidal compounds. The intrinsic resistance provided by an impermeable cell wall contributes to the ability of these pathogenic mycobacteria to establish a niche in resting macrophages. It is tempting to speculate that loss of the MspA porin in pathogenic mycobacteria provides a selective advantage in adverse environments, including the arrested phagosomes of resting macrophages, by reducing membrane permeability. However, this advantage is achieved at the cost of a slower growth rate.
M. smegmatis mutants in mmpL11 and lppS were also isolated in the screen for Ub2-resistant mutants. Like the mspA mutant, these two mutants also had reduced membrane permeability compared to wild type bacteria. M. smegmatis MmpL11 shares 69% protein identity to M. tuberculosis MmpL11. MmpL proteins are members of the RND superfamily of drug exporters. The contribution of individual MmpL proteins to drug efflux was examined by Domenech and colleagues. No significant differences between single mmpL mutants and wild type were found for the antibiotics tested (Domenech et al., 2005). While it is possible that some mycobacterial MmpL proteins are involved in export of bactericidal compounds, others appear to serve a specialized function in transporting fatty acid and lipid constituents. MmpL7 and MmpL8 play a role in phthiocerol dimycocerosate (PDIM) and sulfolipid export, respectively (Converse et al., 2003; Domenech et al., 2004; Jain and Cox, 2005). To date a specific transport substrate or function for MmpL11 in M. tuberculosis has not been described, but the M. tuberculosis mmpL11 mutant was attenuated in a mouse model of infection indicating a role MmpL11 in the host environment (Domenech et al., 2005). M. smegmatis LppS is 71% identical to M. tuberculosis LppS and is a predicted lipoprotein with no known function. Data presented here suggest that both MmpL11 and LppS play a role in the M. smegmatis outer membrane as their absence affects Ub2 resistance and membrane permeability. Further studies in M. smegmatis and M. tuberculosis are necessary to define the function of these proteins and determine if this role is conserved between the two species.
In conclusion, our data suggest a model where the access of bactericidal ubiquitin-derived peptides to the mycobacterial inner membrane depends on outer membrane permeability. Once these peptides gain access to the inner membrane we propose that multimerize, insert into the membrane and form pores or otherwise disrupt membrane integrity of the bacterium. This membrane damage likely expose the bacterium to external conditions such as the low pH of the lysosome, and promote cytoplasmic leakage over time that contributes to bacterial death. The contribution of low membrane permeability to the intrinsic resistance of mycobacteria has long been appreciated (Barry, 2001). Our findings emphasize the contribution of this intrinsic resistance to host antimicrobial compounds and the intracellular environment.
EXPERIMENTAL PROCEDURES
Maintenance of bacterial cultures and cells
M. tuberculosis wild type strain CDC 1551 was provided by Ian Orme (Colorado State University). M. smegmatis mc2155 was obtained from ATCC. M. smegmatis SMR5 is a streptomycin-resistant variant of M. smegmatis mc2155 (Sander et al., 1995). The construction of porin mutants MN01(ΔmspA), ML10 (ΔmspA, ΔmspC) and ML16 (ΔmspA, ΔmspC, ΔmspD) in the SMR5 background has been described previously (Stephan et al., 2005; Stephan et al., 2004). Construction of plasmid pMN013 that carries the mspA gene under control of the constitutive pimyc promoter has been described elsewhere (Mailaender et al., 2004). Mycobacterial strains were maintained in Middlebrook 7H9 liquid media (Difco) or on Middlebrook 7H11 agar (Difco) plates supplemented with OADC (BD). RAW264.7 cells (ATCC) were maintained in the absence of antibiotics in DMEM supplemented with 10% fetal calf serum.
Transposon mutant screen
A transposon mutant library was generated in M. smegmatis mc2 155 using the mariner-based transposon mutagenesis vector pM272B as described (Gao et al., 2003). This library consists of 7,680 individual mutants archived in 96-well plates. For screening purposes, pools were generated by combining individuals from eight 96-well plates, yielding 10 pools total (768 clones each). The mutant pools were sub-cultured to 5×105 bacteria/mL in 7H9 media containing 200 μM Ub2. After 18h incubation, the treated bacteria were recovered by plating to 7H11 agar plates. Transposon insertion sites were identified using Ligation-Mediated PCR (LM-PCR) as described (Prod'hom et al., 1998). Briefly, chromosomal DNA was digested with SalI, ligated with SalI linkers, and PCR performed using the Sal primer (5'-TAGCTTATTCCTCAAGGCACG-3') that binds to the linker sequence and the mariner primer (5'-CGGGGACTTATCAGCCAACC-3') that binds within the transposon. Resulting PCR products were sequenced to identify the junction where transposon DNA disrupts chromosomal DNA.
Isolation and immundepletion of bactericidal soluble fraction
Lysosomes were isolated from BALB/c bone marrow-derived macrophages and soluble fraction (SF) was obtained as described previously (Alonso et al., 2007). For immuno-depletion studies, 50 μg SF was incubated with 10 μL rabbit polyconal anti-Ubiquitin (FL-76, Santa Cruz) or rabbit polyclonal anti-BSA (Sigma) for four hours. The antibody-bound proteins were removed using Protein A resin (Sigma). To quantitate the relative amount of ubiquitin present in SF, an immuno-blot was performed on equivalent protein amounts from SF and SF-Ub. The immuno-blot was probed with monoclonal antibody against ubiquitin (FK2, Biomol) and an HRP anti-mouse secondary antibody. Signals were quantitated using the ImageQuant software.
Bactericidal assays
Ub2 (STLHLVLRLRGG) and the granulysin-derived GranF2 peptide (VCRTGRSRWRDVCRNFMRRYQSR) were synthesized by GenScript Corp. (Piscataway, NJ). LL-37, LL-37 (18-37) fragment, and hepcidin were purchased from Anaspec (San Jose, CA). For bactericidal assays, LL-37 and LL-37 (18-37) were used at 20 μM (Martineau et al., 2007), GranF2 was used at 100 μM (Andreu et al., 1999), and hepcidin was used at 34 μM (Sow et al., 2007). To determine bactericidal activity against M. smegmatis, 5 × 105 cfu/mL bacteria were treated with the indicated concentration of peptide overnight. After treatment, the number of surviving bacteria was determined by plating serial dilutions. To determine pH dependence of Ub2, bactericidal assays were performed as above in 7H9 OADC buffered with HCl to pH 5.0, pH 5.5, pH 6.0, and pH 6.6 (standard conditions).
The standard assay to determine antimycobacterial activity of soluble fraction was as follows: 5×105 cfu of log-phase M. smegmatis or M. tuberculosis was incubated in media containing 50 μg/mL of SF in sodium acetate buffer or an equivalent amount of buffer (control) for the indicated time. After treatment, serial dilutions were plated to determine the cfu/mL of SF-treated bacteria compared to the control.
SDS sensitivity was a modification of the method performed previously (Vandal et al., 2009). Briefly, strains were grown to early log phase, serially diluted from OD600 0.01-0.00001 and 5 μL spotted onto 7H11 OADC plates with or without 0.005% SDS. Sensitivity to hydrogen peroxide was determined as follows: early log cultures were normalized to 5×106 cfu/mL and incubated 4h in the presence or absence of 5mM H2O2. Each sample was serially diluted and plated onto 7H11 OADC agar.
Quantification of ubiquitin in SF
To quantitate the relative amount of ubiquitin present in SF and SF-Ub, an immuno-blot was performed on equivalent lysate (pre- and post-immunodepletion) using mouse monoclonal antibody against ubiquitin and an HRP anti-mouse secondary antibody. Detection by luminescence was carried out using SuperSignal detection reagents (Pierce, Rockford, IL)
Western Analysis of M. tuberculosis whole cell lysates
Whole cell lysates were prepared as follows: M. tuberculosis strains were harvested by centrifugation and washed with PBS. Cells were resuspended at an OD600=10 in PBS containing protease inhibitors (Protease Inhibitor Cocktail III, Calbiochem), transferred to a tube containing 0.1 mm glass beads and broken by bead beater for 1 minute. Unbroken cells and beads were pelleted and the supernatant retained as total lysate. Total lysates were separated by SDS-12%PAGE and transferred to nitrocellulose. Western analysis was performed using polyclonal antibody pAK#13 against MspA (1:2000 dilution). HRP-conjugated anti-rabbit secondary antibody was used at a 1:10,000 dilution.
Spectrofluorimeter assays
M. smegmatis and M. tuberculosis was labeled in PBS, pH 7 with Cell Tracker 5'-chloromethylfluoroscein diacetate (CMFDA) (Molecular Probes, Eugene, OR). For microplate assays, bacteria were washed, then resuspended at OD600=1.2 in PBS, pH 5.5 and transferred to a 96-well microplate. Fluorescence emission was measured at 520 nm using the excitiation wavelengths of 450 nm and 490 nm in a SpectraMax Gemini EM fluorescent plate reader (Molecular Devices, Sunnyvale, CA). The ionophore nigericin was used at 10 μM, Ub2 was used at 100 μM. Conversion of the excitation ratio to pH was achieved through polynomial regression of a standard curve generated by treating CMFDA-labeled bacteria with 10 μM nigericin in buffers of known pH. Kinetic studies using M. smegmatis and M. tuberculosis in a 3 mL cuvette were performed using 100 μM Ub2 and measured by a QMSE4 spectrofluorimeter (Photon Technologies International, Lawrenceville, NJ, USA).
Macrophage infection assays
For each condition, RAW 264.7 macrophages were infected in quadruplicate at an MOI of 2:1, then incubated at 37° C in standard culture media, or either Earle's Basic Salts Solution (starvation media) or 50 μg/mL rapamycin to induce autophagy. The autophagy inhibitor 3-methyl adenine (3MA, Sigma) was added at 10 mM final concentration. The number of viable bacteria was determined by harvesting infected monolayers and plating serial dilutions. For immunofluorescence studies macrophages were infected with M. smegmatis expressing GFP and M. tuberculosis was stained beforehand with CMFDA. Infected macrophages were fixed in 4% paraformaldehyde. Primary rat ID4B antibody against LAMP-1 (Developmental Studies Hybridoma Bank) and Texas Red-conjugated secondary (Jackson ImmunoResearch) were used. Images were captured using an Olympus Fluoview confocal microscope.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health awards AI079399 to GEP, AI057086 and AI067027 to DGR, and AI063432 to MN.
REFERENCES
- Adams LB, Dinauer MC, Morgenstern DE, Krahenbuhl JL. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis. 1997;78:237–246. doi: 10.1016/s0962-8479(97)90004-6. [DOI] [PubMed] [Google Scholar]
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu D, Carreno C, Linde C, Boman HG, Andersson M. Identification of an anti-mycobacterial domain in NK-lysin and granulysin. Biochem J. 1999;344(Pt 3):845–849. [PMC free article] [PubMed] [Google Scholar]
- Barry C.E.r. Interpreting cell wall 'virulence factors' of Mycobacterium tuberculosis. Trends Microbiol. 2001;9:237–241. doi: 10.1016/s0966-842x(01)02018-2. [DOI] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Horwitz MA. The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J Exp Med. 1996;184:1349–1355. doi: 10.1084/jem.184.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun. 2000;68:2671–2684. doi: 10.1128/iai.68.5.2671-2684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci U S A. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Mailaender C, Niederweis M. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2008a;52:2503–2511. doi: 10.1128/AAC.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Pavlenok M, Niederweis M. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother. 2008b;52:3127–3134. doi: 10.1128/AAC.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chastellier C, Thilo L. Cholesterol depletion in Mycobacterium avium-infected macrophages overcomes the block in phagosome maturation and leads to the reversible sequestration of viable mycobacteria in phagolysosome-derived autophagic vacuoles. Cell Microbiol. 2006;8:242–256. doi: 10.1111/j.1462-5822.2005.00617.x. [DOI] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Barry C.E.r. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry C.E.r. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- Fabrino DL, Bleck CK, Anes E, Hasilik A, Melo RC, Niederweis M, Griffiths G, Gutierrez MG. Porins facilitate nitric oxide-mediated killing of mycobacteria. Microbes Infect. 2009 doi: 10.1016/j.micinf.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Gao LY, Groger R, Cox JS, Beverley SM, Lawson EH, Brown EJ. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect Immun. 2003;71:922–929. doi: 10.1128/IAI.71.2.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordao L, Bleck CK, Mayorga L, Griffiths G, Anes E. On the killing of mycobacteria by macrophages. Cell Microbiol. 2008;10:529–548. doi: 10.1111/j.1462-5822.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailaender C, Reiling N, Engelhardt H, Bossmann S, Ehlers S, Niederweis M. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiology. 2004;150:853–864. doi: 10.1099/mic.0.26902-0. [DOI] [PubMed] [Google Scholar]
- Martineau AR, Wilkinson KA, Newton SM, Floto RA, Norman AW, Skolimowska K, Davidson RN, Sorensen OE, Kampmann B, Griffiths CJ, Wilkinson RJ. IFN-gamma- and TNF-independent vitamin D-inducible human suppression of mycobacteria: the role of cathelicidin LL-37. J Immunol. 2007;178:7190–7198. doi: 10.4049/jimmunol.178.11.7190. [DOI] [PubMed] [Google Scholar]
- Murakami M, Lopez-Garcia B, Braff M, Dorschner RA, Gallo RL. Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J Immunol. 2004;172:3070–3077. doi: 10.4049/jimmunol.172.5.3070. [DOI] [PubMed] [Google Scholar]
- Niederweis M. Mycobacterial porins--new channel proteins in unique outer membranes. Mol Microbiol. 2003;49:1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x. [DOI] [PubMed] [Google Scholar]
- Niederweis M. Nutrient acquisition by mycobacteria. Microbiology. 2008;154:679–692. doi: 10.1099/mic.0.2007/012872-0. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Prod'hom G, Lagier B, Pelicic V, Hance AJ, Gicquel B, Guilhot C. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol Lett. 1998;158:75–81. doi: 10.1111/j.1574-6968.1998.tb12803.x. [DOI] [PubMed] [Google Scholar]
- Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. CellHost Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Sander P, Meier A, Bottger EC. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbati-Tehrani S, Meister B, Appel B, Lewin A. The porin MspA from Mycobacterium smegmatis improves growth of Mycobacterium bovis BCG. Int J Med Microbiol. 2004;294:235–245. doi: 10.1016/j.ijmm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Sharbati-Tehrani S, Stephan J, Holland G, Appel B, Niederweis M, Lewin A. Porins limit the intracellular persistence of Mycobacterium smegmatis. Microbiology. 2005;151:2403–2410. doi: 10.1099/mic.0.27969-0. [DOI] [PubMed] [Google Scholar]
- Sow FB, Florence WC, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J Leukoc Biol. 2007;82:934–945. doi: 10.1189/jlb.0407216. [DOI] [PubMed] [Google Scholar]
- Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, Niederweis M. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol Microbiol. 2001;40:451–464. doi: 10.1046/j.1365-2958.2001.02394.x. [DOI] [PubMed] [Google Scholar]
- Stephan J, Bender J, Wolschendorf F, Hoffmann C, Roth E, Mailander C, Engelhardt H, Niederweis M. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol Microbiol. 2005;58:714–730. doi: 10.1111/j.1365-2958.2005.04878.x. [DOI] [PubMed] [Google Scholar]
- Stephan J, Mailaender C, Etienne G, Daffe M, Niederweis M. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob AgentsChemother. 2004;48:4163–4170. doi: 10.1128/AAC.48.11.4163-4170.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. JBacteriol. 2009;191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]