Abstract
Direct and rapid analysis of cholesterol was accomplished in the ambient environment using reactive desorption electrospray ionization (DESI) mass spectrometry. This was achieved by electrospraying reagent solutions in the form of high velocity charged droplets at surfaces such as dried serum samples and animal tissue sections. Betaine aldehyde, incorporated into the spray solvent, reacts selectively and rapidly with the alcohol group of cholesterol by nucleophilic addition, forming a hemiacetal salt. Limits of detection for pure cholesterol and related compounds were ∼1 ng when a solution of cholesterol of 1ug/mL was spotted onto the surface). Quantitative analysis of free cholesterol in serum using reactive DESI was demonstrated using cholesterol-d7 as internal standard. High throughput analysis of small volumes of serum spotted onto a suitable substrate was achieved at an analysis rate of ∼14 seconds per sample, with a relative standard deviation (RSD) of ca. 6%. Use of reactive DESI in the imaging mode allowed 2D spatial distributions of phospholipids and cholesterol to be recorded simultaneously in rat brain tissues.
Introduction
Cholesterol is the major precursor of various steroid hormones whose presence and concentrations might be related to human diseases including Alzheimer's disease1, atherosclerosis2 and Smith-Lemli-Optiz syndrome (SLOS)3. A wide range of methods for the analysis of cholesterol and its metabolites has been reported, ranging from thin layer chromatography4, through enzymatic assays5 to nuclear magnetic resonance (NMR)3. However, the most-widely accepted methods for specific and quantitative analysis remain colorimetric maximal extraction assays6, liquid chromatography-mass spectrometry (LC/MS)7, and gas chromatography-mass spectrometry (GC/MS)8, 9, the latter using electron ionization (EI). Each of these methods requires time-consuming sample preparation. Cholesterol is readily ionized by EI but other MS ionization techniques including electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) are less well-suited for analyzing this molecule because of its low proton affinity and low acidity.7, 10 Cholesterol may be successfully characterized using ESI after offline derivatization10-13 or it may be directly ionized by energetic ionization methods, such as secondary ion mass spectrometry (SIMS)14, but direct ambient analysis, which is highly desirable for fast screening and quantification of cholesterol in biological fluids and for MS imaging of cholesterol in tissue samples, is not available yet.
DESI is a soft ambient ionization technique that can be applied to untreated samples in the ordinary environment.15, 16 Capabilities for quantitative analysis of small molecules using DESI-MS has been demonstrated.17, 18 A variant on the basic DESI experiment - reactive DESI - implements rapid chemical reactions occurring at the spot being sampled concurrently with acquisition of mass spectra to improve sensitivity and selectivity of target molecules.19 The experiment is carried out by dissolving a suitable chemical reagent into the spray solvent; no sample preparation is needed. To be analytically useful, reactions must occur rapidly in the chosen solvent and produce products that are ionized efficiently, often typically because they have a permanent charge20-23.
This paper demonstrates the capability of direct, rapid and quantitative analysis of free cholesterol in human serum under ambient conditions using reactive DESI. In addition, MS imaging of cholesterol in biological tissue using reactive DESI is reported. In these reactive DESI experiments, precharged betaine aldehyde (BA) is used as the reagent. This reagent reacts selectively with alcohols by nucleophilic addition to generate a hemiacetal salt, the precharged nature of which greatly improves analytical performance.
Experimental Section
Chemicals and Biological Samples
Cholesterol, retinol (vitamin A), vitamin D2, vitamin D3, L-ascorbic acid (vitamin C), glucose, fructose and betaine aldehyde chloride were purchased from Sigma-Aldrich Inc. (Milwaukee, WI). Cholesterol-d7 and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC 16:0/18:1) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Vitamin D3 tablets (400 International Units (IU)) with a mass of ∼400 mg, containing 10 μg of vitamin D3, were manufactured by US Nutrition, Inc. (Bohemia, NY). Liquid Cod liver oil was manufactured and distributed by Walgreen Co. (Deerfield, IL), containing ∼2 μg/ml vitamin D and ∼320 μg/ml vitamin A in concentration. Porous polytetrafluoroethylene (PTFE) sheets, of 1/16 inch thickness, were purchased from Small Parts Inc. (Miami Lakes, FL). Microscope glass slides (75×25 mm) of 1 mm thickness were purchased from Gold Seal Products Inc. (Portsmouth, NH). The serum sample, pooled from 10 individuals, was purchased from Innovative Research Inc. (Novi, MI), and stored in -20°C until analysis. The rat brains were purchased from Pel-Freez Inc. (Rogers, AR). They were cut into 11 μm sections at -20°C using a Shandon SME Cryotome Cryostat and thaw mounted onto microscope glass slides. The tissue sections were stored at -80°C until analysis. Prior to analysis the sections were dried in a vacuum dessicator for at least 2 hours.
DESI Experiments
Reactive DESI experiments and tandem mass spectrometry were carried out using a commercial Thermo Scientific LTQ (San Jose, CA) linear ion trap mass spectrometer equipped with a custom-built, automated DESI ion source. XCalibur 2.0 software was used to acquire the data. The home-built DESI ion source is similar to the OmniSpray source from Prosolia Inc. (Indianapolis, IN); it consists of an inner capillary (fused silica, 50 μm i.d., 150 μm o.d.) (Polymicro Technologies, AZ) for delivering the spray solvent and an outer capillary (250 μm i.d., 350 μm o.d.) for delivering the nebulizing gas. A sprayer-to-surface distance of 3 mm, a sprayer-to-inlet distance of 3 mm, a spray impact angle of 52°, and a capillary temperature of 250°C was used. The spray voltage was set to 5 kV, the capillary voltage was set to 5 V, and the tube lens voltage was set to 170 V. The nebulizing gas (N2) pressure was 160 psi. Acetonitrile (ACN)/chloroform (CHCl3) (1:2) containing 50 ppm BA was sprayed at a constant flow rate of 4 μL/min for the reactive DESI experiments. To determine the detection limit of different alcohols, samples were prepared for reactive DESI analysis by depositing 1 uL aliquots of the solution with the appropriate concentration onto a porous PTFE substrate. For the analysis of cod liver oil, 1 μL of the cod liver oil was examined from a microscope glass slide. All the samples were allowed to dry at room temperature prior to reactive DESI experiments. For the analysis of vitamin D3 tablets, the tablet coating was removed with a razor blade before attaching the tablet to a microscope glass slide with double-sided sticky tape. Tandem MS (MS/MS) product ion scans using collision-induced dissociation (CID) were carried out to confirm the identity of the products of reaction with BA. For the (MS/MS) experiments, an isolation window of 1.2 mass/charge units, a normalized collision energy of 20 to 40% (manufacturer's units) and a Mathieu parameter qz value of 0.25 during collisional activation were used.
For the reactive DESI imaging experiments, the spray solvent was ACN:water (H2O): dimethylformamide (DMF) = 8: 3: 1 containing 65 ppm BA and the flow rate was 1.5 μL/min. For the normal DESI imaging experiments methanol (MeOH)/H2O (1:1) was sprayed at the same flow rate.
Quantitative Assays
To create a calibration curve, stock solutions of cholesterol and the internal standard (IS) cholesterol-d7 were prepared in MeOH/CHCl3 (1:1) at concentrations of 3.6 mg/mL and 3.0 mg/mL respectively. Serial dilution of the cholesterol stock solution in MeOH provided calibration solutions with concentrations of 0, 0.6, 1.2, 1.8, 2.4, 3.0, and 3.6 mg/mL. Aliquots of 25 μL of the cholesterol calibration solutions and 25 μL internal standard (IS) solution were added to 100 μL of pooled human serum to make seven calibration serum samples, with added concentrations of cholesterol in serum (excluding the concentration of endogenous cholesterol) of 0, 100, 200, 300, 400, 500, 600 μg/mL. The final concentration of the IS was 500 μg/mL in all 7 calibration samples. Aliquots (1μL) of each calibrant were manually spotted along a straight line on a microscope glass slide using a spot-to-spot spacing of about 7 mm. The amount of IS in each spot was 1.3 × 10-9 moles while the amount of analyte cholesterol (again ignoring the endogenous cholesterol) ranged from 0 to 1.6 × 10-9 moles. This procedure, repeated on four consecutive days, resulted in 4×7 spots. All samples were allowed to dry at room temperature prior to analysis.
The analyte and the IS peaks in the extracted-ion chronograms (plots of mass spectral data vs. time) were picked automatically using Xcalibur's ICIS peak detection algorithm. The ion chronogram peak area obtained for cholesterol at each concentration was divided by the peak area for the IS from the same spot, and these peak area ratios (PARs) were plotted against cholesterol concentration. Fitting calculations were performed in Microsoft Excel using ordinary minimum least-squares linear methods, as described in the literature24.
Image Acquisition
In order to image the tissue samples, mass spectra were acquired with automatic gain control (AGC) turned off. Imaging experiments were performed by continuously scanning the surface in the x-direction and stepping in the y-direction (moving opposite to the direction of the spray) at the end of each line. The spray spot size was ∼300 μm. The ion injection time was set to 50 ms, and 7 microscans were averaged. The time required to record the whole image depended on the size of the tissue and the pixel size selected. Typically, a coronal section of rat brain was imaged using 40 lines and a step size of 200 μm in the y-direction. The lines were scanned at a constant velocity of 233 μm/s, while collecting one mass spectrum every 0.86 seconds over the range m/z 100–1200. The number of scans per line was 70. This procedure resulted in arrays of 70 × 40 pixels, each pixel covering an area of 200 × 200 μm. The total time to record this image was ∼40 minutes (0.86 sec × 70 × 40). Under these conditions, a lateral spatial resolution of ∼200 μm can normally be achieved in DESI MS imaging16, even though the spot size is slightly greater. The surface moving stage included an XYZ integrated linear stage (Newport, Richmond, CA) and a rotary stage (Parker Automation, Irwin, PA). More detailed information about the instrumentation can be found somewhere else.18, 25
Lab-written software was used to convert the XCalibur mass spectral files (.raw) into a format compatible with BioMap.26 BioMap (freeware, http://www.maldi-msi.org/) was used to process the mass spectral data to generate two-dimensional ion images. The total data size of the image is ∼200 megabytes. Different color templates and appropriate maximum and minimum intensity values in the color bar were selected to increase the contrast in the images.
Results and Discussion
Reactivity of Betaine Aldehyde (BA)
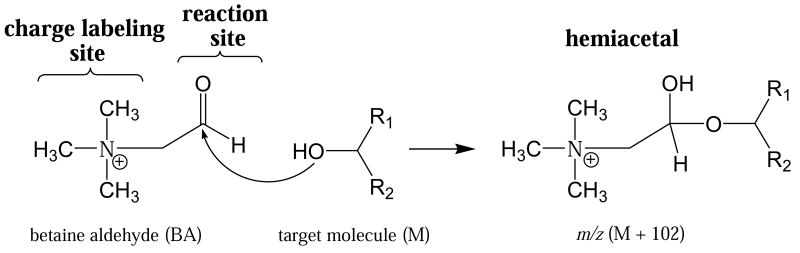
The results show that BA reacts selectively and rapidly with the alcohols of interest by nucleophilic addition to generate a hemiacetal salt (Scheme 1). Nucleophilic addition occurs between the aldehyde group of BA and the hydroxyl group of the target alcohol. BA is chosen as the reagent because it tags the reaction product with a positive charge for improved sensitivity in positive ion mass spectrometry. For a target molecule of molecular weight M, the detected hemiacetal product gives a signal at m/z (M+102), where 102 is the molecular weight of BA.
Scheme 1.
In situ reaction between betaine aldehyde cation and the hydroxyl group of an alcohol
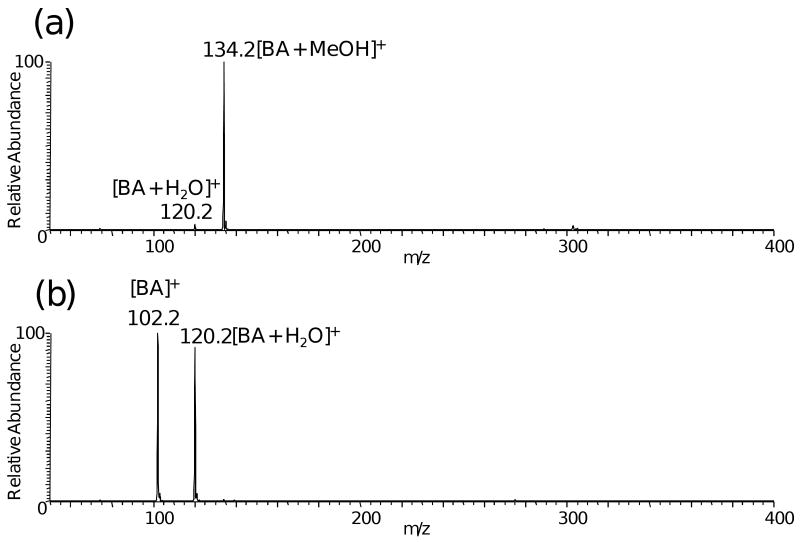
Because methanol, a common solvent in ESI and DESI, reacts efficiently BA, it competes with the target alcohols and cannot be used as solvent. As shown in Figure 1a, an abundant ion, m/z 134.2, is formed in the reaction between BA and methanol, and its identity was confirmed by collision-induced dissociation (data not shown). Water can also react with BA to form a product ion at m/z 120.2, but water is much less reactive than methanol, so some BA survives in DESI experiments using water as spray solvent (Figure 1b). The reactivities of two other derivatization reagents p-dimethylaminobenzaldehyde and (p-formylphenyl) trimethylammonium iodide were tested also. However, neither reagent reacted with cholesterol under the normal DESI conditions using a spray solvent of ACN/H2O (1:1) containing 50 ppm of the reagent. The special reactivity of betaine aldehyde might be due to the activation of the carbonyl group by the positive charge on the quaternary amine through the σ bonds.
Figure 1.
DESI experiments on blank glass slides, using 50 ppm betaine aldehyde in a spray solvent of (a) MeOH/H2O (1:1) (b) ACN/H2O (1:1).
Charge labeling based on reaction with BA greatly improved the detection sensitivity of cholesterol in DESI experiments. The detection limit for cholesterol was reduced from >200ng (conventional non-reactive DESI) to ∼1 ng (reactive DESI); this value is adequate for the complex mixture analysis and imaging experiments discussed below. Not only cholesterol, but other secondary alcohols such as fat-soluble vitamins can be detected in reactive DESI with BA as the reagent (Figure S1, Supporting Information). The limits of detection of various alcohols are summarized in Table 1.
Table 1.
Limits of detection (LOD) of alcohols in reactive DESI[a]
| Compound | Mass-to-charge ratio(m/z) | Corresponding ions | LOD |
|---|---|---|---|
| Retinol (Vitamin A) | 388.3 | [M+BA]+ | 0.5 ng |
| Ergocalciferol (Vitamin D2) | 498.5 | [M+BA]+ | 1 ng |
| Cholecalciferol (Vitamin D3) | 486.4 | [M+BA]+ | 2 ng |
| L-ascorbic acid (Vitamin C) | 278.2 | [M+BA]+ | 2 ng |
| Cholesterol | 488.5 | [M+BA]+ | 0.5 ng |
| Glucose | 282.2 | [M+BA]+ | 1 ng |
| Fructose | 282.2 | [M+BA]+ | 1 ng |
Porous PTFE surfaces were used and the spot size interrogated was 300 μm diameter, the spray solvent was ACN/CHCl3 (1:1) with 50 ppm BA.
[M+BA]+ refers to the hemiacetal shown in Scheme 1.
Phenols and related compounds, such as 2-naphthalol, also react with BA although much less rapidly. This can be explained by the decreased nucleophilicity of the hydroxyl group caused by electron delocalization. Sugars and related polyhydroxy compounds, such as vitamin C (ascorbic acid), also react with BA, but only one BA attachment is generally observed. The reaction is particularly useful for alcohols containing one hydroxyl group and those with low proton affinities and acidities, like cholesterol and its metabolites. Because phenols are acidic, they are better detected in the negative ion mode. Similarly, alcohols containing multiple hydroxyl groups have higher proton affinities and often can be detected using simple cationization with salts in DESI and ESI.27 The results indicate the sensitivity for detecting alcohols with multiple hydroxyl groups using reactive DESI is comparable to that of simple salt cation attachment procedure.
Quantitative Analysis of Cholesterol in Human Serum
The concentration of total cholesterol, consisting of free cholesterol and cholesterol esters, ranges from 1300 to 2500 μg/mL in healthy adult human serum, and the concentration of free cholesterol is in the range of ∼200 to 550 μg/mL.28-30 Reactive DESI-MS allows free cholesterol to be detected selectively in serum because cholesterol esters do not react with BA. Pooled human serum was used without any prior purification or extraction. An isotopically labeled version of the analyte, cholesterol-d7, was selected as internal standard and added to the serum sample. An IS is critical in DESI quantification, as reported previously.17 A relative standard deviation (RSD) under 10% and a relative error (RE) of less than 5% can often be achieved at the 1 ng level in DESI when using an IS.17, 18
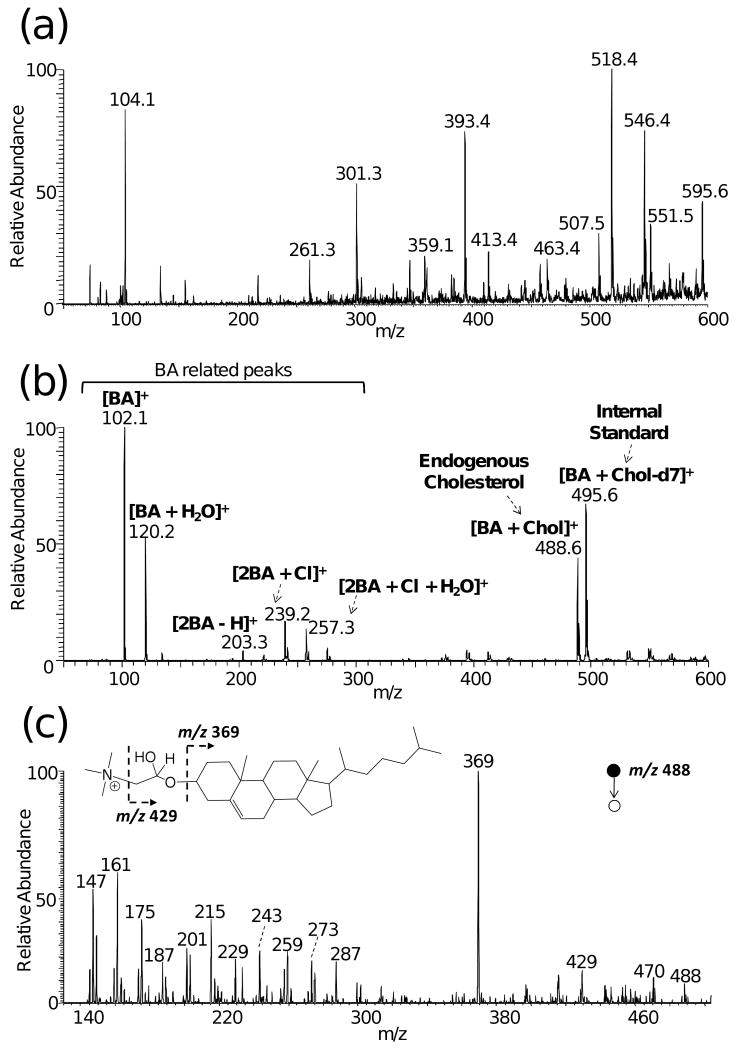
Figure 2a and 2b compare mass spectra recorded using normal and reactive DESI on a dried serum spot. The serum sample was doped with 500 μg/mL cholesterol-d7 before deposition on the glass slide. The normal DESI spectrum provides no information about cholesterol (Figure 2a). None of the possible cholesterol related peaks, including the protonated peak [Chol + H]+ (m/z 387), water loss peak [Chol + H − H2O]+ (m/z 369) and sodium adducts [Chol + Na]+ (m/z 409), is observed.
Figure 2.
(a) Normal DESI on a dried human serum spot, with ACN/CHCl3 (1:2) as the solvent (b) Reactive DESI on a dried human serum spot doped with cholesterol-d7 as internal standard A solvent of ACN/CHCl3 (1:2) with 50 ppm BA was used. (c) Product ion MS/MS spectrum of m/z 488. In each case, an aliquot (1μL) of serum was spotted onto a microscope glass slide.
The reactive DESI spectrum recorded using the same solvent shows a strong cholesterol signal (Figure 2b). The signals at m/z 495.6 and 488.6 correspond to reaction products of cholesterol-d7 and endogenous cholesterol in serum with BA. The peaks observed in normal DESI (Figure 2a) were suppressed in reactive DESI due to the strong signals of cholesterol and BA related peaks. Other compounds with the alcohol functional group which are present in serum, such as estradiol and several vitamins, give ions of negligible abundance, either because their concentrations are too low or because their ionization is suppressed by competition with the much more abundant cholesterol. The peaks in the lower mass region (m/z 102, 120, 203, 239, 257) are BA related peaks. Even with ACN and CHCl3 as the solvent, trace amounts of water in ACN or in the solvent spray system react with BA, producing the peak at m/z 120. However, the presence of trace amounts of water does not affect the quantitative analysis of cholesterol. Product ion tandem MS experiments further confirm that m/z 488 is the reaction product between cholesterol and BA (Figure 2c). The fragment ion m/z 429 is due to the loss of the head group N(CH3)3 and m/z 369 corresponds to protonated cholestadiene. The mass difference of 14 between adjacent peaks in the lower mass region is indicative of the hydrocarbon skeleton of the product ion. The fragmentation pattern of m/z 488 observed in the serum sample is consistent with what was observed in the authentic cholesterol standard. Product ion tandem MS of the internal standard [BA + Chol-d7]+ at m/z 495 shows a similar fragmentation pattern to [BA + Chol]+ (Figure S2, Supporting Information).
The capability of quantitative analysis of cholesterol using reactive DESI was demonstrated using standard addition. Seven calibration samples were prepared with final cholesterol concentrations of (0 + x), (100 + x), (200 + x), (300 + x), (400 + x), (500 + x), (600 + x) μg/mL, where x represents the unknown concentration of endogenous cholesterol. The internal standard concentration in the calibration serum samples was kept constant at 500 μg/mL. An aliquot of 1μL of each serum calibration solution was spotted manually on a glass slide with a spot-to-spot spacing of about 7 mm.
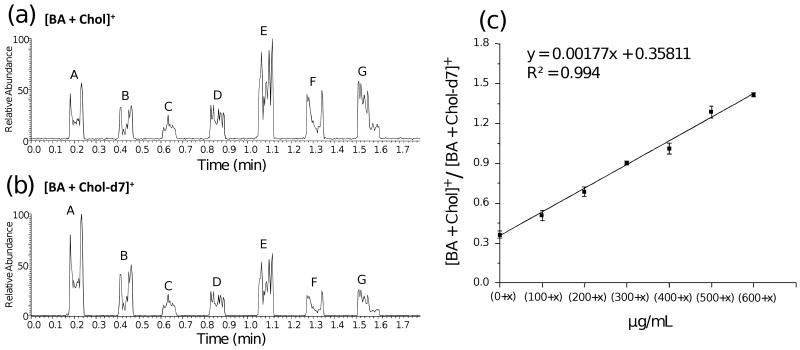
Figure 3a and 3b show the extracted-ion chronograms for [BA + Chol]+ and [BA + Chol-d7]+ obtained from the seven sample spots, which were scanned by the DESI sprayer. The expected increase in signal with increasing concentration of cholesterol is not seen in Figure 3a; similarly, the data set shown in Figure 3b shows significant variation although the concentration of the IS is constant. These results, which could be due to sweet spots, differences in spot position/sprayer position, or signal saturation, demonstrate that an internal standard is essential for quantitative measurements. In fact, the ratio of the cholesterol signals to the labeled cholesterol signals is linear with respect to the amount of cholesterol in the serum sample. Figure 3c shows the calibration curve obtained after repeating the analysis on four consecutive days, resulting in a 4×7 data point set. The calibration curve is linear from (0 + x) to (600 + x) μg/mL cholesterol, where x represents the unknown concentration of endogenous cholesterol in the calibration serum samples. The linearity of this calibration curve is evident from the correlation coefficient (R2) of 0.994. By extrapolating the standard addition calibration curve, the concentration of endogenous cholesterol is measured as 202 μg/mL in calibration serum samples.
Figure 3.
Extracted ion chronograms of (a) [BA + Chol]+ at m/z 488.6 and (b) Internal standard [BA + Chol-d7]+ at m/z 495.6 from seven samples (labeled A to G) spotted on a glass slide. A represents the sample with the lowest cholesterol concentration of (0+x) μg/mL, while G represents the sample with the highest cholesterol concentration of (600+x) μg/mL; where x represents the unknown concentration of endogenous cholesterol. (c) Calibration curve obtained from seven samples with different cholesterol concentrations in serum by plotting peak area ratios (PARs) against the cholesterol concentration. The bars represent the standard deviation of analysis for four replicates at different cholesterol concentrations.
Because the original serum was diluted in the course of the addition of the IS and cholesterol solutions (25 μL IS and 25 μL cholesterol solution were added to 100 μL original serum), the concentration of the endogenous cholesterol in the original pooled human serum is estimated to be 303 μg/mL (after correction for the a dilution factor of 1.5), which falls within the normal free cholesterol range of human serum. The value with confidence interval is 303 ± 36μg/mL for the concentration of the endogenous cholesterol, calculated with Student's t-distribution at 95% confidence level. The precision, represented as the relative standard deviation (RSD), varied from 1.2% to 6.4% for four replicates at different cholesterol concentrations within the linear response range. The performance is comparable to that reported for the much slower GC-MS and ESI-MS cholesterol determinations10, 12, 31, 32 and similar to data reported previously for conventional DESI17, 18.
Reactive DESI not only allows direct and rapid analysis at improved sensitivity (relative to conventional DESI) and with good precision, it also allows high throughput measurements to be made. For the case of seven serum samples with different concentrations of cholesterol, the mass spectrometric analysis time was less than 2 minutes, around 14 seconds per sample (Figure 3a and 3b). This high-throughput feature together with the lack of sample pretreatment is clearly useful for screening large numbers of samples.
The concentration of total cholesterol (cholesterol plus esters) cannot be obtained using this method. However, hydrolysis of cholesterol esters could be performed according to standard procedures33 before using reactive DESI to obtain the concentration of total cholesterol, but this extension of the experiment is beyond the scope of this paper.
Reactive DESI Imaging
Mass spectrometric imaging techniques34-40 provide spatially specific chemical composition information on biomolecules in tissue, data which can increase understanding of physiological processes, disease pathology and the most appropriate course of treatment. Compared to other MS imaging techniques (MALDI and SIMS), DESI imaging is implemented under ambient conditions and can be used to characterize objects their native environment without addition of matrices which might cause interference in the low mass range. Ionization by DESI is soft, occurring without fragmentation or with minimum fragmentation, especially when compared to SIMS. Imaging of cholesterol by MS has previously been performed under vacuum by mapping fragment ions of cholesterol in SIMS.41
Using reactive DESI, cholesterol in rat brain tissue (∼13 mg/g)42 is easily imaged under ambient conditions giving a full 2-D image of 200 μm pixel size within an hour (Figure 4a and b). The ion image of the peak at m/z 488.5, corresponding to [BA+Chol]+, extracted from the full set of data, shows the expected increased concentration of cholesterol in the white matter (e.g. corpus callosum, anterior part of anterior commissure, cerebellum) as compared to the gray matter. The spatial distribution of cholesterol mapped by DESI is consistent with literature results43, 44.
Figure 4.
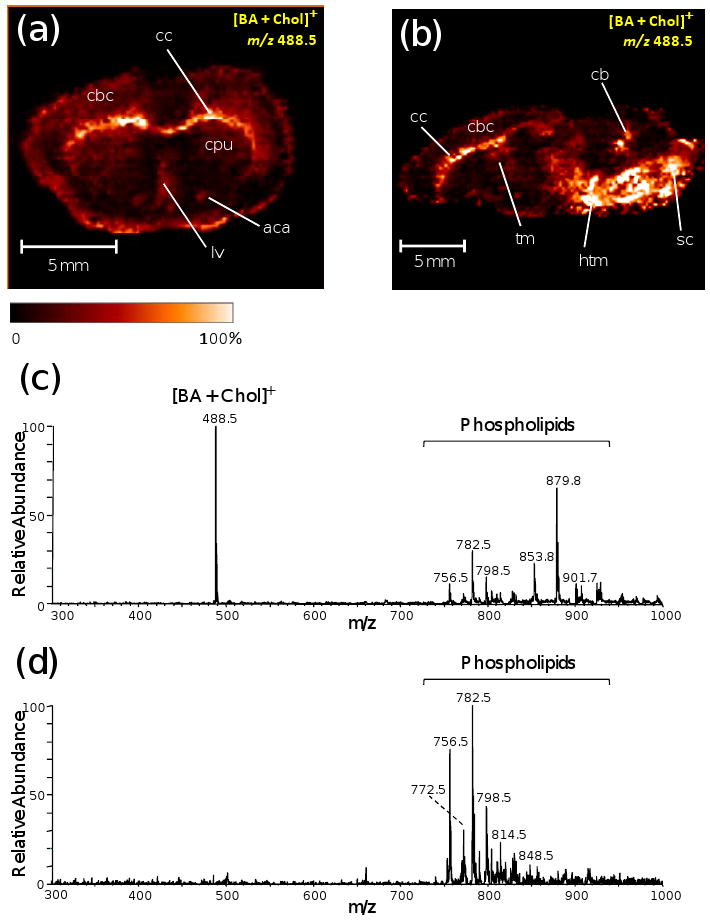
Cholesterol images at m/z 488.5 in (a) 14×8 mm coronal section of rat brain and (b) 22× 8 mm sagittal section of rat brain observed by reactive DESI in the positive ion mode (65 ppm BA in a solvent of ACN/H2O/DMF (8:3:1)). cc=corpus callosum; cpu=striatum; cbc=cerebral cortex; lv=lateral ventricle; aca=anterior part of anterior commissure; cb=cerebellum; sc=spinal cord; tm=thalamus; htm=hypothalamus (c) Reactive DESI mass spectrum (background subtracted) of rat brain tissue using a solvent of ACN/H2O/DMF (8:3:1) doped with 65 ppm BA, showing derivatized cholesterol (with S/N ≫ 100) and various phospholipids. (d) Normal DESI mass spectrum (background subtracted) of rat brain tissue using ACN/H2O/DMF (8:3:1) as the spray solvent without adding the BA reagent.
The solvent system is an important factor dictating the signal intensity when analyzing samples with different polarities. This is because desorption of the sample from the surface is related to the solubility of the sample in the spray solvent. Different solvent systems (ACN, H2O, CHCl3, DMF and their mixtures) were tested in the reactive DESI imaging experiment to optimize the resolution as well as the signal intensity of cholesterol in rat brain tissue. It was found that the solvent system ACN/H2O/DMF (8:3:1) is a good solvent system for imaging of cholesterol, while a mixture of methanol and water gives better signal intensity for the polar lipids in the higher mass range in normal DESI imaging. The same solvent system ACN/CHCl3/DMF (8:3:1), was used for comparison of imaging using reactive DESI (Figure 4c) with normal DESI (Figure 4d). The results show that the distribution of cholesterol in rat brain cannot be obtained using normal DESI solvents(Figure S3, Supporting Information).
Besides the predominant cholesterol peak at m/z 488.5, the lipids observed in the mass range from m/z 750 to 850 (Figure 4c) can also be imaged. These lipids are mostly glycerophosphocholines (PCs), which can also be observed with normal DESI (Figure 4d). Interestingly, some additional peaks occur in the mass range m/z 850 to 1000 compared to normal DESI. As suggested by multi-stage mass spectrometry (MS2 and MS3, Figure S4, Supporting Information), they are reaction products of PCs with BA and water, possibly through reaction at the phosphate group of the PCs (Scheme S1, Supporting Information). However these reactions do not provide new chemical information as compared with normal DESI. The assignments of PC peaks (Table S1, Supporting Information) were made by comparisons with tandem mass spectrometry for the authentic lipid standards and with existing DESI mass spectra45. Although non-ambient imaging MS techniques such as SIMS and MALDI, can reach higher lateral resolution, the long acquisition time and correspondingly large data collections involved during these experiments are not always needed. As demonstrated here, high-lateral resolution is not required to map cholesterol in specific substructures of the brain.
Conclusion
Rapid screening of cholesterol in human serum was accomplished at atmospheric pressure using reactive DESI mass spectrometry. The experiment combines physical desorption by DESI with chemical derivatization in situ to efficiently ionize this non-polar compound. A charged hemiacetal is formed in the course of a heterogeneous phase reaction between the solution-phase BA and solid-phase cholesterol in dried serum spots. BA was demonstrated to greatly improve the selectivity and sensitivity of analysis of cholesterol. This method is to be applicable to the analysis of cholesterol in different materials, including biological fluids and tissues. Using reactive DESI imaging, the distribution of cholesterol was recorded in rat brain tissues. Traditional MS imaging methods, including normal DESI, are much less successful for low polarity compounds. Quantitative analysis of cholesterol in serum by reactive DESI showed good precision at physiological levels.
In addition to human serum and rat brain tissue, rapid detection of cholesterol and vitamin D3 in cod liver oil and vitamin D3 tablets was demonstrated using reactive DESI (Figure S5 and S6, Supporting Information).
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health (NIH grant 1R21EB009459-01) and the Office of Naval Research (N00014-05-1-0405).
References
- 1.Valenza M, Cattaneo E. Prog Neurobiol. 2006;80:165–176. doi: 10.1016/j.pneurobio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Felton CV, Crook D, Davies MJ, Oliver MF. Atertio Thromb Vasc Biol. 1997;17:1337–1345. doi: 10.1161/01.atv.17.7.1337. [DOI] [PubMed] [Google Scholar]
- 3.Ruan BF, Wilson WK, Pang JH, Gerst N, Pinkerton FD, Tsai J, Kelley RI, Whitby FG, Milewicz DM, Garbern J, Schroepfer GJ. J Lipid Res. 2001;42:799–812. [PubMed] [Google Scholar]
- 4.Asmis R, Buhler E, Jelk J, Gey KF. J Chromatogr B. 1997;691:59–66. doi: 10.1016/s0378-4347(96)00436-7. [DOI] [PubMed] [Google Scholar]
- 5.Dehoff JL, Davidson LM, Kritchevsky D. Clin Chem. 1978;24:433–435. [PubMed] [Google Scholar]
- 6.Hoving EB. J Chromatogr B. 1995;671:341–362. doi: 10.1016/0378-4347(95)00223-6. [DOI] [PubMed] [Google Scholar]
- 7.Tian Q, Failla ML, Bohn T, Schwartz SJ. Rapid Commun Mass Spectrom. 2006;20:3056–3060. doi: 10.1002/rcm.2700. [DOI] [PubMed] [Google Scholar]
- 8.Schwertner HA, Johnson ER, Lane TE. Clin Chem. 1990;36:519–521. [PubMed] [Google Scholar]
- 9.Minami Y, Yokoi S, Setoyama M, Bando N, Takeda S, Kawai Y, Terao J. Lipids. 2007;42:1055–1063. doi: 10.1007/s11745-007-3099-4. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DW, ten Brink HJ, Jakobs C. J Lipid Res. 2001;42:1699–1705. [PubMed] [Google Scholar]
- 11.Quirke JME, Van Berkel GJ. J Mass Spectrom. 2001;36:1294–1300. doi: 10.1002/jms.233. [DOI] [PubMed] [Google Scholar]
- 12.Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. Biochim Biophys Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Weiskopf AS, Vouros P, Cunniff J, Binderup E, Bjorkling F, Binderup L, White MC, Posner GH. J Mass Spectrom. 2001;36:71–78. doi: 10.1002/jms.105. [DOI] [PubMed] [Google Scholar]
- 14.Piehowski PD, Carado AJ, Kurczy ME, Ostrowski SG, Heien ML, Winograd N, Ewing AG. Anal Chem. 2008;80:8662–8667. doi: 10.1021/ac801591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 16.Ifa DR, Wiseman JM, Song QY, Cooks RG. Int J Mass spectrom. 2007;259:8–15. [Google Scholar]
- 17.Ifa DR, Manicke NE, Rusine AL, Cooks RG. Rapid Commun Mass Spectrom. 2008;22:503–510. doi: 10.1002/rcm.3377. [DOI] [PubMed] [Google Scholar]
- 18.Manicke NE, Kistler T, Ifa DR, Cooks RG, Ouyang Z. J Am Soc Mass Spectrom. 2009;20:321–325. doi: 10.1016/j.jasms.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Nyadong L, Green MD, De Jesus VR, Newton PN, Fernandez FM. Anal Chem. 2007;79:2150–2157. doi: 10.1021/ac062205h. [DOI] [PubMed] [Google Scholar]
- 20.Busch KL, Unger SE, Vincze A, Cooks RG, Keough T. JACS. 1982;104:1507–1511. [Google Scholar]
- 21.Domon B, Mueller DR, Richter WJ. Org Mass Spectrom. 1994;29:713–719. [Google Scholar]
- 22.Sonsmann G, Romer A, Schomburg D. J Am Soc Mass Spectrom. 2002;13:47–58. doi: 10.1016/S1044-0305(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 23.Depauw E, Pelzer G, Marien J, Piette JL, Pardon MC. Org Mass Spectrom. 1985;20:692–693. [Google Scholar]
- 24.Lavagnini I, Magno F. Mass Spectrom Rev. 2007;26:1–18. doi: 10.1002/mas.20100. [DOI] [PubMed] [Google Scholar]
- 25.Wiseman JM, Ifa DR, Venter A, Cooks RG. Nat Protoc. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 26.Song QY. Ph D thesis. Purdue University; West Lafayette: 2008. [Google Scholar]
- 27.Kauppila TJ, Talaty N, Jackson AU, Kotiaho T, Kostiainen R, Cooks RG. Chem Commun. 2008:2674–2676. doi: 10.1039/b804413a. [DOI] [PubMed] [Google Scholar]
- 28.Kim DH, Lee KJ, Heo GS. J Chromatogr B. 1994;655:1–8. doi: 10.1016/0378-4347(94)00032-8. [DOI] [PubMed] [Google Scholar]
- 29.Leonard PJ, Shaper AG, Jones KW. Am J Clin Nutr. 1965;17:377–380. doi: 10.1093/ajcn/17.6.377. [DOI] [PubMed] [Google Scholar]
- 30.Sperry WM. J Biol Chem. 1936;114:125–133. [Google Scholar]
- 31.Sevanian A, Seraglia R, Traldi P, Rossato P, Ursini F, Hodis H. Free Radic Biol Med. 1994;17:397–409. doi: 10.1016/0891-5849(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 32.Ostlund RE, Hsu FF, Bosner MS, Stenson WF, Hachey DL. J Mass Spectrom. 1996;31:1291–1296. doi: 10.1002/(SICI)1096-9888(199611)31:11<1291::AID-JMS424>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Abell LL, Levy BB, Brodie BB, Kendall FE. J Biol Chem. 1952;195:357–366. [PubMed] [Google Scholar]
- 34.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Nature. 2007;449:1033–1037. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 35.Nemes P, Barton AA, Li Y, Vertes A. Anal Chem. 2008;80:4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 36.McDonnell LA, Heeren RMA. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 37.Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Nat Methods. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiseman JM, Ifa DR, Song QY, Cooks RG. Angew Chem Int Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 39.Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0812020106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz TA, Gamez G, Setz PD, Zhu L, Zenobi R. Anal Chem. 2008;80:6537–6544. doi: 10.1021/ac8005044. [DOI] [PubMed] [Google Scholar]
- 41.Zheng L, McQuaw CM, Ewing AG, Winograd N. JACS. 2007;129:15730–15731. doi: 10.1021/ja0741675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu GR, Servatius RJ, Shefer S, Tint GS, O'Brien WT, Batta AK, Salen G. Metab, Clin Exp. 1998;47:878–882. doi: 10.1016/s0026-0495(98)90130-5. [DOI] [PubMed] [Google Scholar]
- 43.Touboul D, Halgand F, Brunelle A, Kersting R, Tallarek E, Hagenhoff B, Laprevote O. Anal Chem. 2004;76:1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- 44.Sjovall P, Johansson B, Lausmaa J. Appl Surf Sci. 2006;252:6966–6974. [Google Scholar]
- 45.Manicke NE, Wiseman JM, Ifa DR, Cooks RG. J Am Soc Mass Spectrom. 2008;19:531–543. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.