Abstract
Matrix metalloproteinase-2 (MMP-2) is a well known mediator of cancer metastasis, but is also thought to be involved in several aspects of cancer development, including cell growth and inflammation. We comprehensively characterized genetic variation across the MMP-2 gene and evaluated associations with breast cancer risk using a two phase (Phase 1 and Phase 2) study design. A total of 39 polymorphisms were genotyped among 6,066 Chinese women participating in the Shanghai Breast Cancer Study (SBCS), a population based case-control study. Two MMP-2 promoter polymorphisms were found to have consistent results between Phase 1 and Phase 2 participants, and to be significantly associated with breast cancer risk among all genotyped participants. Minor allele homozygotes for rs11644561 (G/A) were found to have a decreased risk of breast cancer (OR: 0.6, 95% CI: 0.3–1.0) compared to major allele homozygotes, as were minor allele homozygotes for rs11643630 (T/G) compared to major allele homozygotes (OR: 0.8, 95% CI: 0.7–1.0). When analyzed together, a rare haplotype (4.4%) with both rs11644561 A and rs11643630 G was found to have a significantly reduced risk of breast cancer (OR 0.6, 95% CI: 0.4–0.8). In addition, rare allele homozygotes for rs243865 (−1306 C/T) tended to have an increased risk of breast cancer (OR: 1.4, 95% CI: 0.9–2.4). Together, these findings support a role for MMP-2 genetic variation in breast cancer susceptibility.
Keywords: matrix metalloproteinase-2, polymorphisms, breast cancer, susceptibility
Introduction
As the enzyme capable of cleaving type 4 collagen, the major structural component of the epithelial basement membrane, matrix metalloproteinase-2 (MMP-2, gelatinase A), is well known to be integral for cancer cell invasion and metastasis 1–3. With many additional extracellular matrix (ECM) and non-ECM substrates, MMP-2 is also involved in a variety of other, potentially pathologic processes, including inflammation, angiogenesis, and cellular proliferation 1–5. Oncogene-mediated cellular transformation was found to induce MMP-2 expression, causing altered cell growth and increased capacity for malignant progression 6;7. In vitro assays of breast cancer cells stably transfected with MMP-2 demonstrated increased invasive properties, while accelerated tumor growth, enhanced metastatic colonization, and increased tumor burden was seen after the transfected cells were injected into mice 8;9. On the other hand, MMP-2 deficient mice were found to have reduced tumor-induced angiogenesis, significantly slower tumor growth rates, and decreased metastatic colonization of the lung after implantation of either melanoma or lung carcinoma cells 10.
In humans, normal breast tissue and benign breast lesions were rarely found to express MMP-2, while expression has been detected in both tumor and surrounding stromal cells 11–15. Further, compared to adjacent breast tissue, a gradual increase in MMP-2 expression was seen from noninvasive to invasive cancers, while MMP-2 activity has also been found to be significantly higher in malignant breast tissue compared to other breast tissues 16;17. In addition, breast cancer patients were found to have significantly higher circulating MMP-2 levels compared to control volunteers 18.
Genetic variation that modulates MMP-2 expression may contribute to individual differences in cancer susceptibility. Two single nucleotide polymorphisms (SNPs) in the MMP-2 promoter have been shown to affect expression in vitro; C to T transitions at −1306 (rs243865) and −735 (rs2285053) both result in lower transcriptional activities 19–21. These two SNPs are reported to be in high linkage disequilibrium (LD) and have an interactive effect on MMP-2 transcription 21. Several epidemiological studies have evaluated these promoter polymorphisms in relation to cancer risk; with inconsistent results. To date, only a few studies have evaluated genetic variation in MMP-2 in relation to breast cancer susceptibility, and all included only one polymorphism (rs243865) 22–25. Preliminary results from a small study (89 cases and 100 controls) among Latin American women indicated that there was no association 22, while a small study among Mexican women (90 cases and 96 controls) found a significantly increased risk of breast cancer associated with −1306 CC 23. A larger study (462 cases and 509 controls) among Chinese women found a significantly decreased risk of breast cancer for T allele carriers 24, while the largest study to date (959 cases and 952 controls), conducted among Swedish women, found no association 25.
As MMP-2 has been shown to contribute not only to cancer invasion and metastasis, but also to cellular transformation and tumor growth, this study was undertaken in order to comprehensively characterize genetic variation across the MMP-2 gene and evaluate associations of MMP-2 polymorphisms and breast cancer susceptibility.
Materials and Methods
Study subjects were participants of the Shanghai Breast Cancer Study (SBCS), a population-based case-control study among Chinese women; detailed information on the study design and data collection procedures have been previously described 26. Briefly, Phase 1 cases were women diagnosed with breast cancer between August 1996 and March 1998, 25–64 years of age, without a previous cancer diagnosis, and alive at the time of interview. Recruitment for Phase 2 occurred between April 2002 and February 2005, and eligibility criteria were expanded to include women 20–70 years of age 27;28. All cases were identified via the population-based Shanghai Cancer Registry; diagnoses were confirmed by two senior pathologists. Controls were randomly selected from the general population using the Shanghai Resident Registry, a population registry of adult residents in urban Shanghai; women with previous cancer diagnoses were excluded. Structured questionnaires were administered by trained interviewers, and were used to obtain detailed information on demographic, reproductive, and behavioral factors; height and weight were also measured. Of eligible participants, 1,459 (91.1%) cases and 1,556 (90.3%) controls in Phase 1 and 1,989 cases (83.7%) and 1,989 controls (70.4%) in Phase 2, completed in-person interviews. In Phase 1, 1,193 cases (81.8%) and 1,310 controls (84.2%) donated blood samples. In Phase 2, 1,932 (97.1%) cases and 1,857 (93.4%) controls donated either blood or buccal cell samples. Genomic DNA was extracted using Puregene’s DNA Purification kits (Gentra Systems, Minneapolis, MN) or Qiagen’s DNA Purification kits (Qiagen, Valencia, CA) according to manufacturers’ instructions. Laboratory staff was blinded to the case-control status of these subjects for all subsequent genotyping described.
Haplotype tagging SNPs (htSNPs) were selected by searching Han Chinese data from the HapMap Project 29 using the Tagger program 30. htSNPs were selected to cover polymorphisms with minimum minor allele frequency (MAF) of 0.05 in the MMP-2 gene ± 5 kb with an r2 of 0.90 or greater. Selection of htSNPs was completed in December 2005 using HapMap Release 19. Genotyping assays for 19 htSNPs were completed for 2,131 Phase 1 participants in 2006 using a Targeted Genotyping System (Affymetrix, Santa Clara, CA) based on an advanced Molecular Inversion Probe (MIP) method 31. Blinded (N=39) and HapMap samples (N=12) were also included; consistency rates averaged 99.6%.
Two SNPs with promising results in Phase 1 (rs1116195 and rs243865) were selected for additional genotyping among Phase 2 participants. Further, a polymorphism reported to be functional that was not genotyped in Phase 1 (rs2285053) was also selected. These three SNPs were genotyped among 2,932 Phase 2 participants using the Sequenom iPLEX MassARRAY platform (Sequenom, Inc., San Diego, CA). Polymerase chain reaction (PCR) and extension primers were designed using Sequenom Assay Design software. PCR and extension reactions were performed according to manufacturer’s instructions as previously described 32. Allele-specific extension products were determined by using matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS). Blinded duplicate samples and negative controls were included in each 96-well plate; concordance rates between duplicate samples were 100% for all three polymorphisms genotyped by this method.
Recently, we completed genotyping for 4,157 cases and controls (2,213 Phase 1 and 1,944 Phase 2 participants) using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix), that includes 906,602 SNPs. From this, data for 19 additional SNPs that are located in MMP-2 ±10kb were included in the current study in order to increase the density of genetic coverage and the statistical power of our analysis.
Hardy-Weinberg equilibrium (HWE) was tested by comparing the observed and expected genotype frequencies of the controls (χ2 test). Characteristics of cases and controls were compared with the χ2 test or t-test for categorical or continuous variables, respectively. Covariates considered included age at diagnosis, age at menarche, age at first live birth among parous women, age at menopause among postmenopausal women, history of breast fibroadenomas, BMI, and leisure physical activity in the decade preceding diagnosis. Associations with breast cancer risk were evaluated by computing odds ratios and corresponding 95% confidence intervals (OR, 95% CI) by logistic regression. Additive models of effect were applied to all SNPs; tests for trend were conducted by coding for the number of variant alleles and reporting the p-value for the beta coefficient. Dominant or recessive effect models and p-values were also calculated when suggested to be appropriate for particular SNPs. Linkage disequilibrium between polymorphisms was assessed by Haploview 33. Associations between haplotypes and breast cancer risk were analyzed with HAPSTAT software 34; additive, dominant, and recessive models of effect were also evaluated. Study phase was adjusted for in analyses that included participants from both phases. All statistical tests were two-tailed, and p-values of ≤ 0.05 were interpreted as statistically significant.
Results
A total of 6,066 Chinese women were included in the present study: 2,291 Phase 1 participants and 3,775 Phase 2 participants. Women in Phase 2 were slightly older, and tended to participate in more regular physical activity than Phase 1 women, but were generally comparable. As expected, breast cancer cases were found to differ from controls in regards to demographic, reproductive, and other known breast cancer risk factors (Table 1). Cases were more likely to have an earlier age at menarche, older age at first live birth, a history of breast fibroadenomas, higher BMI and WHR, and less likely to participate in regular physical activity than controls.
Table 1.
Characteristics of Patients Genotyped for MMP-2, by Study Phase, The Shanghai Breast Cancer Study
| Phase 1 (N=2,291) |
Phase 2 (N=3,775) |
|||||
|---|---|---|---|---|---|---|
| Characteristics |
Cases (N=1,114) |
Controls (N=1,177) |
p-value |
Cases (N=1,925) |
Controls (N=1,850) |
p-value |
| Demographic Factors | ||||||
| Age, mean (years) | 47.6 ± 8.0 | 47.2 ± 8.6 | 0.280 | 50.9 ± 8.3 | 51.7 ± 8.4 | 0.003 |
| Age, range (years) | (28–64) | (25–64) | NA | (20–70) | (23–70) | NA |
| Education (less than middle school) | 138 (12.4%) | 171 (14.5%) | 0.134 | 312 (16.2%) | 209 (11.3%) | <0.001 |
| Reproductive Risk Factors | ||||||
| Age at menarche (years) | 14.5 ± 1.6 | 14.7 ± 1.7 | <0.001 | 14.4 ± 1.7 | 14.7 ± 1.8 | <0.001 |
| Premenopausal | 745 (66.9%) | 758 (64.4%) | 0.213 | 1,084 (56.3%) | 934 (50.5%) | <0.001 |
| Age at menopause (years)1 | 48.1 ± 4.7 | 47.4 ± 5.0 | 0.031 | 48.5 ± 4.4 | 48.3 ± 4.6 | 0.196 |
| Age at first live birth (years)2 | 26.8 ± 4.1 | 26.2 ± 3.8 | 0.001 | 26.2 ± 3.6 | 25.7 ± 3.8 | <0.001 |
| Used oral contraceptives | 244 (21.9%) | 254 (21.6%) | 0.852 | 341 (17.7%) | 356 (19.2%) | 0.226 |
| Used estrogen replacement therapy | 29 (2.6%) | 30 (2.6%) | 0.929 | 89 (4.6%) | 60 (3.2%) | 0.029 |
| Additional Risk Factors | ||||||
| First degree relative with breast cancer | 37 (3.3%) | 30 (2.6%) | 0.273 | 104 (5.4%) | 57 (3.1%) | <0.001 |
| Ever had breast fibroadenomas | 107 (9.6%) | 58 (4.9%) | <0.001 | 192 (10.0%) | 105 (5.7%) | <0.001 |
| Body mass index (kg/m2) | 23.6 ± 3.4 | 23.2 ± 3.4 | 0.013 | 23.7 ± 3.3 | 23.4 ± 3.2 | 0.004 |
| Waist-to-hip ratio | 0.81 ± 0.06 | 0.80 ± 0.06 | 0.002 | 0.83 ± 0.05 | 0.82 ± 0.06 | <0.001 |
| Regular physical activity | 214 (19.2%) | 300 (25.5%) | <0.001 | 562 (34.4%) | 636 (34.4%) | <0.001 |
Continuous variables: mean values ± standard deviation, p-value from t-tests; Categorical variables: numbers and percentages, p-values from χ2 test
Among postmenopausal women
Among parous women
Bold values considered to be significant at p ≤0.05
A total of 39 MMP-2 SNPs were genotyped: 19 htSNPs among Phase 1 participants, 1 selected SNP from the literature among Phase 2 participants, and 19 additional SNPs among participants of both study phases. Of the MMP-2 SNPs genotyped, three had MAFs of less than 1% in our study population, and were excluded from further analyses (rs16955194, rs7189232, and rs11541998). The remaining SNPs were found to have MAFs between 10.7 and 48.9% among genotyped controls; none were found to deviate from Hardy-Weinberg equilibrium. MAFs among SBCS controls were generally comparable to the MAF found among Han Chinese genotyped in HapMap, with two exceptions: SBCS controls had more A alleles (44.6%) for rs1005912 than HapMap (38%), and fewer G alleles (40.4%) for rs243867 than HapMap (46%), although neither of these differences was statistically significant. Associations with breast cancer susceptibility were initially assessed using additive models of effect. When the same SNP was genotyped by two methods, the data source with the higher number of genotyped participants was utilized. One SNP (rs243865) was genotyped by both Sequenom and Affymetrix 6.0 methods for 1,098 samples; the concordance rate between these methods was 100%. Seven SNPs (rs243865, rs1477017, rs865094, rs1053605, rs243847, rs243839, and rs11639960) were genotyped by both Affymetrix Targeted Genotyping and the Affymetrix 6.0 Genome Wide platform for approximately 2,000 participants; concordance rates for these samples ranged between 98.96 and 99.90%, and averaged 99.65%.
SNP information, genotyping method, study population genotyped, and associations with breast cancer risk for the 36 polymorphic MMP-2 SNPs are detailed in Table 2. All available genotyped participants are included in the analyses; when study phases 1 & 2 are listed, the estimate of effect is pooled. Two promoter SNPs were found to confer significant reductions in breast cancer risk among rare allele homozygotes. rs11644561 AA participants were approximately 40% less likely to be breast cancer cases than those with GG, while rs11643630 AA participants were approximately 20% less likely to be breast cancer cases than those with the TT genotype. Additionally, one SNP, rs1005912, was associated with a small increased risk among heterozygotes, although rare allele homozygotes did not exhibit a stronger positive association.
Table 2.
MMP-2 SNPs and Breast Cancer Risk, The Shanghai Breast Cancer Study
| SNP | Alleles * | Region | Method † | Study Phase † | MAF (%) ‡ | Breast Cancer Risk § |
||
|---|---|---|---|---|---|---|---|---|
| AB OR (95% CI) | BB OR (95% CI) | p-value | ||||||
| rs1005912 | T/A | promoter | Affy 6.0 | 1 & 2 | 45.3 | 1.2 (1.0–1.3) | 1.1 (0.9–1.3) | 0.207 |
| rs1116195 | A/T | promoter | Targeted | 1 & 2 | 44.6 | 1.0 (0.9–1.2) | 1.2 (1.0–1.4) | 0.075 |
| rs11644561 | G/A | promoter | Affy 6.0 | 1 & 2 | 13.0 | 0.9 (0.8–1.1) | 0.6 (0.3–1.0) | 0.098 |
| rs243867 | A/G | promoter | Affy 6.0 | 1 & 2 | 40.4 | 1.1 (0.9–1.2) | 1.1 (0.9–1.3) | 0.403 |
| rs11643630 | T/G | promoter | Affy 6.0 | 1 & 2 | 42.9 | 1.0 (0.8–1.1) | 0.8 (0.7–1.0) | 0.046 |
| rs243866 | G/A | promoter | Affy 6.0 | 1 & 2 | 11.0 | 1.0 (0.9–1.2) | 1.2 (0.7–2.1) | 0.602 |
| rs243865 | C/T | promoter | Targeted | 1 & 2 | 11.5 | 0.9 (0.8–1.1) | 1.4 (0.9–2.4) | 0.776 |
| rs243864 | T/G | promoter | Affy 6.0 | 1 & 2 | 10.7 | 1.0 (0.9–1.2) | 1.1 (0.6–2.0) | 0.782 |
| rs2285053 | C/T | promoter | Targeted | Phase 2 | 23.4 | 1.2 (1.0–1.4) | 0.9 (0.6–1.2) | 0.436 |
| rs1477017 | A/G | intron 2 | Both | 1 & 2 | 27.4 | 1.0 (0.9–1.2) | 1.0 (0.8–1.2) | 0.833 |
| rs865094 | A/G | intron 2 | Both | 1 & 2 | 29.1 | 0.9 (0.8–1.0) | 1.1 (0.9–1.4) | 0.838 |
| rs11646643 | A/G | intron 3 | Affy 6.0 | 1 & 2 | 15.5 | 1.0 (0.8–1.1) | 1.1 (0.7–1.6) | 0.726 |
| rs1053605 | C/T | exon 5 | Both | 1 & 2 | 13.0 | 1.1 (0.9–1.2) | 0.8 (0.4–1.3) | 0.862 |
| rs9302671 | G/T | intron 5 | Affy 6.0 | 1 & 2 | 15.3 | 1.0 (0.8–1.1) | 1.1 (0.8–1.6) | 0.936 |
| rs2241145 | G/C | intron 5 | Targeted | Phase 1 | 48.9 | 1.0 (0.8–1.2) | 0.9 (0.8–1.2) | 0.613 |
| rs2241146 | G/A | intron 5 | Targeted | Phase 1 | 21.8 | 1.1 (0.9–1.3) | 1.0 (0.7–1.5) | 0.632 |
| rs243849 | C/T | exon 7 | Affy 6.0 | 1 & 2 | 18.8 | 0.9 (0.8–1.1) | 1.1 (0.8–1.6) | 0.816 |
| rs12599775 | G/C | intron 7 | Targeted | Phase 1 | 11.2 | 1.1 (0.9–1.4) | 0.9 (0.4–1.9) | 0.453 |
| rs243847 | T/C | intron 7 | Both | 1 & 2 | 42.3 | 1.1 (0.9–1.2) | 1.0 (0.8–1.2) | 0.881 |
| rs2192852 | A/G | intron 7 | Targeted | Phase 1 | 38.1 | 1.0 (0.8–1.2) | 0.9 (0.7–1.2) | 0.546 |
| rs12923011 | C/T | intron 7 | Targeted | Phase 1 | 16.4 | 0.9 (0.7–1.1) | 0.7 (0.4–1.3) | 0.118 |
| rs243845 | G/A | intron 8 | Affy 6.0 | 1 & 2 | 31.1 | 1.0 (0.9–1.2) | 1.0 (0.8–1.2) | 0.945 |
| rs243844 | G/A | intron 8 | Targeted | Phase 1 | 30.7 | 1.0 (0.8–1.2) | 1.1 (0.8–1.5) | 0.604 |
| rs2287074 | G/A | exon 9 | Targeted | Phase 1 | 27.4 | 1.0 (0.8–1.2) | 0.8 (0.5–1.1) | 0.276 |
| rs243842 | T/C | intron 9 | Affy 6.0 | 1 & 2 | 31.9 | 1.0 (0.9–1.2) | 1.0 (0.8–1.2) | 0.882 |
| rs183112 | G/A | intron 9 | Targeted | Phase 1 | 18.7 | 1.0 (0.8–1.2) | 0.9 (0.6–1.5) | 0.874 |
| rs243839 | A/G | intron 9 | Both | 1 & 2 | 41.1 | 1.0 (0.9–1.1) | 1.0 (0.8–1.2) | 0.924 |
| rs9923304 | C/T | intron 9 | Affy 6.0 | 1 & 2 | 27.1 | 1.0 (0.9–1.2) | 0.9 (0.7–1.2) | 0.983 |
| rs11639960 | A/G | intron 10 | Both | 1 & 2 | 28.8 | 1.0 (0.9–1.1) | 1.1 (0.8–1.3) | 0.889 |
| rs243831 | T/G | 3′ FR || | Targeted | Phase 1 | 13.6 | 0.8 (0.7–1.0) | 0.8 (0.4–1.6) | 0.113 |
| rs12930259 | T/C | 3′ FR || | Targeted | Phase 1 | 33.6 | 1.0 (0.9–1.2) | 1.0 (0.7–1.3) | 0.899 |
| rs2192853 | A/G | 3′ FR || | Targeted | Phase 1 | 35.8 | 0.9 (0.8–1.1) | 1.0 (0.7–1.3) | 0.607 |
| rs1583587 | G/C | 3′ FR || | Affy 6.0 | 1 & 2 | 35.7 | 1.0 (0.9–1.2) | 1.0 (0.8–1.2) | 0.796 |
| rs8053806 | C/A | 3′ FR || | Affy 6.0 | 1 & 2 | 23.4 | 1.1 (1.0–1.3) | 1.1 (0.9–1.5) | 0.139 |
| rs12708952 | G/C | 3′ FR || | Affy 6.0 | 1 & 2 | 36.0 | 1.0 (0.9–1.2) | 1.0 (0.8–1.2) | 0.874 |
| rs1583585 | G/A | 3′ FR || | Affy 6.0 | 1 & 2 | 23.1 | 1.1 (0.9–1.2) | 1.1 (0.9–1.5) | 0.192 |
Major/minor alleles as determined by allele frequency among genotyped controls
Genotyping Method and SBCS Study Phase: Affymetrix Targeted Genotyping among 1,062 cases and 1,069 controls from SBCS Phase 1 and/or Sequenom Targeted Genotyping among 1,495 cases and 1,437 controls from SBCS Phase 2 (Targeted), or Affymetrix 6.0 genotyping among 1,104 cases and 1,109 controls from Phase 1, and 965 cases and 971 controls from SBCS Phase 2 (Affy 6.0), or genotyped by both methods (Both); study phases 1&2 show pooled estimates
Minor allele freqency among all genotyped controls
Risk of breast cancer, adjusted for age, education, and Study Phase (when appropriate); AA major allele homozygotes (reference group), AB heterozygotes, BB minor allele homozygotes; p-value for trend from additive models
3′ FR: downstream flanking region, 3′ of the MMP-2 gene
Bold values considered to be significant at p≤0.05
Table 3 includes associations with breast cancer stratified by SBCS Phase; included are the two SNPs above as well as two polymorphisms that had promising results after our initial targeted Phase 1 genotyping. Among Phase 1 participants, rs1116195 was significantly associated with an increased risk of breast cancer for minor allele homozygotes (OR: 1.3, 95% CI: 1.0–1.7), while minor allele homozygotes for rs243865 tended to have an increased risk (OR 1.8, 95% CI: 0.8–4.1). Additional genotyping among 1,491 cases and 1,437 controls from Phase 2 did not confirm an association between rs1116195 and breast cancer risk. However, results from Phase 2 for rs243865 were similar to those from Phase 1. In both Phases, minor allele homozygotes for rs243865 tended to have a moderate increased risk of breast cancer, although even in combined analysis, this did not reach statistical significance. The two SNPs with significant results among all genotyped participants (rs11644561 and rs11643630) had consistent results when stratified by SBCS Phase. Minor allele homozygotes compared to major allele homozygotes were significantly less likely to be breast cancer cases for rs11644561 (OR: 0.6, 95% CI: 0.3–1.0) and for rs11643630 (OR: 0.8, 95% CI: 0.7–1.0).
Table 3.
Selected MMP-2 SNPs and Breast Cancer Risk, by Study Phase, The Shanghai Breast Cancer Study
| Phase 1 OR (95% CI) § |
Phase 2 OR (95% CI) § |
Combined OR (95% CI) § |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Alleles * | Method † | MAF (%) ‡ | AB | BB | p-value | AB | BB | p-value | AB | BB | p-value |
| rs1005912 | T/A | Affy 6.0 | 45.3 | 1.2 (1.0–1.5) | 1.2 (0.9–1.5) | 0.187 | 1.1 (0.9–1.4) | 1.0 (0.8–1.4) | 0.642 | 1.2 (1.0–1.3) | 1.1 (0.9–1.3) | 0.207 |
| rs1116195 | A/T | Targeted | 44.6 | 1.1 (0.9–1.4) | 1.3 (1.0–1.7) | 0.038 | 1.0 (0.8–1.2) | 1.1 (0.9–1.3) | 0.574 | 1.0 (0.9–1.2) | 1.2 (1.0–1.4) | 0.075 |
| rs11644561 | G/A | Affy 6.0 | 13.0 | 0.9 (0.8–1.2) | 0.5 (0.2–1.1) | 0.189 | 0.9 (0.7–1.1) | 0.6 (0.3–1.4) | 0.262 | 0.9 (0.8–1.1) | 0.6 (0.3–1.0) | 0.098 |
| rs11643630 | T/G | Affy 6.0 | 42.9 | 0.9 (0.7–1.1) | 0.8 (0.7–1.1) | 0.155 | 1.0 (0.9–1.3) | 0.8 (0.6–1.0) | 0.191 | 1.0 (0.8–1.1) | 0.8 (0.7–1.0) | 0.046 |
| rs243865 | C/T | Targeted | 11.5 | 0.9 (0.7–1.1) | 1.8 (0.8–4.1) | 0.512 | 1.0 (0.8–1.2) | 1.3 (0.7–2.5) | 0.824 | 0.9 (0.8–1.1) | 1.4 (0.9–2.4) | 0.776 |
Major/minor alleles as determined by allele frequency among genotyped controls
Genotyping Method: Affymetrix Targeted Genotyping among 1,062 cases and 1,069 controls from SBCS Phase 1 and Sequenom Targeted Genotyping among 1,495 cases and 1,437 controls from SBCS Phase 2 (Targeted), or Affymetrix 6.0 genotyping among 1,104 cases and 1,109 controls from Phase 1, and 965 cases and 971 controls from SBCS Phase 2 (Affy 6.0)
Minor allele frequency among all genotyped controls
Breast Cancer Risk Odds Ratios and 95% Confidence Intervals from additive models, adjusted for age, education, and study phase (when appropriate), p-value for trend from additive effect models
Bold values considered to be significant at p≤0.05
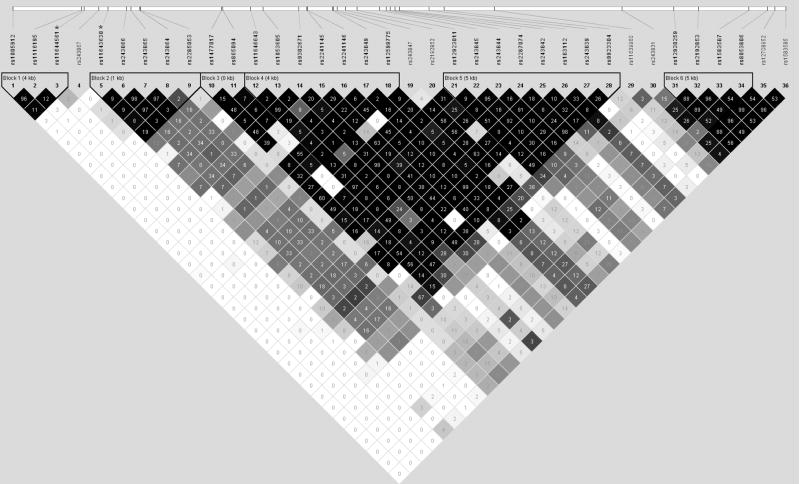
The linkage disequilibrium (LD) structure of the 36 polymorphic MMP-2 SNPs was constructed by combining all available genotyping data from 3,027 controls (Figure 1). The two SNPs with significant risk reductions for homozygotes (rs11644561 and rs11643630) were not found to be in high LD (D′=0.21, r2=0), and so haplotype analysis was performed to assess the effects of these SNPs in concert (Table 4). Compared to the common reference haplotype with both major alleles (H1: GT, 48.5%), the haplotype with minor alleles for both rs11644561 and rs11643630 (H4: AG, 4.4%) was associated with a significantly reduced risk of breast cancer in both additive and dominant models (OR: 0.6, 95% CI: 0.4–0.8). As single SNP analysis resulted in significant risk reductions only for the homozygotes of these variants, and haplotype analysis indicated that it was the two SNPs in combination that best captured a decreased risk of breast cancer, these two SNPS were analyzed further. In logistic regression models that included both rs11644561 and rs11643630, homozygotes for both SNPs were found to have significantly reduced risks; further, an interaction term for the two polymorphisms was not found to reach statistical significance. Finally, haplotype analysis was also conducted for the two MMP-2 SNPs previously reported in the literature; no significant haplotype effects for rs243865 or rs2285053 were observed.
Figure 1. LD Structure of MMP-2 Polymorphisms among 3,027 Shanghai Breast Cancer Study Controls.
Legend: LD structure of 36 MMP-2 SNPs among 3,027 Shanghai Breast Cancer Study controls; value shown is r2. Two SNPs of interest, rs11644561 and rs11643630 are marked with asterisks (*), and are in positions 3 and 5, respectively.
Table 4.
Haplotype Analysis of Selected MMP-2 Polymorphisms, The Shanghai Breast Cancer Study
| Additive Models |
Dominant Models |
Recessive Models |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype1 |
Frequency 2 |
OR 3 |
95% CI 3 |
p-value |
OR 3 |
95% CI 3 |
p-value |
OR 3 |
95% CI 3 |
p-value |
| SNPs with replicated results: rs11644561 and rs11643630 | ||||||||||
| H1: GT | 48.5 | 1.0 | reference | 1.0 | reference | 1.0 | reference | |||
| H2: GG | 38.5 | 0.9 | 0.9–1.0 | 0.302 | 1.0 | 0.9–1.1 | 0.670 | 1.0 | 0.8–1.1 | 0.534 |
| H3: AT | 8.6 | 1.0 | 0.8–1.2 | 0.929 | 1.0 | 0.9–1.3 | 0.641 | 0.9 | 0.5–1.6 | 0.830 |
| H4: AG | 4.4 | 0.6 | 0.4–0.8 | 0.002 | 0.6 | 0.4–0.8 | 0.003 | 0.4 | 0.1–2.8 | 0.341 |
| Literature SNPs: rs243865 and rs2285053 | ||||||||||
| H1: CC | 65.2 | 1.0 | reference | 1.0 | reference | 1.0 | reference | |||
| H2: CT | 23.4 | 1.1 | 0.9–1.2 | 0.347 | 1.1 | 1.0–1.3 | 0.176 | 0.9 | 0.7–1.2 | 0.527 |
| H3: TC | 11.4 | 1.0 | 0.9–1.1 | 0.999 | 1.0 | 0.9–1.1 | 0.674 | 1.2 | 0.9–1.7 | 0.218 |
Bold letters indicate less common alleles
Frequency of haplotype among genotyped controls
Estimates of effect adjusted for age, education, and Study Phase
Bold values considered to be significant at p≤0.05
Discussion
A two-phase case-control study was conducted to first comprehensively evaluate MMP-2 genetic variants in relation to breast cancer risk, and then to validate any promising associations among a second independent sample population. Two MMP-2 SNPs (rs11644561 and rs11643630) were found to have associations with breast cancer risk that were consistent between Phase 1 and Phase 2 study populations as well as significant in combined analyses. While the effects of these SNPs were found to be independent, a rare haplotype that included both minor alleles was associated with significant risk reduction. To the best of our knowledge, neither of these two MMP-2 polymorphisms have been previously evaluated for associations with cancer susceptibility.
Two promoter SNPs, rs243865 (−1306) and rs2285053 (−735), have been previously reported to affect MMP-2 transcription in vitro; both C to T transitions result in reduced expression due to the ablation of specificity protein (Sp)1 transcription factor binding sites 19–21. Only one of these SNPs (rs243865) has been previously evaluated in relation to breast cancer risk, and results have been inconsistent. Two studies found no effect 22;25, while two studies (N=186 and N=971) found significantly decreased risks of breast cancer associated with the T allele 23;24. In contrast, in the current study, rs243865 TT homozygotes tended to have an increased risk of breast cancer, although this effect was not significant. Similarly, results from this study for rs2285053 were not compelling; while heterozygotes had a marginally significant increased risk of breast cancer, homozygotes tended to have a diminished risk. Notably, these two SNPs were not found to be in high LD in this study population (D′=1, r2=0.03), and no haplotype effects on breast cancer risk were found.
While MMP-2 has traditionally been thought of as a mediator of metastasis, a growing body of evidence has connected MMP-2 to earlier aspects of carcinogenesis, including cell growth, inflammation, and angiogenesis 1–5. In addition to a wide range of extracellular matrix (ECM) substrates, including gelatin, elastin, fibronectin, laminin, and collagens, MMP-2 has many non-ECM substrates that include growth factors modulators and cytokines 1;2;4. For example, hydrolysis of membrane bound fibroblast growth factor receptor type 1 (FGF-R1) by MMP-2 releases the soluble ectodomain of the active receptor, thereby influencing the mitogenic and angiogenic activities of FGF 4;35. Additionally, MMP-2 was shown to contribute to the inflammatory response by being an alternative activator of pro-interleukin 1-beta (pro-IL1-β) in the absence of the cytokines’ favored activator caspace-1 36. Finally, degradation of ECM substrates may also contribute to cancer development and progression, as MMP-2 cleavage of the proteoglycan decorin releases transforming growth factor-beta 1 (TGF-β1) from its extracellular reservoir 37.
Studies of the MMP-2 promoter have identified several putative regulatory regions and transcription factor binding sites within 2 kb of the transcription initiation site, including those for Sp1, p53, S1, S2, AP-1, AP-2, Ets-1, C/EBP, CREB, GCN-His, and Pea3 38;39. Some of these elements have been shown to be critical for MMP-2 expression in different cell types or due to different chemical or oncogenic stimuli 38–40. To our knowledge, further upstream MMP-2 promoter sequences do not seem to have been previously characterized. In the current study, rs11644561 and rs11643630 were both found to confer decreased risk for homozygotes; these SNPs are located approximately 4 kb and 2.6 kb upstream of the MMP-2 transcription initiation site, respectively. Using available bioinformatics tools, we tried to evaluate these regions further, but our analysis was uninformative. Therefore, with current evidence, we can not determine whether these loci represent novel functional SNPs that may affect MMP-2 expression, or else, if together, they best tag another, as yet ungenotyped, variation. As their effects were found to be independent, they may each tag this other loci to different degrees, explaining why the haplotype with both SNPs resulted in capturing the risk reduction more than the recessive effects of either haplotype with only one of the alleles alone.
In summary, we identified two MMP-2 promoter polymorphisms that were associated with modest decreases in breast cancer risk. Homozygotes of the minor alleles for rs11644561 and rs11643630 were 40% and 20% less likely to have breast cancer, respectively. Although a two phase study design was used to reduce type I error, we cannot rule out the possibility that our findings could be due to chance. Further, neither association remains significant after adjusting for the number of SNPs evaluated. However, this is the largest and most comprehensive analysis of MMP-2 polymorphisms conducted to date, and our results are consistent with both in vitro and in vivo evidence that demonstrate a role for MMP-2 in breast cancer development. Therefore, additional studies to evaluate these MMP-2 polymorphisms in population studies are warranted.
Acknowledgments
This research was supported by USPHS grants R01CA64277, R01CA908999, and R01CA124558. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The authors wish to thank the participants and research staff of the Shanghai Breast Cancer Study for their contributions and commitment to this project and Brandy Venuti for assistance in the preparation of this manuscript. DNA extraction, sample preparation, and genotyping assays using Affymetrix arrays were conducted at the Vanderbilt Survey and Biospecimen and Microarray Shared Resources that are supported in part by the Vanderbilt Ingram Cancer Center (P30 CA68485). Sequenom assays were performed by Proactive Genomics (Winston-Salem, North Carolina).
References
- 1.Bjorklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta. 2005;1755:37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and -9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–97. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 4.Duffy MJ, Maguire TM, Hill A, McDermott E, O’Higgins N. Metalloproteinases: role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000;2:252–7. doi: 10.1186/bcr65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen M, Arkell J, Jackson CJ. Human endothelial gelatinases and angiogenesis. Int J Biochem & Cell Bio. 2001;33:960–70. doi: 10.1016/s1357-2725(01)00007-3. [DOI] [PubMed] [Google Scholar]
- 6.Moon A, Kim MS, Kim TG, et al. H-ras, but not N-ras, induces an invasive phenotype in human breast epithelial cells: a role for MMP-2 in the H-ras-induced invasive phenotype. Int J Cancer. 2000;85:176–81. doi: 10.1002/(sici)1097-0215(20000115)85:2<176::aid-ijc5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Baruch RR, Melinscak H, Lo J, Liu Y, Yeung O, Hurta RAR. Altered matix metalloproteinase expression associated with oncogene-mediated cellular transformation and metastasis formation. Cell Bio Int. 2001;25:411–20. doi: 10.1006/cbir.2000.0647. [DOI] [PubMed] [Google Scholar]
- 8.Cockett MI, Murphy G, Birch ML, et al. Matrix metalloproteinases and metastatic cancer. Biochem Soc Symp. 1998;63:295–313. [PubMed] [Google Scholar]
- 9.Tester AM, Waltham M, Oh SJ, et al. Pro-Matrix Metalloproteinase-2 Transfection Increases Orthotopic Primary Growth and Experimental Metastasis of MDA-MB-231 Human Breast Cancer Cells in Nude Mice. Cancer Res. 2004;64:652–8. doi: 10.1158/0008-5472.can-0384-2. [DOI] [PubMed] [Google Scholar]
- 10.Itoh T, Tanioka M, Yoshida H, Yoshioka T, Nishimoto H, Itohara S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998;58:1048–51. [PubMed] [Google Scholar]
- 11.Brummer O, Athar S, Riethdorf L, Loning T, Herbst H. Matrix-metalloproteinases 1, 2, and 3 and their tissue inhibitors 1 and 2 in benign and malignant breast lesions: an in situ hybridization study. Virchows Arch. 1999;435:566–73. doi: 10.1007/s004280050442. [DOI] [PubMed] [Google Scholar]
- 12.Baker EA, Stephenson TJ, Reed MW, Brown NJ. Expression of proteinases and inhibitors in human breast cancer progression and survival. Mol Pathol. 2002;55:300–4. doi: 10.1136/mp.55.5.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebeau A, Muller-Aufdemkamp C, Allmacher C, et al. Cellular protein and mRNA expression patterns of matrix metalloproteinases-2, -3 and -9 in human breast cancer: correlation with tumour growth. J Mol Histol. 2004;35:443–55. doi: 10.1023/b:hijo.0000045943.26251.24. [DOI] [PubMed] [Google Scholar]
- 14.Pellikainen JM, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. Expression of Matrix Metalloproteinase (MMP)-2 and MMP-9 in Breast Cancer with a Special Reference to Activator Protein-2, HER2, and Prognosis. Clin Cancer Res. 2004;10:7621–8. doi: 10.1158/1078-0432.CCR-04-1061. [DOI] [PubMed] [Google Scholar]
- 15.Fujiwara A, Shibata E, Terashima H, et al. Evaluation of matrix metalloproteinase-2 (MMP-2) activity with film in situ zymography for improved cytological diagnosis of breast tumors. Breast Cancer. 2006;13:272–8. doi: 10.2325/jbcs.13.272. [DOI] [PubMed] [Google Scholar]
- 16.Garbett EA, Reed MW, Stephenson TJ, Brown NJ. Proteolysis in human breast cancer. Mol Pathol. 2000;53:99–106. doi: 10.1136/mp.53.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanemaaijer R, Verheijen JH, Maguire TM, et al. Increased gelatinase-A and gelatinase-B activities in malignant vs. benign breast tumors. Int J Cancer. 2000;86:204–7. doi: 10.1002/(sici)1097-0215(20000415)86:2<204::aid-ijc9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 18.LaRocca G, Pucci-Minafra I, Marrazzo A, Taormina P, Minafra S. Zymographic detection and clinical correlations of MMP-2 and MMP-9 in breast cancer sera. Br J Cancer. 2004;90:1414–21. doi: 10.1038/sj.bjc.6601725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price SJ, Greaves DR, Watkins H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J Biol Chem. 2001;276:7549–58. doi: 10.1074/jbc.M010242200. [DOI] [PubMed] [Google Scholar]
- 20.Vasku V, Vasku A, Tschoplova S, Izakovicova HL, Semradova V, Vacha J. Genotype association of C(−735)T polymorphism in matrix metalloproteinase 2 gene with G(8002)A endothelin 1 gene with plaque psoriasis. Dermatology. 2002;204:262–5. doi: 10.1159/000063355. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Zhou Y, Miao X, Xiong P, Tan W, Lin D. Functional Haplotypes in the Promoter of Matrix Metalloproteinase-2 Predict Risk of the Occurrence and Metastasis of Esophageal Cancer. Cancer Res. 2004;64:7622–8. doi: 10.1158/0008-5472.CAN-04-1521. [DOI] [PubMed] [Google Scholar]
- 22.Roehe AV, Frazzon AP, Agnes G, Damin AP, Hartman AA, Graudenz MS. Detection of polymorphisms in the promoters of matrix metalloproteinases 2 and 9 genes in breast cancer in South Brazil: preliminary results. Breast Cancer Res Treat. 2007;102:123–4. doi: 10.1007/s10549-006-9273-1. [DOI] [PubMed] [Google Scholar]
- 23.Delgado-Enciso I, Cepeda-Lopez FR, Monrroy-Guizar EA, et al. Matrix metalloproteinase-2 promoter polymorphism is associated with breast cancer in a Mexican population. Gynecol Obstet Invest. 2008;65:68–72. doi: 10.1159/000108282. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Yu C, Miao X, et al. Substantial reduction in risk of breast cancer associated with genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes. Carcinogenesis. 2004;25:399–404. doi: 10.1093/carcin/bgh020. [DOI] [PubMed] [Google Scholar]
- 25.Lei H, Hemminki K, Altieri A, et al. Promoter polymorphisms in matrix metalloproteinases and their inhibitors: few associations with breast cancer susceptibility and progression. Breast Cancer Res Treat. 2007;103:61–9. doi: 10.1007/s10549-006-9345-2. [DOI] [PubMed] [Google Scholar]
- 26.Gao YT, Shu XO, Dai Q, et al. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Beeghly-Fadiel A, Long JR, Gao YT, et al. Common MMP-7 Polymorphisms and Breast Cancer Susceptibility: A Multistage Study of Association and Functionality. Cancer Res. 2008;68:6453–9. doi: 10.1158/0008-5472.CAN-08-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye C, Dai Q, Lu W, et al. Two-stage case-control study of common ATM gene variants in relation to breast cancer risk. Breast Cancer Res Treat. 2007;106:121–6. doi: 10.1007/s10549-006-9473-8. [DOI] [PubMed] [Google Scholar]
- 29.The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 30.de Bakker PIW, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardenbol P, Yu F, Belmont J, et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–75. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang G, Gao YT, Cai QY, Shu XO, Cheng JR, Zheng W. Modifying effects of sulfotransferase 1A1 gene polymorphism on the association of breast cancer risk with body mass index or endogenous steroid hormones. Breast Cancer Res Treat. 2005;94:63–70. doi: 10.1007/s10549-005-7280-2. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29:299–312. doi: 10.1002/gepi.20098. [DOI] [PubMed] [Google Scholar]
- 35.Levi E, Fridman R, Miao HQ, Ma YS, Yayon A, Vlodavsky I. Matrix metalloproteinase 2 releases active soluble ectodomain of fibroblast growth factor receptor 1. PNAS USA. 1996;93:7069–74. doi: 10.1073/pnas.93.14.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schonbeck U, Mach F, Libby P. Generation of Biologically Active IL-1{beta} by Matrix Metalloproteinases: A Novel Caspase-1-Independent Pathway of IL-1{beta} Processing. J Immunol. 1998;161:3340–6. [PubMed] [Google Scholar]
- 37.Imai K, Hiramatsu A, Fukushima D, Pierschbacher MD, Okada Y. Degradation of decorin by matrix metalloproteinases: identification of the cleavage sites, kinetic analyses and transforming growth factor-beta1 release. Biochem J. 1997;322( Pt 3):809–14. doi: 10.1042/bj3220809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin H, Sun Y, Benveniste EN. The Transcription Factors Sp1, Sp3, and AP-2 Are Required for Constitutive Matrix Metalloproteinase-2 Gene Expression in Astroglioma Cells. J Biol Chem. 1999;274:29130–7. doi: 10.1074/jbc.274.41.29130. [DOI] [PubMed] [Google Scholar]
- 39.Kim ES, Sohn YW, Moon A. TGF-[beta]-induced transcriptional activation of MMP-2 is mediated by activating transcription factor (ATF)2 in human breast epithelial cells. Cancer Letters. 2007;252:147–56. doi: 10.1016/j.canlet.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Song H, Ki SH, Kim SG, Moon A. Activating Transcription Factor 2 Mediates Matrix Metalloproteinase-2 Transcriptional Activation Induced by p38 in Breast Epithelial Cells. Cancer Res. 2006;66:10487–96. doi: 10.1158/0008-5472.CAN-06-1461. [DOI] [PubMed] [Google Scholar]