Abstract
The developing embryos of the South African (Xenopus laevis) and Western (Xenopus tropicalis) clawed frogs provide an experimentally tractable and easily visualized model for vertebrate cardiovascular development. Most of the genes used to execute the cardiac developmental program are the same in frogs and humans. Experiments using Xenopus provide an underutilized but valuable complement to studies on the molecular, cellular, physiological and morphological consequences of genetic and environmental influences on cardiac disease.
Introduction
Nearly one out of every hundred children is born with congenital heart defects caused by either genetic or environmental influences on cardiovascular development. More often than not, the causal incidents are difficult or impossible to observe during fetal development. The relatively recent ability to identify the genes associated with a variety of congenital heart defects provides predictive power. However, identification of the gene is only one step in a careful analysis of the misregulated molecular and cellular features of heart development. The widespread adaptation of mouse transgenic models for congenital disease has provided a great deal of information on heart development, but other animals may be equally valuable for the investigation of particular questions. Fruit flies, zebrafish, chick, quail and frogs have all contributed to our understanding of heart development. In this short review, we'll highlight some of the opportunities for asking experimental questions on heart development well suited to Xenopus as a model system. Rather than list all of the excellent work being done in the field we have chosen examples that we feel highlight the techniques that make Xenopus an outstanding model system. A broader list resources available for studies using Xenopus can be found at Xenbase on the world wide web[1].
The advantages of using Xenopus embryos to study heart development center around five principle attributes. First, it is easy to obtain and raise large numbers of embryos from a single female via in vitro fertilization. Thus, the experimenter has control over the time of fertilization and the synchronous development of hundreds or even a thousand siblings. The development of the frog embryos can occur in a simple salt solution (essentially artificial pond water) at temperatures between 16-22 degrees. This particular feature of development allows a relatively straightforward system for studies on environmental influences on cardiac development. Second, the early embryos are large; a fertilized egg is approximately 1mm in diameter making microinjection very simple. As we'll see, microinjection is the basis for mRNA reduction, introduction of mutant forms of protein via mRNA, and transgenesis. Third, the embryonic cells that give rise to the heart are well mapped making both targeted injection meaningful and experimental manipulation of a part of the embryo (often called animal cap assays) possible. Fourth, the free living embryos are accessible for physiologic examination and are small enough through early development for whole mount analysis, leaving the whole embryo intact as developmental changes are examined. And fifth, the early embryos are capable of relatively normal development in the absence of a working heart, allowing even ultimately lethal scenarios to be examined for more specific information on early disruption of cardiovascular pathways.
Introduction to spatial and temporal organization of cardiac development
Unlike their mammalian counterparts, Xenopus embryos need not dedicate cells for placental interactions. Even prior to fertilization, under normal conditions, different areas of the egg predictably give rise to ectoderm, mesoderm or endoderm. Mesoderm formation results from inductive signals generated by endodermal cells. Early studies using vital dyes identified the cells of the early embryo whose descendants normally contribute to the heart. More detailed studies made clear regional control of cardiac fields[2]. Like mammals, the cardiac fields are bilateral, coming principally from lateral dorsal mesoderm.
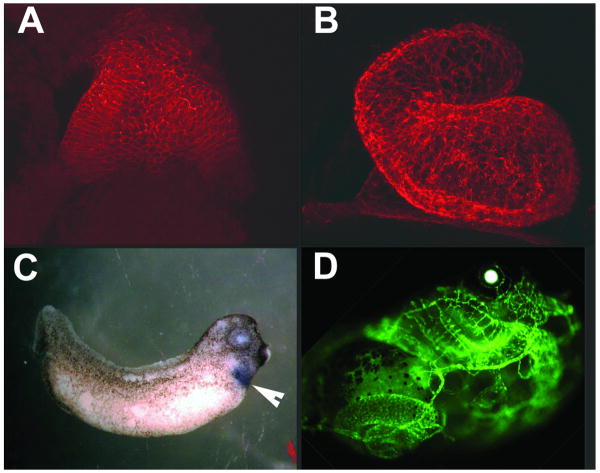
In Xenopus the organization of the embryonic fields, up until the 4000 cell stage, occurs with minimal zygotic transcription. Cell division is rapid and synchronous, with the first 12 cell cycles taking about 30 minutes at room temperature. After the first 12 divisions, the cell cycles slow down, become asynchronous and region specific activation of gene transcriptions starts[3]. About 9 hours after fertilization, at the earliest stages of gastrulation, the precardiac fields are forming. Over the next ten hours the cardiac primordia are found in the lateral plate mesoderm and move to the ventral midline. The fusion of the right and left cardiac primordia at the midline is followed by formation of the heart tube. The atrial region is initially caudal to the ventricle but repositions to a posterior and cranial position during looping. Cardiac development continues with valve formation, trabeculation, septation and establishment of the conduction system. A functioning three-chambered heart is well established in 4 or 5 days[4]. Confocal images of the heart 32 hours (stage 28), 46 hours (stage 35) and 106 hours (stage 46) hours after development are shown in figure 1A and B and figure 2B.
Figure 1.
Morphological and spatial analysis of cardiovascular development. Panels A and B show whole mount confocal images of the Xenopus heart at stage 28 and 35. Heart tissue was detected by an anti-tropomyosin antibody. Panel C shows the in situ hybridization detection of Nkx2-5 mRNA. Cardiac regions (indicated by the arrowhead) expressing Nkx2-5 appear purple. Panel D shows an embryo transgenic for the flk promoter driving a GFP reporter. This transgenic embryo expresses GFP in the vascular system and was used to delineate the important regulatory elements of the flk promoter.
Figure 2.
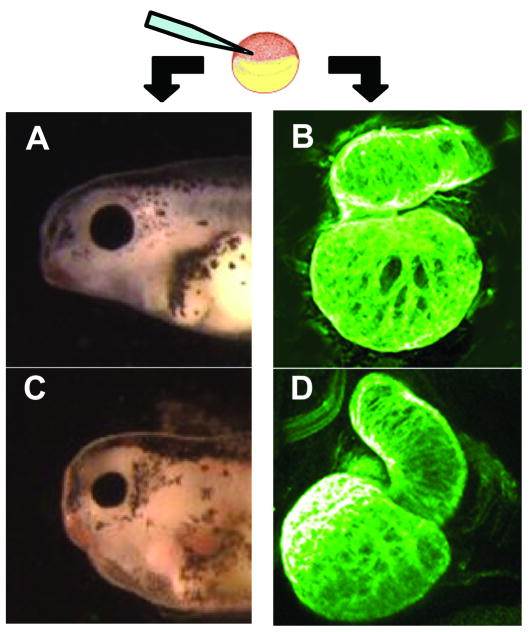
Monitoring disruption of normal gene expression in Xenopus embryos. Microinjection of antisense oligonucleotides, mutant forms of cardiac genes or exposure to environmental toxins can lead to heart abnormalities. In panel A is a lateral view the normal head and cardiac region of a stage 42 embryo. In panel B is a whole mount confocal image of a normal stage 46 heart. In panel C is a stage 42 embryo displaying the ventral edema characteristic of many treatments that cause heart defects. In panel D is an abnormally looped stage 46 heart of an embryo that was injected with an antisense oligonucleotide targeting the PitX2c mRNA. The heart in figure D shows how loss of PitX2c disrupts normal right/left patterning.
Using Xenopus to understand a cardiac gene of interest. There are several basic questions that arise when a gene is implicated in disrupted development. These include: 1) Is the gene expressed in a spatial and temporal manner consistent with the function hypothesized? 2) Does the loss of the gene produce the disruption? 3) Does overexpression or misexpression have a phenotype? 4) Do mutations in the gene lead to defective development? If it does, what is the mechanism and can a correction be made?
Question 1 can be answered using whole mount in situ hybridization or immunohistochemistry. For example, Pollet et al. have published an atlas of differential gene expression that includes genes expressed in the heart[5]. Their study and others serve as the starting point for a more complete compilation for gene expression in embryos which is a work in progress. In situ hybridization showing the spatial expression of the Nkx2-5 gene is shown in figure 1C. The harvest and analysis of essentially any stage of embryonic development[4] is straightforward. More structural information can be addressed by either whole mount immunohistochemistry using confocal microscopy[6] or more traditional immunostaining and sectioning[7].
An alternative take on question 1 would ask about the minimal promoter requirements for cardiovascular gene expression. This has been approached by generating transgenic embryos with reporter constructs[8-10]. Although there are now several options for making transgenic embryos[11-14], most promoter analysis has been carried out by a variation of the restriction enzyme mediated insertion (REMI) of linear plasmid into permeabilized sperm nuclei followed by injection of the sperm nuclei into unfertilized eggs[15]. For example, Sparrow et al. identified the minimal elements for cardiac expression of the Nkx2-5 promoter[9] and the Krieg lab has mapped the regions atrial natriuretic factor promoter responsible for regulated expression[8]. In both cases the analysis was carried out in the Fo generation. In figure 1D is an example of establishing the control regions of the flk promoter using transgenesis. When all the appropriate regulatory regions of the flk promoter are present the flk:GFP reporter construct expresses in and marks the vascular system of a transgenic tadpole (provided by P.A. Krieg, unpublished). Laboratories practiced in generating transgenic embryos can test the faithful temporal and spatial expression of a promoter construct extremely rapidly. In addition there are multiple examples of promoters from mammalian models being appropriately regulated in Xenopus.
In model systems like Drosophila with rapid generation times questions 2-4 are often approached by making mutants and analyzing generations of progeny. Xenopus based approaches more commonly use direct analysis of embryos that are experimentally altered through microinjection. Embryos with compromised heart function can often be visually identified by a ventral edema that worsens during development (figure 2A and C). The loss of function or developmental null question can be addressed through the injection of antisense oligonucleotides that result in near complete loss of the specific expression of a protein[16]. For example, Brown et al.[17] used antisense oligonucleotides to reduce the levels of the transcription factors Tbx5 (the causal gene of Holt-Oram syndrome) and Tbx20 (a causative agent of diverse anatomic defects including atrial septal and mitral valve disease). In their study, they found that reduction of either lead to morphological defects but did not affect either specification or migration of cardiac precursors. Two other examples of this approach include studies on the establishment of the right/left program by Dagle et al.[18] and on the role of transcription factor Nkx2-6 (mutations in this gene are implicated in persistent truncus arteriosis[19]) by Allen et al.[20]. Dagle et al. found that the loss of PitX2c (Reigers syndrome gene and involved in early laterality decisions) results in not just reversal of heart laterality but a general inability to establish normal right left positional information in the heart. An example of a heart from a tadpole that has been depleted of Pitx2c is shown compared to a normal heart in figure 2D and B. Allen et al. showed that reduction of the Xenopus homologue of Nkx2-6 (in Xenopus called Nkx2-10) diminishes heart size and causes outflow tract defects. These findings are especially interesting because loss of Nkx2-6 in the mouse apparently had no cardiac defect[19].
One of the most dramatic examples of mislocalization and overexpression studies on cardiac genes comes from a study done by Cleaver et al.[21] on Nkx2-5. In humans mutations in Nkx2-5 cause a spectrum of heart defects. Overexpression of Nkx2-5, accomplished by injection of mRNA during development, expands the number of cells in the heart field transiently giving rise to an enlarged heart. Restricting injected Nkx2-5 mRNA to regions of the embryo outside the area that normally gives rise to the heart had no noticeable consequence. These experiments established the very specific action of Nkx2-5 on the heart field.
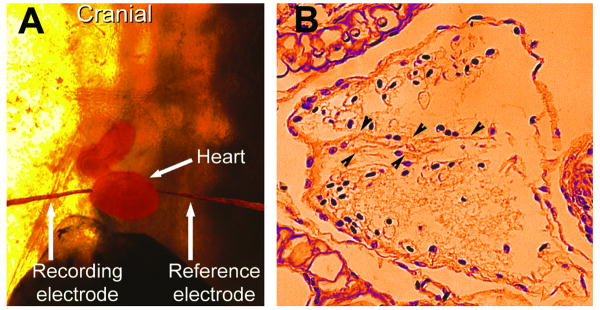
Similar progress has been made using both synthetic mutations and testing naturally occurring mutations for the effects on early development. Synthetic constructs that fused the engrailed repressor to Nkx2-5[22] resulted in embryos with very small, malformed hearts. Injections of Nkx2-5 transcripts based on some of the early mutations found in human phenocopied not only defects in atrial septal and atrioventricular valve formation but also established that Xenopus tadpoles can be used to evaluate mutations that lead to conduction system anomalies[23,24]. Direct measurement of a tadpole electrocardiogram and histology showing a defective atrial septal development are shown in figure 3. One advantage utilized in these studies was the opportunity to analyze many embryos. Thus, even subtle or low penetrance phenotypes can be usefully analyzed in Xenopus embryos.
Figure 3.
How mutations that cause human mutations affect Xenopus heart development. Mutations in the Nkx2-5 gene that lead to conduction defects and atrial septal defects in human also cause those defects in Xenopus embryos. Panel A shows how recording and reference electrodes were placed in live embryos that had been injected with mRNA encoding mutant forms of Nkx2-5 to discover if treated embryos developed conduction defects. In Panel B, a cross section of a heart with an abnormal atrial septum in an embryo that had expressed the Nkx2-5. Arrowheads point at the septum.
Environmental influences on heart development
Model systems like Xenopus offer an opportunity to carry out large scale screening for defects caused by chemical agents. Tomlinson et al.[25] described a number of small molecule screens that resulted in ventral edema typical of interruption of normal cardiac function. Another specific example was reported by Chen et al. who in the course of examining inhibitors of mitochondrial electron transport chain I with rotenone uncovered a specific effect on heart development that tracks back to an inhibition of Nkx2-5 expression[26]. Controlled exposure of Xenopus embryos to suspected toxins have tremendous potential for unraveling environmental risk factors for congenital heart defects. Controlled exposure of Xenopus embryos manipulated in a manner to generate heart defects may also prove valuable in screening for drugs that suppress the defects. For example, the rotenone related defects in heart development center around a failure to properly regulate calcineurin activity. Calcineurin, which has also been implicated in cardiac hypertrophic growth, could be targeted in a screen for activating compounds. Normal heart development in the presence of this environmental toxin would be a reflection of successful intervention.
Genetics Genomics and in silico experiments with Xenopus
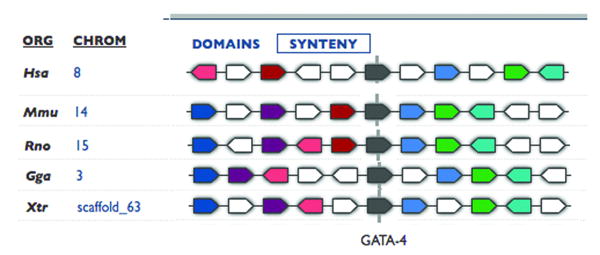
The large (3 × 109 base pairs), pseudo tetraploid composition of the genome of Xenopus laevis and the relatively long time to sexual maturity (under optimal conditions 6 months[4] but often longer) make genetic studies cumbersome and has played a role in the inability to secure a commitment to fully sequence the Xenopus laevis genome. A practical compromise has been a program to sequence the closely related, diploid, and faster developing species, Xenopus tropicalis. So far promoter regulation and expression analysis of mRNA indicate a remarkable level of functional conservation between these species. The partially completed sequence of the Xenopus tropicalis genome allows a comparison of gene sequence conservation between Xenopus and other organisms but also the syntenic analysis of gene organization. Shown is figure 4 is the syntenic comparison of the GATA4 gene in frog mouse and human. The inescapable conclusion is that not only is the GATA4 gene sequence conserved between species, so is its position among other genes on the chromosome. This type of analysis may help indicate potential homologs even when there is less conservation in nucleotide sequence.
Figure 4.
Syntenic analysis of the GATA4 gene. Sufficient genomic sequence is available to compare the sequence and the genomic positioning of genes across species, including genes in Xenopus. Syntenic analysis of the GATA 4 gene (in grey) from Human (Hsa), mouse (Mmu), rat (Rno), chicken (Gga) and frog (Xtr). Block arrows of the same color indicate the same gene. This type of analysis indicates the conservation of genomic positioning of genes between species. This figure was adapted from and analysis using the Metamorph program sponsored by the Joint Genomics Institute.
In silico experiments with Xenopus take advantage of microarrays that allow a global analysis of expression at different stages of development or embryos that have been experimentally manipulated. Completion and annotation of the Xenopus tropicalis genomic sequence will provide new opportunities for analysis of epigenetic changes that affect development. The advantages of using Xenopus in studies relevant to cardiac and other human disorders should stimulate the funding and effort required to bring sequence databases featuring Xenopus to the levels seen with other model systems.
Model Comparison
The conservation of regulatory and structural genes among all organisms that form hearts provides a rich resource for understanding heart disease. Xenopus embryos group together with Drosophila and zebrafish as organisms where experimenters have easy and direct access to embryos starting with a fertilized egg. Of these three Xenopus is the least well developed for genetic studies, but may be the simplest to use for injection based experiments. Recently a growing interest in developing Xenopus tropicalis as a tractable genetic system and the establishment of transgenic stock centers promise to open new opportunities using Xenopus. Xenopus forms a three chambered heart in contrast to the single chamber of the fly or the two chambers of the fish. This allows questions about complex looping, septation and valve development to be addressed. In comparison to chicken, mouse or rat models for heart disease Xenopus does not form two ventricles. The considerably larger size of the chick embryonic heart makes it attractive for use in explant based studies. The remarkable investment in developing the transgenic mouse model dwarfs the investment made in all other models combined. The tools that have been developed, and the mammalian status it shares with human is a good justification for taking advantage of this model. However, in experiments that feature simplest direct manipulation or require survivorship of embryos with serious cardiac defects it may be far easier to use Xenopus embryos.
Model Translation to Humans
Experiments using Xenopus can serve to identify how cardiac development responds in the absence of genes identified a candidates for human heart defects. Queries can also be made into how embryos respond early in development to the presence of different mutations in those genes. Xenopus embryos can play an important role in understanding the molecular pathways that are involved, perhaps suggesting interventional therapies. Xenopus embryos are well suited to better understand the connections between environmental and genetic causes of heart defects.
Conclusions
Xenopus embryos have historically proven to be a hardy and versatile model for animal development. It provides a relatively inexpensive, abundant, and experimentally malleable organism for studies on heart development and defects. Xenopus can provide a direct approach to answering questions on gene function as well as complement and promote the more thoughtful design of experiments in other systems.
Table 1.
Exploring the Impact of a Gene on Heart Development in Xenopus
| Spatiotemporal pattern of expression | Gene loss | Addition of a gene product | |
|---|---|---|---|
| Pros | Comparative analysis of sequential developmental stages | Antisense oligonucleotide mediated reduction of gene expression is simple and reversible using microinjection. | Simple injection protocol for normal or mutant mRNA injections. |
| Cons | Xenopus laevis genome is not sequenced (but coding regions are very similar to those sequenced only in Xenopus tropicalis). | Efficacy of antisense compounds is temporally limited. Antisense compounds will be inherited in cells outside the heart field. | Injected mRNA has a restricted half life. Timing and spatial expression of protein from mRNA will likely be broader than endogenous mRNA. |
| Best use of model | Understanding developmental appearance of mRNA in the embryo and comparison with other mRNAs and morphological features. Defining regulatory regions of promoters by transgenesis. | Testing loss of expression of genes critical for cardiac development through tube closure. | Testing gene misexpression or the effects of mutant protein on early heart development. |
Acknowledgments
The authors acknowledge and thank former lab members Sandra Kolker, Bryan Allen, Abi Struck-Marcell, John Dagle and Ben Sutti for some or the figures used in this review. We thank Paul Krieg (Univ. of Arizona) for allowing the use of his unpublished transgenic analysis of the flk promoter. We also thank Elesa Wedemeyer for editorial comments. We acknowledge support from the American Heart Association (HLB) and the National Institutes of Health (DLW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Heather L. Bartlett, Department of Pediatrics, Roy J. and Lucille Carver College of Medicine, University of Iowa
Daniel L. Weeks, Department of Biochemistry, Roy J. and Lucille Carver College of Medicine, University of Iowa
References
- 1.Bowes JB, et al. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36(Database):D761–767. doi: 10.1093/nar/gkm826. LInk at: http://www.xenbase.org/common/ [DOI] [PMC free article] [PubMed]
- 2.Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol. 1987;119(2):560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- 3.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North Holland publishing Company; 1967. [Google Scholar]
- 5.Pollet N, et al. An atlas of differential gene expression during early Xenopus embryogenesis. Mech Dev. 2005;122(3):365–439. doi: 10.1016/j.mod.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Kolker SJ, et al. Confocal imaging of early heart development in Xenopus laevis. Dev Biol. 2000;218(1):64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohun TJ, et al. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218(1):74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- 8.Small EM, Krieg PA. Transgenic analysis of the atrial natriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261(1):116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- 9.Sparrow DB, et al. Regulation of the tinman homologues in Xenopus embryos. Dev Biol. 2000;227(1):65–79. doi: 10.1006/dbio.2000.9891. [DOI] [PubMed] [Google Scholar]
- 10.Latinkic BV, et al. Distinct enhancers regulate skeletal and cardiac muscle-specific expression programs of the cardiac alpha-actin gene in Xenopus embryos. Dev Biol. 2002;245(1):57–70. doi: 10.1006/dbio.2002.0639. [DOI] [PubMed] [Google Scholar]
- 11.Ogino H, et al. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123(2):103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Pan FC, et al. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn. 2006;235(1):247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- 13.Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2(12):975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesneau A, et al. Transgenesis procedures in Xenopus. Biol Cell. 2008;100(9):503–521. doi: 10.1042/BC20070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122(10):3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 16.Dagle JM, Weeks DL. Oligonucleotide-based strategies to reduce gene expression. Differentiation. 2001;69(23):75–82. doi: 10.1046/j.1432-0436.2001.690201.x. [DOI] [PubMed] [Google Scholar]
- 17.Brown DD, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132(3):553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagle JM, et al. Pitx2c attenuation results in cardiac defects and abnormalities of intestinal orientation in developing Xenopus laevis. Dev Biol. 2003;262(2):268–281. doi: 10.1016/s0012-1606(03)00389-0. [DOI] [PubMed] [Google Scholar]
- 19.Heathcote K, et al. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet. 2005;14(5):585–593. doi: 10.1093/hmg/ddi055. [DOI] [PubMed] [Google Scholar]
- 20.Allen BG, et al. Reduction of XNkx2-10 expression leads to anterior defects and malformation of the embryonic heart. Mech Dev. 2006;123(10):719–729. doi: 10.1016/j.mod.2006.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cleaver OB, et al. Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development. 1996;122(11):3549–3556. doi: 10.1242/dev.122.11.3549. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, et al. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125(22):4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 23.Bartlett HL, et al. Characterization of embryonic cardiac pacemaker and atrioventricular conduction physiology in Xenopus laevis using noninvasive imaging. Am J Physiol Heart Circ Physiol. 2004;286(6):H2035–2041. doi: 10.1152/ajpheart.00807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartlett HL, et al. Transient early embryonic expression of Nkx2-5 mutations linked to congenital heart defects in human causes heart defects in Xenopus laevis. Dev Dyn. 2007;236(9):2475–2484. doi: 10.1002/dvdy.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomlinson ML, et al. Xenopus as a model organism in developmental chemical genetic screens. Mol Biosyst. 2005;1(3):223–228. doi: 10.1039/b506103b. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, et al. The mitochondrial respiratory chain controls intracellular calcium signaling and NFAT activity essential for heart formation in Xenopus laevis. Mol Cell Biol. 2007;27(18):6420–6432. doi: 10.1128/MCB.01946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]