Abstract
S-adenosylmethionine (AdoMet) is a methyl donor used by a wide variety of methyltransferases, and it is also used as the source of an α-amino-α-carboxypropyl (“acp”) group by several enzymes. tRNA-yW synthesizing enzyme-2 (TYW2) is involved in the biogenesis of a hypermodified nucleotide, wybutosine (yW), and it catalyzes the transfer of the “acp” group from AdoMet to the C7 position of the imG-14 base, a yW precursor. This modified nucleoside yW is exclusively located at position 37 of eukaryotic tRNAPhe, and it ensures the anticodon-codon pairing on the ribosomal decoding site. Although this “acp” group has a significant role in preventing decoding frame shifts, the mechanism of the “acp” group transfer by TYW2 remains unresolved. Here we report the crystal structures and functional analyses of two archaeal homologs of TYW2 from Pyrococcus horikoshii and Methanococcus jannaschii. The in vitro mass spectrometric and radioisotope-labeling analyses confirmed that these archaeal TYW2 homologues have the same activity as yeast TYW2. The crystal structures verified that the archaeal TYW2 contains a canonical class-I methyltransferase (MTase) fold. However, their AdoMet-bound structures revealed distinctive AdoMet-binding modes, in which the “acp” group, instead of the methyl group, of AdoMet is directed to the substrate binding pocket. Our findings, which were confirmed by extensive mutagenesis studies, explain why TYW2 transfers the “acp” group, and not the methyl group, from AdoMet to the nucleobase.
Keywords: modification, S-adenosylmethionine, X-ray crystallography
Post-transcriptional modifications of RNA molecules have significant biological roles in maintaining life. More than 100 different modifications exist throughout the three phylogenic domains (1). Most of these modifications are found in transfer RNA (tRNA), and they confer various chemical properties to the RNA residues. Especially, the anticodon loop of tRNA contains many modified nucleosides, which are considered to facilitate the correct codon-anticodon pairing on the ribosome (2).
Wybutosine (yW) is a hypermodified nucleoside located at position 37, adjacent to the 3′ position of the anticodon, of eukaryotic tRNAPhe. The chemical structure of yW is characterized by the tricyclic 1H-imidazo[1,2-α]purine core with a large side chain (3) (Fig. 1A). Its bulky, hydrophobic structure is considered to stabilize not only the conformation of the anticodon loop itself but also the codon-anticodon pairing on the ribosome, thereby preventing a potential −1 frame shift at a phenylalanine codon (4).
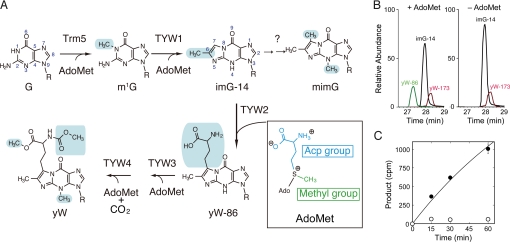
Fig. 1.
Acp-group transfer catalyzed by PhTYW2. (A) Biosynthetic pathway of wybutosine. The chemical structures of the wyosine derivatives are shown. Modification enzymes, Trm5 and TYW1–4, are involved in each modification step. The chemical groups included in each reaction are shadowed in blue. The chemical structure of AdoMet is shown in the box. The methyl and acp groups are colored green and blue, respectively. (B) In vitro reconstitution of yW-86 synthesis using recombinant PhTYW2. RNaseT1-digested yeast tRNAPhe with the imG-14 intermediate (black), obtained from the ΔTYW2 strain, was reacted with PhTYW2 in the presence (left panel) or absence (right panel) of AdoMet and was analyzed by LC/MS. Mass chromatographs for anticodon-containing fragments including imG-14 (m/z 661, black), yW-86 (m/z 678, green), and yW-173 (m/z 664, red) are overlaid. (C) Kinetic analysis of the acp-group transfer by PhTYW2. The α-carboxyl-14C labeled AdoMet and the imG-14 containing tRNAPhe were incubated at 50 °C with recombinant PhTYW2 (closed circles) and without PhTYW2 (open circles), and the incorporation of α-carboxyl-14C into the tRNAPhe was quantified by scintillation counting. Data are represented as mean ± SE (n = 3).
Recent studies revealed the biosynthetic pathway of yW in yeast, in which five enzymes catalyze successive S-adenosylmethionine (AdoMet)-dependent reactions to form yW from the G37 residue (5, 6) (Fig. 1A). In the first step, the AdoMet-dependent tRNA methyltransferase (MTase) Trm5 forms N1-methylated G37 (m1G) (6, 7). In the next step, tRNA-yW synthesizing enzyme-1 (TYW1) catalyzes the radical reaction, involving two [4Fe-4S] clusters, to form the tricyclic ring structure (4-demethylwyosine, imG-14), using an adenosyl radical derived from AdoMet (Fig. 1A) (8). Furthermore, three enzymes, TYW2, 3, and 4, catalyze AdoMet-dependent modifications to form the yW nucleoside. In this pathway, TYW2 catalyzes the transfer of the α-amino-α-carboxypropyl (acp) group, instead of the methyl group, of AdoMet to the C7 position of the tricyclic core structure of imG-14, forming yW-86 (yW minus 86 Da) (Fig. 1A) (5). A recent study indicated that this acp group of the yW base has a significant negative impact on −1 frame shifting at a phenylalanine codon (9), suggesting the biological importance of TYW2 for the maintenance of translational fidelity. In addition, the enzymes that transfer the acp group of AdoMet to substrates are also present in the biosynthetic pathways for diphthamide (10), nocardicin (11), and diacylglyceryl-O-4′-(N,N,N,-trimethyl) homoserine (12). However, the detailed mechanism of the acp group transfer by these enzymes remains elusive.
Previous studies demonstrated the existence of wyosine (Y) derivatives in several archaea (13–15). Moreover, homologous genes to eukaryotic TYW1 and TYW2 exist in archaeal genomes, suggesting that these Y derivatives are formed by a similar modification pathway to that in eukaryotes. Therefore, these Y derivatives may play a similar role in maintaining the translational fidelity of the phenylalanine codon, as in eukaryotes (15). However, there is no experimental evidence showing that the archaeal Y derivatives are located in the tRNAPhe anticodon loop. In addition, previous studies reported the archaea-specific Y derivative, 7-methylwyosine (mimG, Fig. 1A), in which the C7 position is modified by the methyl group, rather than the acp group (13). Thus, the in vivo functions of these archaeal TYW2 homologues remain elusive.
Here, we report the functional characterizations and structural analyses of two archaeal TYW2 enzymes from Pyrococcus horikoshii and Methanococcus jannaschii. An in vitro reconstitution assay by mass spectrometry and kinetic analyses with radioisotope-labeled AdoMet confirmed that the archaeal TYW2 enzymes catalyze the acp group transfer reaction in an AdoMet-dependent manner. The crystal structures of the archaeal TYW2s, in conjunction with the extensive mutational analyses, provide the structural basis for the transfer of the acp group of AdoMet to the substrate.
Results and Discussion
Acp-Group Transfer Activity of Archaeal TYW2.
The P. horikoshii PH0793 (278 amino acids) and M. jannashii MJ1557 (249 amino acids) proteins share 22% and 25% sequence identities to yeast TYW2, respectively. To determine whether these archaeal TYW2 homologs possess an activity to transfer the acp group to imG-14 at position 37 of tRNAPhe, we performed in vitro reconstitution assays using these recombinant proteins (Fig. 1B and Fig. S1). As a substrate, we used tRNAPhe purified from the yeast ΔTYW2 strain, which contains imG-14 at position 37 (Fig. S1A). In the reconstitution assay with the PH0793 and MJ1557 proteins, a mass spectrometric analysis detected AdoMet-dependent products corresponding to yW-86 (Fig. 1B and Fig. S1 B and D). We confirmed our assignment of the peaks by the positive control experiment with purified yeast TYW2 (Fig. S1C). To further confirm the activity of the archaeal TYW2 homologs, we assayed the acp-group transfer by monitoring the incorporation of the acp group from α-carboxyl-14C labeled AdoMet into the imG-14 containing tRNAPhe (Fig. 1C and Fig. S1E). The results consistently indicated that these archaeal enzymes have an activity to incorporate the radio-labeled acp group into tRNA. Therefore, these data strongly indicate that the PH0793 and MJ1557 proteins transfer the acp group of AdoMet to imG-14, to form yW-86 at position 37 of tRNAPhe.
In the reconstitution assays using mass spectrometry, a considerable amount of imG-14 remained with either the PH0793 or MJ1557 protein, while imG-14 was completely converted to yW-86 by yeast TYW2. Consistently, the acp-transfer activity of PhTYW2 measured with the radio-labeled substrate was lower than that of yeast TYW2 (Fig. S1E). This is probably due to their low catalytic efficiency under the experimental conditions, in which the substrate was a heterologous yeast tRNA and the reaction temperature was 50 °C. In addition, in the mass-spectrometric analyses, a trace amount of yW-173 (MW 335) was detected in all products as well as in the substrate, regardless of the presence of the enzyme or AdoMet (Fig. 1B and Fig. S1). This minor peak may stem from a methylated product of imG-14 at position N4, formed by endogenous TYW3 in yeast cells. Altogether, our results indicate that the PH0793 and MJ1557 proteins have an activity to form yW-86 from imG-14 at position 37 of tRNAPhe. Therefore, we hereafter refer to PH0793 and MJ1557 as PhTYW2 and MjTYW2, respectively.
Structure Determination.
To understand the mechanism of the modification by TYW2, we determined the crystal structures of PhTYW2 in the AdoMet-bound and 5′-deoxy-5′-methylthioadenosine (MeSAdo)-bound forms, and MjTYW2 in the AdoMet-bound form (Fig. 2). The initial phases were obtained by the molecular replacement method, using the apo-form PhTYW2 structure as a search model (PDB ID: 2FRN), which was previously determined by the structural genomics group as a function-unidentified protein. The AdoMet- and MeSAdo-bound forms of PhTYW2 were determined at 2.3-Å and 2.5-Å resolutions, respectively (Table S1). The crystals contain one PhTYW2 monomer with one AdoMet or MeSAdo molecule per asymmetric unit, and belong to the space group R3, which is different from the space group (P21) of the previous PhTYW2 structure (PDB ID: 2FRN). Consequently, the crystal packing of our structures differs from that of the previous structure, which resulted in the lower B factor for the N-terminal domain in our structures. Therefore, we could model the N-terminal domain including residues 6–18, which were disordered in the previous structure. The final models of the PhTYW2 structures contain residues 6–69 and 79–278.
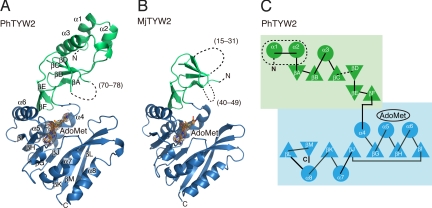
Fig. 2.
Overall structure of archaeal TYW2. (A) Crystal structure of AdoMet-bound PhTYW2. (B) Crystal structure of AdoMet-bound MjTYW2. In (A and B), the N-terminal and C-terminal domains are colored green and blue, respectively. AdoMet is shown in a stick model. The disordered regions are depicted by dashed lines. 2Fo−Fc annealed omit maps around AdoMet contoured at 1.0 σ are shown. (C) Topology diagram of the PhTYW2 structure with the same color code as in (A). The PhTYW2-specific extension region is enclosed by a dashed circle, and the disordered region is indicated by dashed lines. The AdoMet-binding site is indicated with a circle.
The AdoMet-bound form of MjTYW2 was determined at 2.0-Å resolution (Fig. 2B and Table S1). The crystal contains one MjTYW2 monomer per asymmetric unit, and belongs to the space group P212121. The final model of the MjTYW2 structure contains residues 4–14, 32–39, and 50–249.
Overall Structure of Archaeal TYW2.
The overall structure of PhTYW2 can be divided into an N-terminal domain and a C-terminal MTase domain. The N-terminal domain (residues 6–106) consists of six β-strands (βA–F) and three α-helices (α1–3) (Fig. 2 A and C). A three-stranded anti-parallel β-sheet (βA–C) forms the core of the N-terminal domain, while strands βE and βF form a β-hairpin structure. The loop connecting strands βC and βD is partially disordered (residues 70–78). The MTase domain (residues 107–278) consists of a seven-stranded β-sheet (βG–M) flanked by five α-helices (α4–8). These β-strands have a parallel orientation, except for the C-terminal strand βM (Fig. 2C). The AdoMet- and MeSAdo-bound structures are essentially identical (RMSD of 0.2 Å over 264 Cα atoms). In addition, there is no significant structural difference between the apo form (PDB ID: 2FRN) and the cofactor-bound forms (RMSD of 1.1 Å over 245 Cα atoms), with the exception of the aforementioned N-terminal extension (residues 6–18).
The structure of MjTYW2 is very similar to that of PhTYW2 (RMSD of 1.5 Å for 218 Cα atoms), with several exceptions (Fig. 2B). The N-terminal domain of MjTYW2 (residues 4–77) lacks two helices (α1 and α2 of PhTYW2: Fig. 2C) as compared to PhTYW2, reflecting the shorter amino acid sequence of its N-terminal domain as compared to that of PhTYW2. Moreover, although the structure of MjTYW2 was determined at higher resolution than that of PhTYW2, the N-terminal domain of MjTYW2 has considerably higher B factors than that of PhTYW2, and the helix α3 of MjTYW2 is completely disordered. In contrast to PhTYW2, the N-terminal domain of MjTYW2 is not involved in crystal packing interactions. Therefore, its poor electron density suggests the intrinsic flexibility of the N-terminal domain of TYW2.
A DALI search (16) revealed that PhTYW2 and MjTYW2 share structural similarity with other AdoMet-dependent class-I MTases (Fig. S2). The C-terminal MTase domain of PhTYW2 shares high structural similarity to M. jannaschii tRNA(m1G37) MTase Trm5 (7), P. horikoshii putative MTase PH1915 (17), E. coli 23S rRNA(m5C) MTase Rlm1 (18), and P. horikoshii tRNA(m22G26) dimethytransferase Trm1 (19). Furthermore, the N-terminal domain of PhTYW2 shares structural similarity with the D2 domains of Trm5, Ph1915, and Rlm1 (Fig. S2 A–C). Therefore, PhTYW2 has a very similar domain composition to those of Trm5, Ph1915, and Rlm1. However, the precise spatial arrangements of the MTase and D2 domains are different in all four of the enzymes, which may reflect the differences in their substrate RNAs. In addition, PhTYW2 lacks the N-terminal OB-fold (D1) domain, which is shared among Trm5, PH1915, and Rlm1.
AdoMet-Binding Pocket.
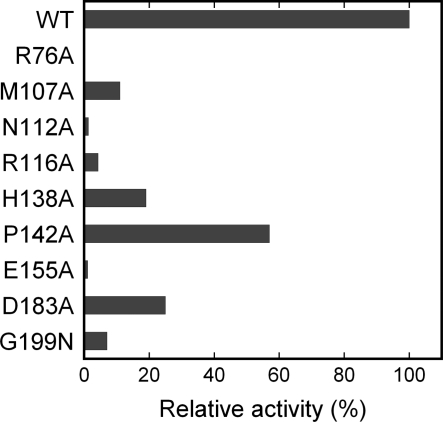
In the AdoMet-bound form of PhTYW2, we clearly observed the electron density of AdoMet in the MTase domain (Fig. 2A and Fig. S3A). Previous studies on MTases identified the conserved motifs (motifs I to VIII and X), of which three (motifs I–III) are especially important for AdoMet binding (20–23). In the present crystal structure, AdoMet is also recognized by motifs I–III of PhTYW2 (Fig. 3A and Fig. S4A). The adenine moiety of AdoMet is recognized by the hydrophobic residues (Phe-133, Tyr-200, and Phe-207) and the conserved Asp-183 residue. The N6 amino and N1 imino groups of the adenine moiety hydrogen bond with the β-carboxyl group of Asp-183 and the main-chain amino group of Asn-184, respectively (Fig. 3A). These hydrogen-bonding interactions between motif III and AdoMet provide the base-specific recognition and are conserved among other class-I MTases (Fig. 3B) (22). The 2′- and 3′-hydroxyl groups of the AdoMet ribose hydrogen bond with the γ-carboxyl group of the conserved Glu-155 residue (Fig. 3A). This interaction manner between motif II and AdoMet is also conserved in other class-I MTases (Fig. 3B) (22), and provides the specific recognition for the ribose moiety. To confirm the importance of these interactions, we measured the in vitro activity of Ala mutants of Asp-183 and Glu-155 by monitoring the incorporation of the [14C]acp group donated from AdoMet (Fig. 4 and Fig. S5). The D183A and E155A mutations decreased the enzymatic activities as compared to the wild-type enzyme, indicating the importance of these interactions.
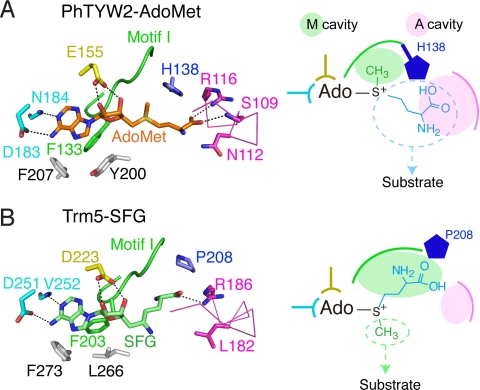
Fig. 3.
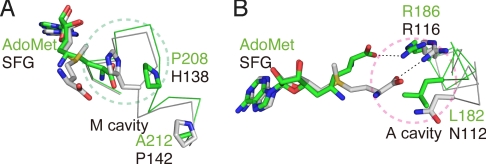
Structural comparison of the cofactor-binding pockets in (A) PhTYW2 with AdoMet and (B) Trm5 with sinefungine (SFG) (PDB ID 2YX1). AdoMet and SFG are colored orange and light green, respectively. Residues involved in the cofactor recognition are shown by stick models (left panels). Interactions are indicated with black dashed lines. Motifs I, II, and III are colored green, yellow, and cyan, respectively. His-138 (A) and Pro-208 (B) are colored blue, and the hydrophobic residues that recognize the adenine moiety of AdoMet are colored gray. Schematic diagrams of the cofactor binding pocket are shown in the right panels. The M and A cavities are shown in green and magenta circles, respectively. In the schematic diagram of (B), AdoMet is shown instead of the inhibitor SFG.
Fig. 4.
Acp-group transfer activity of the wild-type and mutants of PhTYW2. The relative activities to the wild-type enzyme were evaluated from the initial velocities calculated from data in Fig. S5B.
In contrast to the recognition manner for the adenine and ribose moieties of AdoMet, that for the acp group is quite different from those of the bona fide class-I MTases. As observed in the class-I MTases, motif I of PhTYW2 also has a Gly-rich conserved sequence (DXXXGXG), which forms a loop structure lining the inside of the AdoMet-binding pocket (Fig. 3A). In the class-I MTases, the acp group of AdoMet is specifically recognized by a cavity formed by the C terminus of motif I and the adjacent helix α5. For example, in Trm5, the acp group of AdoMet interacts with this cavity by water-mediated interactions (Fig. 3B). Consequently, the methyl group of AdoMet is directed toward the substrate-binding pocket. Hereafter, we refer to this bona fide MTase-specific cavity as the “M cavity” (Fig. 3B, right panel). In contrast, in PhTYW2, the “M cavity” is occupied by the bulky side chain of His-138 in helix α5, which directs the acp group of AdoMet toward the substrate-binding pocket (Fig. 3A, right panel). This bulky residue at the “M cavity” is conserved as His or Tyr in both the archaeal and eukaryotic TYW2s (Fig. S4A). In turn, the acp group of AdoMet is bound to the cavity formed by Met-107, Ser-109, Asn-112, and Arg-116 (Fig. 3A). Especially, the carboxyl moiety of the acp group interacts with the main-chain amide group of Ser-109 and the guanidinium group of Arg-116. These residues are located on the loop connecting strand βF and helix α4, and are strictly conserved among the archaeal and eukaryotic TYW2s, forming a sequence motif, MXSX2NX3R/K (“motif A,” Fig. S4A). Hereafter, we refer to this TYW2-specific cavity for accommodating the acp-group as the “A cavity” (Fig. 3A, right panel). These specific features in the “M cavity” and the “A cavity” are conserved among the archaeal and eukaryotic TYW2 enzymes (Fig. S4A). Therefore, a similar structural mechanism may be used in the acp transfer reaction from AdoMet to tRNA in eukaryotic TYW2.
In the complex structure with MeSAdo, which is a product of the acp group transfer reaction, we observed the clear electron density of MeSAdo (Fig. S3B). As expected, MeSAdo is bound in the same mode as that of AdoMet. Its adenine and ribose moieties are recognized by the same residues in motifs I-III, as described above. In contrast, no electron density corresponding to the acp group is observed in the “A cavity” (Fig. S3), supporting the validity of our assignment of the acp group in the AdoMet-bound structure.
Structural Determinant for the acp-Group Transfer.
Trm5 catalyzes the transfer of the methyl group from AdoMet to the N1 nitrogen of tRNA-G37 (Fig. 1A). The D2 and MTase (D3) domains of archaeal Trm5 (7) have strikingly similar structures to those of archaeal TYW2. PhTYW2 shares 33% sequence identity to M. jannaschii Trm5 (MjTrm5) (Fig. S4B), and the core β sheet (βG–βM) in the MTase domain of PhTYW2 superimposes quite well on that of MjTrm5 (Fig. S2A). The residues around the catalytic site are conserved (Fig. S4B), and their side chain conformations are also structurally conserved. This indicates a close phylogenetic relationship between TYW2 and Trm5, and may reflect their recognition of similar substrates (i.e., tRNA anticodon stem-loop). However, there are several local structural differences between PhTYW2 and MjTrm5. Therefore, a precise structural comparison of these enzymes will provide insight into the structural basis that defines the specificity of their chemical reactions (i.e., methyl or acp group transfer from AdoMet to RNA).
One major difference between TYW2 and Trm5, and the other class-I MTases as well, is the conserved His/Tyr residue in the “M cavity” (His-138 in PhTYW2, Fig. S4B). In addition, in archaeal TYW2, helix α5 is kinked at the Pro residue (Pro-142 in PhTYW2), which pushes the His/Tyr residue up toward the “M cavity” (Fig. 5A). As a consequence, the “M cavity” of PhTYW2 is smaller than that of MjTrm5, and thereby excludes the acp group of AdoMet, which is instead accommodated in the “A cavity” (Fig. 5A).
Fig. 5.
Structural comparison of TYW2 and Trm5. (A) Comparison of the M cavities of PhTYW2 and MjTrm5. (B) Comparison of the A cavities of PhTYW2 and MjTrm5. PhTYW2-AdoMet and Trm5-SFG (PDB ID 2YX1) are colored gray and green, respectively. The M and A cavities are enclosed by dashed circles in panels (A and B), respectively.
Another structural difference between TYW2 and Trm5 is the location of helix α4 and its preceding loop (residues 107–116) surrounding the “A cavity,” which consists of the conserved “motif A” (MXSX2NX3R/K, Fig. S4). The Asn residue (Asn-112 in PhTYW2) in this “motif A” is strictly conserved among the TYW2s, but is replaced with Leu (Leu-182 in MjTrm5) in Trm5 (Fig. 5B). Thus, as compared to that of Trm5, the “A cavity” of TYW2 is hydrophilic, which is suitable for binding the hydrophilic acp group of AdoMet. In contrast, the last Arg residue in “motif A” is commonly conserved in both TYW2 and Trm5 (Arg-116 in PhTYW2 and Arg-186 in MjTrm5, Fig. S4B), and its guanidinium group forms a similar interaction with the carboxyl group of AdoMet (Fig. 3). However, the guanidinium group of this Arg in PhTYW2 is farther away from the catalytic site as compared to that in MjTrm5, resulting in the larger “A cavity” than that in MjTrm5 (Fig. 5B). Consequently, the “A cavity” of TYW2 can accommodate the acp group of AdoMet.
Altogether, we have defined the following differences: (i) His/Tyr at the M cavity, (ii) Pro in helix α5, and (iii) Asn and Arg in motif A, which are likely to define the distinct enzymatic activities between TYW2 and Trm5. To further confirm the acp group-recognizing mechanism, we prepared Ala mutants of Asn-112, Arg-116, His-138, and Pro-142, and analyzed their acp-transfer activities (Fig. 4 and Fig. S5). The N112A and R116A mutations both abolished the activity, supporting our hypothesis that the Asn and Arg residues in the A cavity have critical roles for the acp-group transfer activity. Although the H138A and P142A mutations also displayed lower activity as compared to the wild-type enzyme, their effects were relatively moderate as compared to the A-cavity mutants. These M-cavity mutations could enlarge the M cavity but not directly affect the binding ability of the acp group at the A cavity, which may explain the moderate effects of these mutants.
tRNA and imG-14 Recognition Mechanism.
Recently, the M. jannaschii Trm5·tRNACys complex structure was determined (24). The catalytic domains of PhTYW2 and MjTrm5 share very high sequence similarity, as described above, and tertiary structural similarity as well (RMSD of 1.3 Å over 160 Cα atoms, Fig. S2A). These features allowed us to create a plausible docking model of PhTYW2 and tRNA containing imG-14 at position 37 (Fig. 6).
Fig. 6.
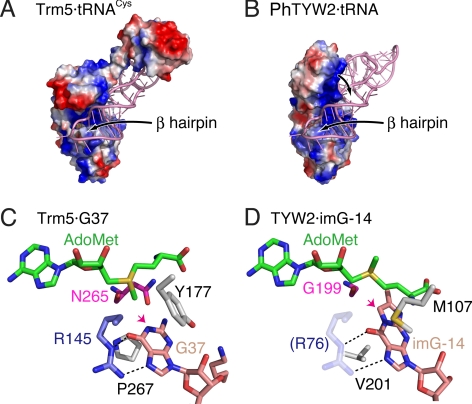
tRNAPhe and imG-14 recognition mechanism by PhTYW2. (A) Overview of the Trm5·tRNACys complex structure. (B) Overview of the PhTYW2·tRNA docking model. The tRNA molecules are colored pink. The electrostatic potential distribution is presented on the contact surface of PhTYW2, with positively and negatively charged regions colored blue and red, respectively. The contact surface and the electrostatic potential were calculated by PyMOL (http://pymol.sourceforge.net/). (C and D) Active sites of the Trm5·tRNACys complex and the PhTYW2 docking model. The G37 and imG-14 residues and AdoMet are shown by stick models. Hydrogen bonds are indicated by black dashed lines. The acceptor atoms of the chemical groups transferred from AdoMet are indicated by arrows.
The Trm5·tRNACys complex structure revealed that the substrate tRNA is bound to the positively-charged cleft formed between the core β-sheet in the N-terminal domain and the MTase domain (Fig. 6A). TYW2 also has a cleft between the core β-sheet of the N-terminal domain and the catalytic site of the MTase domain, and the conserved basic residues around this cleft (Arg-116 and Lys-260 in PhTYW2) form a positively-charged patch. Our docking model suggests that this basic cleft would also be involved in the recognition of the tRNA anticodon stem (Fig. 6B). Furthermore, there is a large positively-charged patch on the N-terminal domain, which is not in direct contact with the modeled tRNA in our docking model (Fig. 6B). A structural comparison between MjTYW2 and PhTYW2 suggested the intrinsic flexibility of the N-terminal domain. Therefore, we can speculate that tRNA binding induces an inter-domain motion between the N-terminal and MTase domains, so that this positively-charged patch on the N-terminal domain would fit onto the phosphate backbone of tRNA (Fig. 6B). In addition, in the Trm5·tRNACys complex structure, a β-hairpin structure is inserted into the tRNA anticodon loop, deforming its canonical U-turn structure (Fig. 6A). Consequently, the target G37 residue of tRNA is flipped into the catalytic pocket of Trm5. This β-hairpin structure (βL-βM) is also present in TYW2, suggesting that it could be involved in the recognition of the tRNA anticodon loop, as observed in the Trm5·tRNACys complex structure (Fig. 6 A and B).
Our docking model provides further insights into the imG-14 recognition mechanism by TYW2 (Fig. 6D). In the Trm5·tRNACys complex structure, the guanine base at position 37 is sandwiched by hydrophobic residues, Tyr-177 and Pro-267 (Fig. 6C). The size and the properties of these residues are also conserved in TYW2 (Met-107 and Val-201 in PhTYW2, respectively), and thus we can imagine that the imG-14 base would be sandwiched by these hydrophobic residues (Fig. 6D). Actually, the M107A mutation disrupted the enzymatic activity (Fig. 4 and Fig. S5), supporting our docking model. Moreover, in the Trm5·tRNACys complex structure, the O6 and N7 atoms of G37 are recognized by the guanidinum group of Arg-145 (Fig. 6C), which is also conserved in TYW2 (Arg-76 in PhTYW2, Fig. S4). Although the βC-βD β-hairpin (residues 70–78) including this Arg is disordered in the PhTYW2 and MjTYW2 structures (Fig. 2), this β hairpin would become ordered upon tRNA binding, thus enabling the recognition of the O9 and N1 atoms of the imG-14 base by this Arg residue (Fig. 6D). The R76A mutation drastically reduced the acp-group transfer activity (Fig. 4 and Fig. S5), strongly supporting our hypothesis on the role of this β hairpin and Arg-76 for the recognition of imG-14. In contrast, Asn-265 in MjTrm5, which recognizes the N2 atom of G37, is replaced with Gly in TYW2 (Gly-199 in PhTYW2). This TYW2-specific Gly residue would generate a larger space to accommodate imG-14 (Fig. 6 C and D), which has a bulkier structure than that of a guanine base (Fig. 1A). The G199N mutation of PhTYW2, which reduces the size of the imG-14 binding pocket, significantly impaired the enzymatic activity (Fig. 4 and Fig. S5), again supporting the validity of our docking model.
Biological Implications for Y Derivatives in Archaea.
Although previous studies indicated the existence of several Y derivatives in archaea (13, 14), the locations of the archaeal Y derivatives within the tRNA have remained elusive. This is a non-trivial question, because some modifications of similar chemical structures are found in different positions in the tRNAs from different phylogenetic domains and they have diverse roles, as exemplified by the case of the 7-deazaguanine derivatives (25). Our in vitro reconstitution assay demonstrated that archaeal TYW2 can cross-react with yeast tRNAPhe containing imG-14 at position 37. The result strongly suggests that Y derivatives are also located exclusively at position 37 of tRNAPhe from archaea, as well as eukaryotes. Therefore, even though the frame shifting caused by the Y-modification deficiency has not been reported in archaea, it is possible that archaeal Y derivatives also play an essential role in preventing the –1 frame shifting at a phenylalanine codon, thus ensuring translational fidelity.
In addition, our in vitro reconstitution assays also demonstrated that archaeal TYW2 possesses an enzymatic activity to form yW-86 from imG-14 in tRNA. Although a previous study indicated that archaeal tRNA contains imG-14, imG, mimG, and imG2 (13), no report has shown the existence of the Y base derivatives with a large side chain at position C7, including yW-86. Interestingly, McCloskey and coworkers reported the presence of an unknown nucleotide, N422, in M. maripaludis, M. vannielii, and M. jannaschii, which exhibits UV absorption spectra characteristic of Y derivatives (14). Since the molecular mass of N422 is identical to that of yW-86 (Mr of 422 Da), it is feasible that this N422 nucleoside is yW-86 formed by archaeal TYW2. On the other hand, archaeal tRNA also contains Y derivatives with a methyl group at position C7 (i.e., imG2 and mimG; Fig. 1A), which are mostly found in Crenarchaeota (13). Sequence comparisons around the “M cavity” and the “A cavity” allowed us to discriminate between TYW2 and Trm5 (Fig. S4B), which suggests that the genome sequences from Crenarchaeota lack an ORF corresponding to TYW2. Therefore, it is likely that TYW2 and its product, yW-86, are specific to Euryarchaeota, while an uncharacterized MTase catalyzes the methylation at position C7 in Crenarchaeota.
The present structure not only revealed the structural basis for acp group transfer to tRNA by TYW2, but also explains a general mechanism for other enzymes catalyzing acp or aminoalkyl group transfer. The detailed and general mechanisms will be discussed in the SI Text and Fig. S6.
Materials and Methods
Sample Preparation and Structure Determination.
The P. horikoshii PH0793 and M. jannashii MJ1557 genes were cloned into a pET28a derivative vector. The proteins were overexpressed in Escherichia coli BL21-CodonPlus(DE3)-RIL cells (Stratagene), and were purified by heat treatment, followed by chromatography on Ni-NTA, heparin, and size-exclusion columns. Crystals of the PhTYW2-AdoMet and PhTYW2-MeSAdo complexes were grown from the reservoir solution containing 0.1 M Mes-NaOH, pH 6.0, and 14% PEG10,000. Crystals of the MjTYW2-AdoMet complex were grown from the reservoir solution containing 0.1 M Mes-NaOH, pH 6.5, 0.1 M ammonium sulfate, and 15% PEG5,000. The crystal structures were determined by the molecular replacement method. Detailed procedures for sample preparation and structure determination are described in the SI Text.
Measurements of acp-Transfer Activity of TYW2.
The tRNA modification activities of archaeal TYW2s were measured using yeast tRNAPhe containing imG-14 at position 37, obtained from the yeast ΔTYW2 strain, as a substrate. The incorporation of the acp group from AdoMet to tRNAPhe was measured by an LC/MS analysis and a standard filter binding assay. Detailed procedures for the activity measurements are described in the SIText.
Supplementary Material
Acknowledgments.
We thank S. Yokoyama (RIKEN, Japan) for kindly providing the coordinates of the archaeal Trm5·tRNA complex structures. We thank Y. Sakaguchi, T. Ohira, and K. Miyauchi (The University of Tokyo, Japan) for tRNA preparation and mass spectrometry experiments; and the beam-line staffs at BL41XU of SPring-8 and NW12A of KEK PF-AR for assistance in data collection. This work was supported by a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to T.S. and O.N.); by a Grant-in-Aid for Scientific Research of MEXT (to H.N., R.I., and O.N.); by Kurata Memorial Hitachi Science and Technology Foundation grants (to O.N.); by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.S.); and by JSPS Fellowships for Japanese Junior Scientists (to M.U. and A.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3A25, 3A26, and 3A27).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905270106/DCSupplemental.
References
- 1.Czerwoniec A, et al. MODOMICS: A database of RNA modification pathways 2008 update. Nucleic Acids Res. 2009;37:D118–121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishitani R, Yokoyama S, Nureki O. Structure, dynamics, and function of RNA modification enzymes. Curr Opin Struct Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K. Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry. 1973;12:188–193. doi: 10.1021/bi00726a002. [DOI] [PubMed] [Google Scholar]
- 4.Konevega AL, et al. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waas WF, de Crecy-Lagard V, Schimmel P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J Biol Chem. 2005;280:37616–37622. doi: 10.1074/jbc.M506939200. [DOI] [PubMed] [Google Scholar]
- 7.Goto-Ito S, et al. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, et al. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 10.Liu S, et al. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeve AM, Breazeale SD, Townsend CA. Purification, characterization, and cloning of an S-adenosylmethionine-dependent 3-amino-3-carboxypropyltransferase in nocardicin biosynthesis. J Biol Chem. 1998;273:30695–30703. doi: 10.1074/jbc.273.46.30695. [DOI] [PubMed] [Google Scholar]
- 12.Klug RM, Benning C. Two enzymes of diacylglyceryl-O-4′-(N,N,N,-trimethyl)homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proc Natl Acad Sci USA. 2001;98:5910–5915. doi: 10.1073/pnas.101037998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou S, et al. Structures of two new “minimalist” modified nucleosides from archaeal tRNA. Bioorg Chem. 2004;32:82–91. doi: 10.1016/j.bioorg.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 14.McCloskey JA, et al. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjork GR, et al. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 17.Sun W, et al. The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii. Protein Sci. 2005;14:3121–3128. doi: 10.1110/ps.051821805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunita S, et al. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J Mol Biol. 2008;383:652–666. doi: 10.1016/j.jmb.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 19.Ihsanawati, et al. Crystal structure of tRNA N2,N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J Mol Biol. 2008;383:871–884. doi: 10.1016/j.jmb.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 20.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 23.Martin JL, McMillan FM. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 24.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon-loop modification in tRNA functional maturation. Nat Struct Mol Biol in press. 2009 doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe M, et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem. 1997;272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.