Abstract
A striking and widespread observation is that higher-order folding for many RNAs is very slow, often requiring minutes. In some cases, slow folding reflects the need to disrupt stable, but incorrect, interactions. However, a molecular explanation for slow folding in most RNAs is unknown. The specificity domain of the Bacillus subtilis RNase P ribozyme undergoes a rate-limiting folding step on the minute time-scale. This RNA also contains a C2′-endo nucleotide at A130 that exhibits extremely slow local conformational dynamics. This nucleotide is evolutionarily conserved and essential for tRNA recognition by RNase P. Here we show that deleting this single nucleotide accelerates folding by an order of magnitude even though this mutation does not change the global fold of the RNA. These results demonstrate that formation of a single stacking interaction at a C2′-endo nucleotide comprises the rate-determining step for folding an entire 154 nucleotide RNA. C2′-endo nucleotides exhibit slow local dynamics in structures spanning isolated helices to complex tertiary interactions. Because the motif is both simple and ubiquitous, C2′-endo nucleotides may function as molecular timers in many RNA folding and ligand recognition reactions.
Keywords: RNA folding, RNA SHAPE chemistry
To function properly inside the cell, RNA molecules undergo complex folding transitions to form specific, biologically active, three-dimensional structures (1). These essential structures involve both local base pairing and also complex higher-order tertiary interactions. A persistent and incompletely explained observation is that many RNAs fold very slowly, on timescales requiring minutes or longer. RNAs that fold slowly span all sizes including small riboswitch and catalytic RNAs, medium-sized catalytic introns, and the large RNAs in ribosomes (2–6).
Slow folding has important consequences both because correct folding ultimately governs the rate at which an RNA can perform its biological function and because bottlenecks in assembly potentially require cellular mechanisms to overcome slow folding. In some cases, slow folding results from formation of stable, non-native, and kinetically trapped states (7, 8). Such misfolding is remedied, in part, by cellular chaperone activities (9–11). In many cases, the molecular basis for slow folding is unknown.
The intricate three-dimensional structures formed by RNA molecules ultimately reflect the underlying contributions of individual nucleotides. Most nucleotides in an RNA molecule exist in the C3′-endo conformation. Although less frequent, the C2′-endo conformation is highly overrepresented in catalytic active sites and in critical tertiary structures (12, 13). At the static structure level, the C2′-endo conformation induces greater helical twist in an A-form helix (14, 15) and spans a much longer 5′-to-3′ distance (12, 16). At the level of local motion, some C2′-endo nucleotides exhibit extraordinarily slow conformational dynamics with half-lives on the 10–100-s timescale (17).
Here we show that the slow conformational dynamics that characterize a single C2′-endo nucleotide can ultimately govern the folding of a large ribozyme with numerous constituent tertiary interactions. This dramatic effect on folding can be attributed to a single RNA position because deletion of one C2′-endo nucleotide accelerates RNA folding by an order of magnitude. We envision that other functionally critical rate-limiting steps in RNA biology will eventually be shown to reflect slow conformational dynamics at single C2′-endo nucleotides.
Results
A130 Has Distinct Local Dynamics and a Critical Role in RNase P Function.
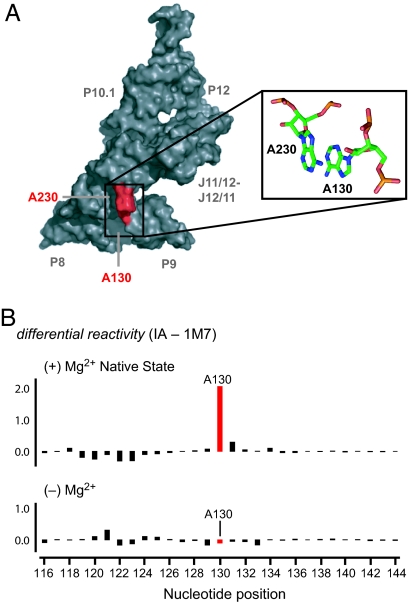
RNase P is a conserved ribonucleoprotein enzyme found in all kingdoms of life. This ribozyme catalyzes site-specific cleavage of the 5′ end of precursor tRNAs to yield the mature tRNA (18, 19). Bacterial RNase P RNAs are composed of two independently folding domains, a catalytic and a specificity domain (20). One critical and conserved tertiary interaction in the specificity domain is a stacking interaction between A130 and A230 (in the Bacillus subtilis numbering) (21, 22). A130 forms the C2′-endo conformation (17) and, together with its stacking partner at A230, are the only two nucleotides in the B-type RNase P specificity domain shown to be directly involved in recognizing and binding the tRNA substrate (23–25) (in red, Fig. 1A). Mutation of A130 severely compromises RNase P function (25). Both A130 and A230 experience a significant decrease in local nucleotide flexibility when tRNA binds (Fig. S1).
Fig. 1.
Structural context and slow local nucleotide dynamics of A130. (A) Three dimensional surface representation of the RNase P Bacillus subtilis specificity domain (21) illustrating the location, in red, and conformation, in stick models, of A130 and its stacking partner A230. (B) Differential SHAPE reactivities between slow (IA) and fast (1M7) 2′-hydroxyl selective reagents for the A130 region in the presence and absence of Mg2+. A large difference in reactivity (emphasized in red) indicates that a nucleotide experiences slow conformational dynamics (17).
Slow conformational dynamics at individual nucleotides can be measured in favorable cases using SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) experiments that measure the differential reactivity of the ribose 2′-hydroxyl group toward fast versus slow-reacting reagents (17). SHAPE reagents selectively form 2′-O-adducts at flexible nucleotides in RNA (26, 27). Sites of 2′-O-adduct formation are identified as stops to primer extension using fluorescently labeled DNA primers, resolved by capillary electrophoresis. In the context of the folded RNase P specificity domain, the 2′-hydroxyl group at A130 shows low reactivity toward a fast SHAPE reagent (1M7, reactive half-time, t1/2, = 14 s) but is highly reactive toward a structurally similar but slow reacting reagent (IA, t1/2 = 2,220 s; see red bar in top panel, Fig. 1B). This strong differential reactivity with the slowly reacting reagent indicates that this C2′-endo nucleotide undergoes an extraordinarily slow conformational change to reach the state in which the 2′-hydroxyl is reactive toward acylation (t1/2 = 10–100 s) (17). When a similar experiment is performed in the absence of Mg2+, where the RNA does not form a well-defined tertiary structure (20, 28), the fast and slow SHAPE reagents show equivalent reactivities at A130 (red bar in bottom panel, Fig. 1B). Thus, slow conformational dynamics at A130 only occur in the context of the native RNA tertiary structure. The full length RNase P RNA also exhibits a strong differential reactivity toward fast and slow SHAPE reagents at A130 (Fig. S2).
In sum, A130 is critical to RNase P function (24, 25), interacts directly with the tRNA substrate (24) (Fig. S1), forms the C2′-endo conformation (17, 21), and exhibits unusual and very slow local nucleotide dynamics (Fig. 1B).
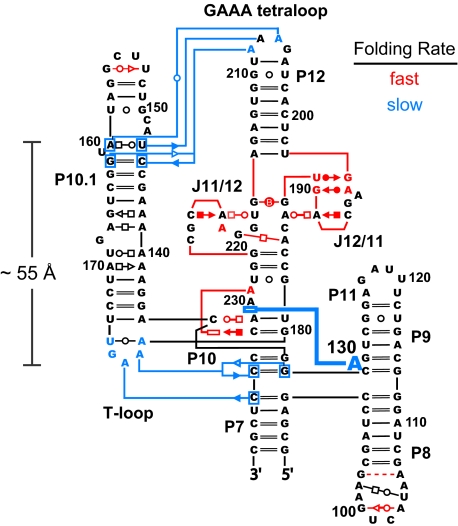
The RNase P specificity domain folds slowly (0.004 s−1) and in two kinetically distinct steps (29). The first phase occurs with a rate constant of 0.06 s−1 (in red, Fig. 2); the second, even slower, phase folds with a rate constant of 0.004 s−1 (in blue, Fig. 2). The rate-limiting step involves formation of tertiary interactions that are approximately 55 Å distant in the RNA. One of the interactions that occurs in this slow rate-determining step is the stacking of A130 on A230, which raises the possibility that slow conformational dynamics at a single nucleotide, A130, slows folding of the entire RNA. To test the hypothesis that slow conformational dynamics at A130 gate folding of the RNase P RNA, we characterized folding for an RNA in which this nucleotide was deleted (termed the ΔA130 RNA).
Fig. 2.
Two-step mechanism for tertiary folding of the RNase P specificity domain (29). Tertiary interactions that form in the fast versus slow steps are illustrated in red and blue, respectively. Slow folding at A130 is emphasized with the heavy blue line representing the stacking interaction with A230.
The ΔA130 RNA Has the Same Global Fold as the Native RNA.
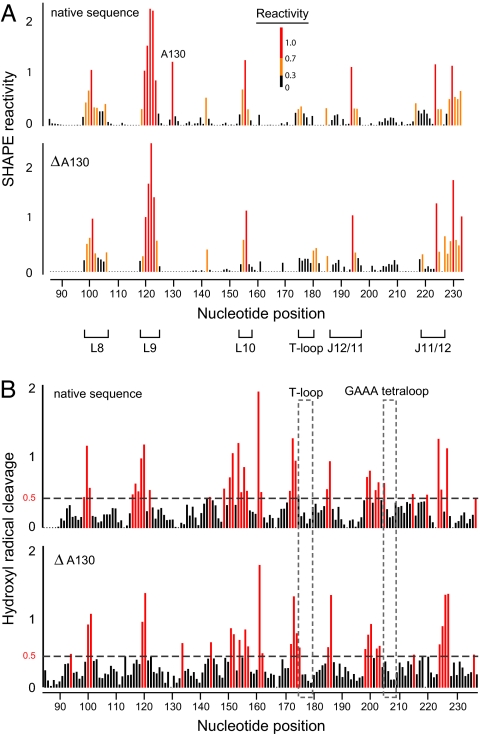
We first confirmed that the equilibrium structures of the native and ΔA130 RNAs are the same despite disruption of the A130–A230 stacking interaction. We compared the structures of the two RNAs by SHAPE using the 2′-hydroxyl-selective reagent benzoyl cyanide (BzCN) (29) under conditions that stabilize the native tertiary fold. Absolute SHAPE reactivities are virtually identical for the two RNAs, in both the starting and final states. RNA loops are reactive while nucleotides constrained by base pairing and tertiary interactions are unreactive in both RNAs (Fig. 3A and Fig. S3). The single exception is that the bulged A130 is reactive in the native sequence but, as expected, this reactivity disappears when the nucleotide is deleted.
Fig. 3.
Native sequence and ΔA130 mutant RNase P specificity domain RNAs form similar secondary and tertiary structures. (A) SHAPE reactivity histograms for the native (top) and ΔA130 (bottom) RNase P RNAs. (B) Hydroxyl radical cleavage intensities, which provide a measure of nucleotide solvent accessibility, for native (top) and ΔA130 (bottom) RNase P RNAs. Solvent accessible nucleotides (normalized nucleotide reactivity ≥0.5) are red and protected, solvent inaccessible, nucleotides are black. Protections due to tertiary interactions involving the T-loop and the GAAA tetraloop (that form in the slow step for the native sequence) are emphasized with gray dashed boxes.
We assessed tertiary structure in the native and mutant RNase P RNAs in a second way using hydroxyl radical footprinting. Hydroxyl radicals react preferentially with solvent accessible regions of the RNA backbone (30). The native and ΔA130 RNAs were exposed to hydroxyl radicals, generated from H2O2 in the presence of Fe(II)-EDTA, under conditions that stabilize the tertiary fold. Cleavage patterns, and thus solvent accessibility, are highly similar in both RNAs, indicating the global fold is the same (Fig. 3B). Critically, the two sets of tertiary interactions that form in the rate-limiting folding step for the native RNA, docking of the T-loop into stacked helices and formation of the GAAA tetraloop-receptor interaction (in blue, Fig. 2) have identical low cleavage intensities in both RNAs (dashed boxes, Fig. 3B).
Finally, we evaluated the net thermal stability of the native sequence versus the ΔA130 mutant. The native sequence and mutant RNAs have similar thermal melting profiles. Tertiary structure, as judged from an intermediate transition, appears to be slightly more stable in the ΔA130 mutant (Fig. S4).
In sum, SHAPE, hydroxyl radical, and thermal melting experiments (Fig. 3 and Figs. S3 and S4) indicate that the native and ΔA130 mutant RNase P RNAs form the same, stable, secondary and tertiary structures and also have the same starting structures for their folding reactions. Any differences in folding rates for the native sequence versus mutant RNA will therefore reflect a change in transition state for structural biogenesis, rather than a difference in the final folded state or the requirement to disrupt some misfolded state.
Fast Folding Kinetics of the ΔA130 RNA.
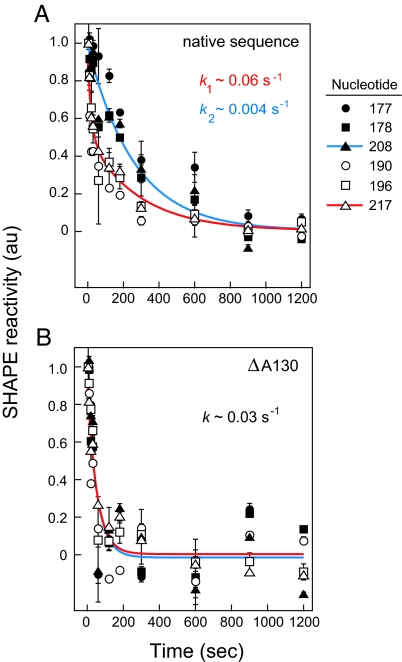
The folding behavior of the native sequence RNase P specificity domain was analyzed by time-resolved SHAPE (29, 31). Time-resolved SHAPE takes advantage of the discovery that BzCN reacts with RNA 2′-hydroxyl groups and simultaneously degrades completely in water in approximately 1 s. Thus, addition of an RNA to this reagent makes it possible to observe RNA folding at single nucleotide resolution in 1-s snapshots. RNA folding was initiated by addition of Mg2+ to an RNA preequilibrated in buffer and monovalent ions to yield final conditions that stabilize the native RNase P structure. Consistent with prior work, tertiary folding of the native sequence RNase P RNA occurs in two kinetically distinct steps: the first step is a fast phase characterized by a rate constant of 0.06 s−1. The second step is a slow phase that occurs with a rate constant of 0.004 s−1 (in red and blue, Fig. 4A).
Fig. 4.
Deletion of A130 accelerates RNA folding. Folding rate constants for the RNase P specificity domain were monitored by time-resolved SHAPE (29). (A) The native sequence folds in two kinetically distinct steps. Nucleotides folding in the fast and slow steps are indicated by open and closed symbols, respectively. (B) All nucleotides in the ΔA130 RNA fold at the same fast rate (compare open and closed symbols). For clarity, these data have been normalized to a uniform scale (spanning 0–1); however, absolute amplitudes for nucleotides exhibiting two kinetically significant steps (in red, panel A) are generally larger than for the other time-progress curves.
We then analyzed the folding behavior of the ΔA130 mutant, also using time-resolved SHAPE. In strong contrast to the native RNA, the mutant RNA folds in a single step characterized by a rate constant of 0.03 s−1 (Fig. 4B). The unnormalized changes in amplitude for time-progress curves at a given nucleotide were similar for slow folding positions in the native sequence and for fast folding positions in the mutant (in blue, Fig. 4). In contrast, amplitudes for nucleotides exhibiting the combined fast and slow phases in the native sequence (in red, Fig. 4A) were generally larger than for the (fast folding) positions in the mutant. These data, plus the observation that each phase falls cleanly into specific structural motifs within the RNA (see Fig. 2), strongly support the interpretation that native sequence folding reflects a single population that folds in two consecutive steps rather than folding of two distinct populations. The ΔA130 mutant thus folds 10-fold faster than the rate-limiting step that governs folding for the native RNA.
We confirmed this striking result using an independent method, time-resolved fluorescence spectroscopy. The native sequence and ΔA130 RNase P RNAs were labeled at their 5′ ends with an environmentally sensitive fluorophore (Oregon green). The fluorescence emission of both 5′-fluorescently labeled RNAs is quenched approximately 40% upon the addition of Mg2+. Mg2+-induced changes in fluorescence reflect RNA conformational changes because the free fluorophore shows no change in fluorescence emission upon addition of Mg2+ (Fig. S5).
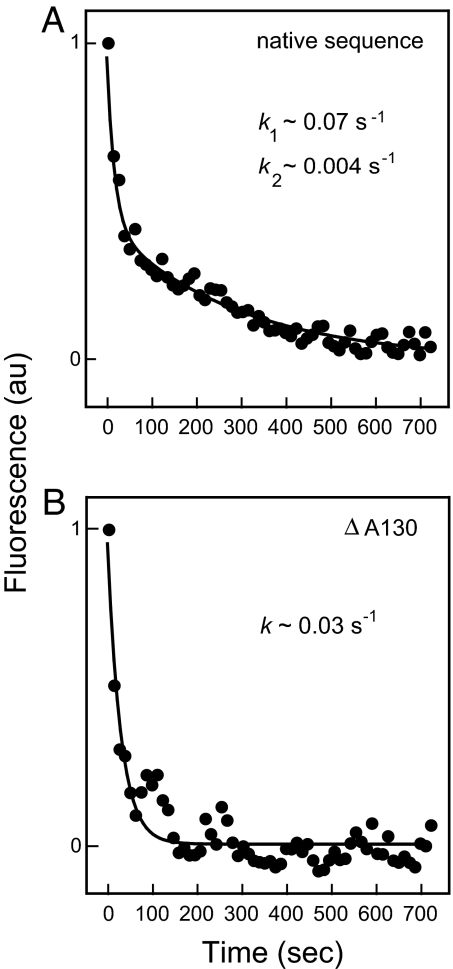
Folding of the native and mutant RNase P RNAs was followed as the decrease in fluorescence emission as a function of time after addition of Mg2+. Native RNA folding exhibits a clear double exponential, indicative of two folding rates (Fig. 5A), with rate constants of 0.07 s−1 and 0.004 s−1. These rates agree exactly with those established by time-resolved SHAPE.
Fig. 5.
Folding rate constants for the RNase P specificity domain monitored by fluorescence spectroscopy. (A) The native sequence folds in two kinetically distinct steps. (B) The ΔA130 mutant RNA folds in a single kinetic step.
When otherwise identical experiments were performed with the ΔA130 point mutant, the fluorescently detected folding profile changed significantly. The mutant RNA folds in a single step (Fig. 5B), with a rate constant of 0.03 s−1, and 10-fold faster than the rate determining step for the native RNA. The observed rate constant of 0.03 s−1 agrees exactly with the time-resolved SHAPE experiment.
Time-resolved SHAPE and fluorescence experiments thus both reveal that the ΔA130 RNA folds an order of magnitude faster than the native RNA (Figs. 4 and 5) despite the fact that both RNAs have similar starting states and fold to the same final tertiary structure (Fig. 3 and Fig. S3). Deletion of a single C2′-endo nucleotide that exhibits slow conformational dynamics has a dramatic effect on tertiary folding in a large RNA.
Discussion
A surprisingly large number of RNAs appear to fold slowly, on the minute timescale or even more slowly. In most cases, the molecular basis for slow RNA folding is unknown. Here we show that the structural requirement to form an interaction involving a nucleotide in the C2′-endo conformation governs folding of a large ribozyme. Deletion of the C2′-endo nucleotide at A130 accelerates RNA folding by an order of magnitude but does not detectably change the final global fold for this RNA.
Nucleotides in the C2′-endo conformation are relatively rare but occur overwhelmingly in specialized, functionally important, structures in RNA. Examples include small RNA motifs, the spliceosome, and the ribosome (13, 32, 33). The A130 nucleotide in RNase P is typical in this respect because its interaction with A230 creates the local structure necessary for recognizing and binding the pre-tRNA substrate (Fig. S1) (23, 25, 28).
Although other simple RNA motifs that undergo slow local conformational changes may be discovered, the C2′-endo nucleotide motif is likely to be especially important. First, the C2′-endo conformation is a simple, and common, variation on RNA structure. Second, slow local conformational dynamics at C2′-endo nucleotides have now been identified in three different classes of RNAs, simple duplexes containing G-A mismatches (17), the RNase P specificity domain (Fig. 1), and at additional sites in the intact multidomain RNase P RNA (Fig. S2). Third, as shown in this work, slow folding at a single C2′-endo nucleotide is sufficient to create a rate-determining bottleneck for folding a large RNA with a complex tertiary structure.
We envision that C2′-endo nucleotides characterized by slow local conformational dynamics have three potential consequences for the function of RNase P and of large RNAs in general.
First, bulged C2′-endo nucleotides are well suited for bridging a helix with a second RNA motif. For example, in the RNase P specificity domain, A130 and A168 are C2′-endo nucleotides characterized by slow nucleotide dynamics and both function to stabilize tertiary interactions between a helix and other RNA elements (17, 21). In the complete RNase P ribozyme, two extrahelical C2′-endo nucleotides, A130 and A374, make direct interactions with the tRNA substrate (Fig. S2). A corollary of the structural role that C2′-endo nucleotides play in stabilizing RNA tertiary structure is that slow dynamics may be a “side-effect” of this useful conformation.
Second, slow conformational dynamics at C2′-endo nucleotides will likely be exploited directly as molecular timers in RNA folding reactions. This molecular timer function could play a structural role on both long-range and local levels. On a long-range level, A130 gates folding in the specificity domain, likely causes this domain to fold more slowly than other structural domains in the RNase P RNA, and therefore potentially functions to cause the specificity domain to fold later than the rest of the RNA.
Third, on a local level, we find it striking that two C2′-endo nucleotides, A130 and A374 (Fig. S2), both contact the tRNA substrate recognized by RNase P (Fig. S1). These nucleotides have the potential to function as molecular timers to facilitate selection of tRNA structures or to coordinate binding with tRNA cleavage.
Finally, because slow conformation dynamics at C2′-endo nucleotides have dramatic consequences for RNA folding and ribonucleoprotein assembly reactions, there may exist RNA chaperone-like proteins (or RNAs) with ribose isomerase activity that function to accelerate or regulate this slow step.
Because the motif is simple and highly overrepresented in functionally critical RNA motifs, we postulate that slow dynamics at C2′-endo nucleotides may be conspicuously exploited to time RNA folding, riboswitch, ligand recognition, and ribonucleoprotein assembly reactions. Nucleotides exhibiting slow conformational dynamics can be readily detected using a very simple differential SHAPE reactivity experiment (Fig. 1) (17). We encourage the RNA community to evaluate their favorite RNAs for the critical biological consequences of slow conformational dynamics at C2′-endo nucleotides.
Methods
Synthesis of Bacillus subtilis RNase P RNAs.
All RNAs were synthesized by in vitro transcription using PCR-generated templates (28). RNAs analyzed by SHAPE and hydroxyl radical footprinting were embedded in 5′ and 3′ structure cassette sequences (34). For fluorescence-detected folding, 5′-monophosphorothioate-labeled RNase P specificity domain RNAs were transcribed in the presence of 10 mM guanosine 5′-O-monophosphorothioate (BioLog), flanking 5′ and 3′ structure cassette sequences were omitted, and the RNAs contained an initial G86-C240 base pair instead of the U86-A240 base pair. All RNAs were purified by denaturing polyacrylamide gel electrophoresis, excised from the gel, and recovered by passive elution and ethanol precipitation. Purified RNAs were resuspended in TE [10 mM Tris (pH 8.0) and 1 mM EDTA] at concentrations of about 40 μM and stored at −20 °C.
SHAPE Analysis.
For equilibrium experiments, native and mutant RNase P specificity domain RNAs (5 pmol in 6 μL H2O) were heated at 95 °C for 2 min, cooled on ice, treated with 3 μL of 3.3× folding buffer [333 mM HEPES (pH 8.0), 333 mM NaCl, and 33.3 mM MgCl2], and incubated at 37 °C for 20 min. The RNA solution was added directly to 1 μL of 10× BzCN (600 mM in anhydrous DMSO). For time-resolved experiments, the procedure was identical to that described previously (29, 31). tRNA binding to the full length RNase P was measured as described (35). Briefly, the full length RNA (20 pmol) and mature tRNAPhe (60 pmol, if present) were heated in buffer [200 mM HEPES (pH 8.0)] at 85 °C for 2 min, incubated at room temperature for 3 min, supplemented with 1 μL of 10× MgCl2 (1 M) and 3 μL of 6.6× KCl (4 M). The RNase P RNA and the tRNAPhe were incubated at 50 °C and 37 °C, respectively, for 10 min. The RNase P solution was divided in half and added to tRNAPhe or a no-tRNA solution and incubated at 37 °C for 15 min. RNA solutions were then added directly to 1 μL of 10× BzCN (200 mM in anhydrous DMSO). The differential reactivity experiment, used to assess local conformational dynamics (17), was performed similarly except the 10× [MgCl2] was 100 mM and no tRNA or additional ions were added. The RNA solution was treated with 1-methyl-7-nitroisatoic anhydride (1M7) or isatoic anhydride (IA) (1 μL, 50 mM in anhydrous DMSO), and allowed to react for 70 s (equal to five 1M7 hydrolysis half-lives) or 36 min (equal to five IA hydrolysis half-lives). No-reagent controls contained 1 μL neat DMSO. Modified RNA was recovered by ethanol precipitation and resuspended in 10 μL TE.
Hydroxyl Radical Probing.
RNA (5 pmol) in 4 μL sterile water was heated at 95 °C for 2 min, cooled on ice, treated with 3 μL of 3.3× folding buffer [66 mM HEPES (pH 7.0) and 33 mM MgCl2], and incubated at 37 °C for 20 min. Freshly prepared solutions of sodium ascorbate (1 μL; 50 mM) and Fe(II)-EDTA (1 μL; 10 mM ferrous ammonium sulfate, 20 mM EDTA, pH 8.0) were mixed and, along with H2O2 (1 μL; 0.03%), were spotted on the lid of a reaction tube (30). Control reactions contained 3 μL H2O. Reactions were initiated by briefly centrifuging the tubes and were then incubated at 37 °C for 2 min. Reactions were stopped by ethanol precipitation and resuspended in 10 μL TE.
Primer Extension and Data Analysis for Modified RNAs.
The general procedure was that outlined previously (28). Briefly, fluorescently labeled DNA primers were annealed to the RNA (10 μL, from the previous modification steps) and extended by reverse transcription. cDNA extension products, along with dideoxy sequencing markers (28), were separated by capillary electrophoresis using an Applied Biosystems 3130 instrument. Raw traces were processed using ShapeFinder (36). All data sets were normalized after excluding the 2% most reactive nucleotides and dividing by the average intensity of the next 8% most reactive nucleotides. On this scale, 1.0 is thus the average intensity of highly reactive positions. Folding rates for individual nucleotides were obtained by normalizing intensities (I) to the first time point and fitting to either a single [I = A + (1 − A)e−k2t] or double [I = A + (1 − A − B)e−k1t + Be−k2t] exponential.
Analysis of Structural Stability by Thermal Melting.
The native and mutant RNase P specificity domain RNAs (1.6 μM, 1 mL) were heated from 25 to 90 °C in an Applied Photophysics Pistar-180 spectrometer at 1 °C/min in folding buffer [100 mM HEPES (pH 8.0), 100 mM NaCl, and 10 mM MgCl2]. After subtracting background from a sample omitting RNA, the denaturing profile, monitored at 260 nm, was geometrically smoothed and differentiated with respect to temperature.
Folding Kinetics Monitored by Fluorescence Spectroscopy.
A fluorescently labeled RNase P RNA was created by incubating a 5′-phosphorothioate RNA (6.28 nmol) in 60 μL buffer (10 mM HEPES, pH 7.0) with Oregon green 488 maleimide (2 μL, 20 mg/mL, Invitrogen) in the dark at room temperature for 3 h. RNA was recovered by ethanol precipitation and fluorescently labeled RNA (≈1,500 pmol) was purified by denaturing polyacrylamide gel electrophoresis and stored in TE at −20 °C in the dark. Folding was initiated by adding a solution of RNA [400 pmol in 522 μL; 100 mM HEPES (pH 8.0) and 100 mM NaCl] at 37 °C to MgCl2 (58 μL, 100 mM), equilibrated in a 400 μL quartz cuvette (at 37 °C for 15 min) in a Varian Cary Eclipse Fluorescence Spectrophotometer. Measurements were averaged over 3 s and taken every 12 s for 30 min; λex = 491 ± 10 nm and λem = 515 ± 10 nm.
Supplementary Material
Acknowledgments.
This work was supported by National Science Foundation Grant MCB−0416941 (to K.M.W.). We thank Dorothy Erie for helpful discussions. Thermal melting experiments were performed at the Macromolecular Interactions Facility at the University of North Carolina-Chapel Hill with help from Ashutosh Tripathy. The full length Bacillus subtilis RNase P construct was generously provided by B. Wayne Huggins.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901319106/DCSupplemental.
References
- 1.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd Ed. New York: Cold Spring Harbor; 2004. [Google Scholar]
- 2.Furtig B, et al. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 3.Webb AE, Weeks KM. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat Struct Biol. 2001;8:135–140. doi: 10.1038/84124. [DOI] [PubMed] [Google Scholar]
- 4.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR. Biophysical studies of bacterial ribosome assembly. Curr Opin Struct Biol. 2008;18:299–304. doi: 10.1016/j.sbi.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodson SA. RNA folding and ribosome assembly. Curr Opin Chem Biol. 2008;12:667–673. doi: 10.1016/j.cbpa.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 8.Thirumalai D, Woodson SA. Maximizing RNA folding rates: A balancing act. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 10.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 12.Richardson JS, et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien KR, Sumita M, Chen PH, Laird-Offringa IA, Hoogstraten CG. Conformationally restricted nucleotides as a probe of structure-function relationships in RNA. RNA. 2008;14:1632–1643. doi: 10.1261/rna.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SantaLucia JJ, Turner DH. Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry. 1993;32:12612–12623. doi: 10.1021/bi00210a009. [DOI] [PubMed] [Google Scholar]
- 15.Heus HA, Wijmenga SS, Hoppe H, Hilbers CW. The detailed structure of tandem GA mismatched base-pair motifs in RNA duplexes is context dependent. J Mol Biol. 1997;271:147–158. doi: 10.1006/jmbi.1997.1158. [DOI] [PubMed] [Google Scholar]
- 16.Wyatt JR, Puglisi JD, Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 17.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 19.Walker SC, Engelke DR. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995;34:902–909. doi: 10.1021/bi00003a024. [DOI] [PubMed] [Google Scholar]
- 21.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 22.Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 23.LaGrandeur TE, Huttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994;13:3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2′-Hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc Natl Acad Sci USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 26.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 27.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 29.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 30.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Struct Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: Quantitative RNA structure analysis in one second snapshots and at single nucleotide resolution. Nat Protoc 4. 2009 doi: 10.1038/nprot.2009.126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolev NG, Steitz JA. In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat Struct Mol Biol. 2006;13:347–353. doi: 10.1038/nsmb1075. [DOI] [PubMed] [Google Scholar]
- 33.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16S and 23S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nature Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 35.Odell L, Huang V, Jakacka M, Pan T. Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res. 1998;26:3717–3723. doi: 10.1093/nar/26.16.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.