Abstract
Transforming growth factor-β (TGF-β) elicits its signals through two transmembrane serine/threonine kinase receptors, type II (TβRII) and type I receptors. It is generally believed that the initial receptor dimerization is an essential event for receptor activation. However, previous studies suggested that TGF-β signals by binding to the preexisting TβRII homodimer. Here, using single molecule microscopy to image green fluorescent protein (GFP)-labeled TβRII on the living cell surface, we demonstrated that the receptor could exist as monomers at the low expression level in resting cells and dimerize upon TGF-β stimulation. This work reveals a model in which the activation of serine-threonine kinase receptors is also accomplished via dimerization of monomers, suggesting that receptor dimerization is a general mechanism for ligand-induced receptor activation.
Keywords: serine/threonine kinase receptor, subunit stoichiometry
Transforming growth factor-β (TGF-β) and related growth factors regulate a variety of important cellular processes such as cell proliferation, differentiation, motility, and apoptosis (1–4). Two cell-surface receptors, type II (TβRII) and type I (TβRI) receptors, are required for TGF-β signal transduction. These receptors belong to the serine/threonine kinase family with a cysteine-rich extracellular domain and the kinase-containing intracellular region. TGF-β signaling is initiated by the binding of TGF-β to TβRII, which leads to the recruitment of TβRI to form a heteromeric complex of TβRI-TβRII on the cell surface. In the complex, TβRI is activated by TβRII via phosphorylation in the GS domain and the signal is transduced to the downstream mediators Smad proteins, which are then accumulated in the nucleus and regulate the expression of target genes (3, 5–8). TβRII is the primary TGF-β-binding receptor and TβRI can interact with TGF-β in the presence of TβRII in the TβRI-TβRII complex (9).
Since the binding of TGF-β to TβRII is the initial and essential event for TGF-β signaling, much effort has been made to understand the physical interaction of this ligand-receptor binding and the molecular nature of the signaling complex formation. One of the important issues is the stoichiometry of TβRII and its oligomerization status before and after ligand binding and receptor complex formation. Previous studies have been mainly carried out with three approaches: double immunoprecipitation of differently tagged-TβRII that were transiently expressed and isotope-metabolically labeled in cells, sedimentation velocity of the metabolically labeled receptors on sucrose gradients, and antibody-mediated immunofluorescence co-patching of the receptors tagged with different epitopes (10–12). These studies have indicated that TβRII exists as a ligand-independent homomeric complex, and the binding of TGF-β to preformed homomeric TβRII leads to the formation of a heteromeric TβRI-TβRII complex (mainly tetramer with two TβRII and two TβRI). This receptor activation mode is in contrast to the well-documented one for tyrosine kinase receptors that exist as monomers in resting cells and dimerize upon ligand stimulation (13, 14). As previous studies are based on in vitro biochemical assays with overexpressed proteins, whether these results reflect the signaling process under the physiological condition remains unclear.
Recent advances in single-molecule fluorescence imaging with living cells have offered a new way to probe the structure and the dynamic behavior of membrane signaling proteins under or near physiological conditions (15–18). Single molecule techniques enable not only the ultrasensitive detection of the molecular events, but also the discovery of spatial and temporal molecular heterogeneity that is hidden in the conventional ensemble measurements of the whole population of the molecules (19, 20). For example, single-molecule study on the membrane proteins such as tyrosine kinase receptors, small G proteins and ion channel receptors have yielded new information on receptor stoichiometry and activation (16, 21–24).
In this work, we applied the single-molecule fluorescence imaging approach to investigate the serine/threonine kinase receptor TβRII. By coupling the receptor with the green fluorescent protein, we observed the existence of individual TβRII molecules on the cell membrane. We further investigated the oligomeric status of TβRII both at resting state and after TGF-β1 treatment in living cells, and found that the monomeric TβRII were dimerized upon ligand stimulation. Our results reveal a model in which the activation of serine-threonine kinase receptors is also accomplished via monomer dimerization upon ligand binding.
Results and Discussion
TβRII Exists as Monomer at Low Density in Resting Cells.
To investigate the oligomerization status of TGF-β receptors, we tagged TβRII at its C terminus with the enhanced green fluorescent protein (GFP). GFP-coupled TβRII was tested to be functional like unlabeled TβRII in activating the expression of the TGF-β-responsive reporter CAGA-luciferase in the presence of TGF-β in the TβRII-deficient cells. Single-molecule fluorescence imaging of the transfected TβRII-GFP was first examined in HeLa cells using an objective-type total internal reflection microscope (TIRFM). Cells were imaged at 3–4 h after transfection, so that TβRII-GFP molecules were expressed at low density (20 to 100 molecules in a 20 × 20 μm area), and individual TβRII-GFP molecules could be distinguished within the spatial resolution of fluorescence microscopy. This expression level of TβRII-GFP was similar to that of the endogenous TβRII molecules (Fig. S1). As shown in the typical TIRFM image (Fig. 1A and Movie S1), most TβRII-GFP molecules appeared as well-dispersed diffraction-limited fluorescent spots (3 × 3 pixels, 690 × 690 nm), and maintained their fluorescence mostly for less than 5 s and then suddenly disappeared.
Fig. 1.
Monomeric TβRII molecules imaged in resting cells. (A) A typical single-molecule image of TβRII-GFP on the living HeLa cell membrane. After transfected with TβRII-GFP for 3–4 h, HeLa cells were imaged with TIRFM. The image is a section (20 × 20 μm) of the first frame from a stack of images (Movie S1) with background subtracted. The diffraction-limited spots (3 × 3 pixel regions) enclosed with green circles represented the signals from individual TβRII-GFP molecules, and were chosen for intensity analysis. (Scale bar, 2 μm.) (B) Distribution of the fluorescence intensity of diffraction-limited TβRII-GFP spots (n = 258) from the living cell imaging. The solid curves show the fitting of Gaussian function and the two peaks represented TβRII-GFP monomers and dimers, respectively. Correlation coefficient (R) of the Gaussian fitting is 0.999. The arrowheads indicate the peak positions of the fitting curves. Numbers in the parentheses are the percentage of the fractions. The experiments have been repeated for more than five times.
To investigate whether these diffraction-limited spots represented monomeric TβRII tagged with one GFP molecule, we first analyzed the fluorescence intensity distribution of the spots. It exhibited a sum of two Gaussian distributions (Fig. 1B). The first population which covering the majority of the spots had the peak intensity (752 counts) close to that of single purified GFP molecules on coverslips (805 counts, Fig. S2A), indicating the signal of single TβRII–GFP molecules. It is understandable that the peak intensity of TβRII–GFP was a little lower than that of GFP on coverslips, as the GFP molecules tagged to the cytoplasmic C-teminal of TβRII were farther away from the total internal reflection interface than those immobilized directly on glass (16). The second population had a peak value about twice in intensity as the first one, suggesting that they were homo-oligomeric TβRII-GFP, likely dimers in most cases. Within 258 spots counted from five cells, 86% were monomers and 14% were dimers. The result suggested that most of the TβRII-GFP molecules existed in the monomeric state.
To further confirm the monomeric state of TβRII-GFP, we then counted the photobleaching steps of individual fluorescent TβRII-GFP molecules. It has been demonstrated that the subunit number and stoichiometry of membrane-bound proteins can be determined by the statistical analysis of bleaching steps of GFP fused to the proteins (22, 24). To reduce the signal fluctuation due to the diffusion of TβRII-GFP on living cell surfaces, cells were fixed before imaging for TβRII-GFP tracking (Fig. 2A and Movie S2). The fluorescence intensity distribution in the fixed cells was very similar to that obtained in the living cells. From the photobleaching plots, within 262 traces of TβRII-GFP fluorescent spots from five cells, we found 93.5% (245 of 262 spots) bleached in one step, 6.1% (16 of 262) bleached in two steps, and 0.4% (1 of 262) bleached in three steps (Fig. 2 B and C). This dominance of one-step bleaching was consistent with the expectation that TβRII was monomer instead of dimer. Meanwhile, the fluorescent dwell time of the spots with one bleaching step was fitted with a single exponential decay function (Fig. 2D). The decay time constant was 3.1 ± 0.6 s and close to that of single GFP proteins (2.4 ± 0.3 s) imaged on coverslips (Fig. S2C), also suggesting that TβRII-GFP receptor was monomer in the TβRII-low-expressing cells.
Fig. 2.
Analysis of bleaching steps of single TβRII-GFP molecules imaged with the fixed HeLa cells. (A) A typical image shows the diffraction-limited fluorescent spots of TβRII-GFP on the fixed cell membrane. The spots enclosed with the green circles (3 × 3 pixels) were chosen for single molecule bleaching analysis. (Scale bar, 2 μm.) (B) Two representative time courses of GFP emission after background correction show one step bleaching. (C) Frequency of one-, two-, and three-step bleaching events (n = 262) revealed TβRII-GFP monomer. (D) Distribution of the fluorescence dwell time of individual fluorescent spots (n = 130). The decay time constant is 3.1 ± 0.6 s determined by the single exponential decay fitting function.
As the subunit counting of TβRII-GFP might be interfered by the endogenous TβRII on HeLa cells, we further examined MCF7 cells which are lack of endogenous TβRII to rule out the effect of the endogenous receptors (25). Single molecule imaging of TβRII-GFP transfected in resting MCF7 cells were obtained under similar conditions mentioned for HeLa cells. The statistical analysis of the fluorescence intensity, bleaching steps and fluorescence dwell time were all in agreement with those in HeLa cells (Fig. S3), indicating that TβRII-GFP molecules were also monomers in MCF7 cells. Thus these data suggested that endogenous TβRII in HeLa cells had little effect on our single-molecule observation of TβRII-GFP.
Monomeric TβRII Molecules Undergo Dimerization After TGF-β1 Stimulation.
Receptor dimerization is regarded essential for growth factor receptor activation, but the serine/threonine kinase receptors, like TβRII, are suggested to be unique as they exist as ligand-independent homo-oligomeric complexes in previous signaling model (10–12). As most of the TβRII–GFP molecules observed by our single-molecule fluorescence microscopy were monomers, we investigated whether the monomeric TβRII molecules undergo dimerization in the present of ligands. The cells were stimulated with TGF-β1 at 3.5 h after transfection and were kept at 4 °C for 15 min to avoid receptor internalization. Then the cells were imaged directly by the TIRF microscope or fixed before imaging.
First, we analyzed fluorescence intensity distribution of the diffraction-limited spots in the living HeLa cells after TGF-β1 stimulation. Histograms of the intensity distribution were also fitted with a sum of two Gaussian distributions, representing monomers and dimers (Fig. 3A). However, within 254 spots from six cells, 58% were monomers and 42% were dimers, which was in contrast to 86% monomers and 14% dimmers in the absence of ligand. Therefore, TGF-β1 stimulation resulted in a significant increase in the second population representing dimers in the distribution.
Fig. 3.
TGF-β-induced TβRII dimerization. (A) Distribution of the fluorescence intensity of diffraction-limited TβRII-GFP spots (n = 254) from the living cell imaging. After transfected with TβRII-GFP for 3.5 h, HeLa cells were treated with 200 pM TGF-β1 for 15 min at 4 °C before TIRFM imaging. The solid curves show the fitting of Gaussian function and the two peaks represented TβRII-GFP monomers and dimers, respectively. Correlation coefficient of the Gaussian fitting is 0.999. The arrowheads indicate the peak positions of the fitting curves. Numbers in the parentheses are the percentage of the fractions. (B) Two representative time courses of GFP emission after background correction show two-step bleaching. (C) Frequency of one-step and multistep bleaching events for TβRII-GFP (n = 252) in fixed HeLa cells.
Furthermore, according to the bleaching analysis of 8 fixed cells which were treated with TGF-β1, 69.4% (175 of 252 spots) bleached in one step, 29.8% (75 of 252) bleached in two steps, 0.8% (2 of 252) bleached in three steps (Fig. 3 B and C). The fraction of two-step bleaching dimers increased obviously (about 24%). The results from both the intensity and the bleaching analysis suggested that monomeric TβRII-GFP dimerizes upon TGF-β1 stimulation.
Comparison with Ligand-Induced Dimerization of EGFR Receptors.
It has been well established that the typical activation process of tyrosine kinase receptors, such as EGFR, is started with ligand-induced dimerization of the monomeric receptors existing on the cell membrane (26). We then imaged the GFP-tagged EGFR in the living or fixed cells for a comparison with TβRII activation. The expression level of EGFR-GFP was also kept low to observe the individual fluorescent spots (20–100 spots per 20 × 20 μm). The results showed that the fluorescence intensity distribution of EGFR-GFP in the living cells had a similar pattern as TβRII-GFP and contained mainly two populations for monomer and dimer respectively (Fig. 4A). After EGF stimulation, the dimer population increased from 13% to 43%, while the monomer population decreased from 87% to 57%. For the fixed resting cells, 91.5% (226 of 247 spots from six cells) bleached in one step, 8.5% (21 of 247) bleached in two steps (Fig. 4B). After EGF treatment, 66.3% (177 of 267 spots from eight cells) bleached in one step, 33.7% (90 of 267) bleached in two steps (Fig. 4B). Therefore, ligand-induced EGFR dimerization was demonstrated by our single-molecule fluorescence imaging, confirming our method was valid. The similar results of intensity distribution and photobleaching step counting between TβRII and EGFR supported that TβRII also exists as monomer like EGFR in the absence of the ligand, and dimerizes upon ligand stimulation.
Fig. 4.
Monomeric EGFR-GFP molecules in resting HeLa cells and their dimerization upon EGF stimulation. (A) Distribution of the fluorescence intensity of individual EGFR-GFP spots in living cells before (left, n = 246 from five cells) and after (right, n = 260 from six cells) EGF stimulation. Histograms of intensity fitted to a sum of two Gaussian distributions. Multiple correlation coefficients of the Gaussian fitting are 0.980 (left) and 0.999 (right). The arrowheads indicate the peak positions of the fitting curves. Numbers in the parentheses are the percentage of the fractions given by the Gaussian fitting. (B) Frequency of one-step and multistep bleaching events for EGFR-GFP without (gray bar, n = 247 from six cells) and with EGF stimulation (light gray bar, n = 267 from eight cells) in fixed HeLa cells.
In the previous reports of subunits counting of the GFP fused membrane proteins in living cells by single molecule technique, it has been found that the distribution of photobleaching steps for the tetrameric protein containing 1, 2, 3 or 4 GFP tags fitted to the binomial distribution with a probability of about 80% GFP to be fluorescent (24). Therefore, for the protein with one GFP-labeled subunit, the populations for one-, and two-step bleaching were 96% and 4%, respectively, while for the protein with two GFP-labeled subunits, the populations for one-, and two-step bleaching were changed to about 25% and 75% respectively. Our result of more than 90% of one-step photobleaching for the single TβRII or EGFR molecules in resting cells was consistent with that reported for the protein with one GFP-labeled subunit, further confirming the monomeric status of both TβRII and EGFR. However, in the ligand-treated cells, the population of the two-step bleaching for either TβRII or EGFR (about 30%) was significantly lower than that observed for the protein with two GFP-labeled subunits. A possible reason is that ligands only induced a portion of receptors to form complexes (27), and this is different from the constitutively assembled proteins with two GFP-labeled subunits.
TGF-β1-Induced Dimerization of TβRII Is TβRI-Independent.
According to the existing TGF-β signaling model, TGF-β1 initiates signaling by binding to the type II receptor homodimers and sequential recruiting type I receptors (1–4, 7). To test whether TGF-β1 could induce the formation of stable TβRII dimers without TβRI, we imaged TβRII-GFP molecules in R1B cells which are deficient in TβRI receptors (1). Fluorescence intensity analysis and subunit counting were carried out with living cells and fixed cells, respectively (Fig. S4). Similar results were obtained with R1B cells comparing to those with HeLa cells, suggesting that TGF-β1 induced TβRII dimerization was independent of TβRI receptors. The result was in agreement with the sequential binding mode of TGF-β1 to TβRII and TβRI (7).
TβRII Oligomerize in Resting Cells When Highly Expressed.
The above results obtained by single-molecule microscopy provide insight into TGF-β receptors oligomerization. Our finding that TβRII exists in the monomeric form in the absence of ligand is different from what have been reported before with conventional biochemical methods. We therefore attempted to test whether the previously reported homodimeric TβRII in resting cells was resulted from high protein expression.
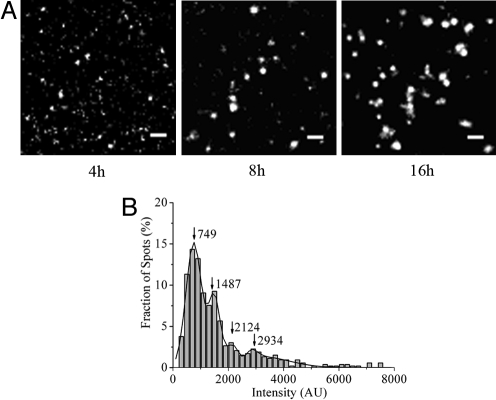
With a longer expressing time (e.g., at 8 h after transfection), expression of TβRII-GFP increased significantly (Fig. S1). When imaging the cells with extended expression time, we found that the intensity of individual TβRII-GFP fluorescent spots increased, and discrete puncta appeared with much longer fluorescence dwell time (more than 20 s) than monomers (Fig. 5A and Movie S3). For example, after 8 h transfection (the expression level increased 2–3 fold compared to the one at 4 h), TβRII-GFP molecules were observed as individual spots with a broad range of fluorescence intensities and the monomer population decreased dramatically (Fig. 5B). This suggested that TβRII-GFP molecules form oligomers with various numbers of TβRII monomers. As these puncta diffused on the membrane surface as one particle and didn't divide into more spots, they were most likely protein aggregates instead of several protein molecules which were accidentally colocalized within diffraction-limited spots. When the expression time was extended to 16 h, the size and density of TβRII-GFP aggregates further increased (Fig. 5A and Movie S4). These results indicated that TβRII molecules would self-assemble and oligomerize at high concentration on the cell membranes.
Fig. 5.
Oligomerization of TβRII-GFP on the cell membranes with the increased expression time. (A) Typical images (after background subtraction) showing the density and size of TβRII-GFP monomers, oligomers and aggregates on cell membranes imaged at 4 h (Left), 8 h (Center), and 16 h (Right) after transfection. (Scale bar, 2 μm.) (B) The histogram of the fluorescence intensities of individual diffraction-limited fluorescent spots (n = 530) on living cell membranes imaged at 8 h after transfection. Correlation coefficient of the Gaussian fitting is 0.995.
Several studies have shown that protein overexpression often leads to the formation of aggregates in both prokaryocytes and eukaryocytes (28, 29). That is true even for the tyrosine kinase receptors EGFR, where a model of EGF-induced dimerization of monomeric receptors has been well demonstrated with the endogenous receptors in the cell (26). In the studies of the transfected cells with overexpressed exogenous receptors, the ligand-independent formation of dimeric or oligomeric EGFR was found to be a step separable from EGF-induced EGFR dimerization (30–32). These studies strongly suggest that the protein expression level of membrane proteins determine their various oligermerization status.
As for TGF-β signal transduction, although homo-oligomeric interactions of TβRII in the absence of ligand have been demonstrated in overexpression systems (10–12), recombinant human TβRII extracellular domain (TβRII-ECD) was reported as monomer, and homodimerization of TβRII-ECD was induced by TGF-β (11, 33). Therefore, it is possible that TβRII exists as monomer at low density or under the physiological condition on the cell membranes.
With the developed single-molecule fluorescence microscopy, we have provided the evidence of the existence of the TβRII monomers in living cells at low expression levels close to the endogenous ones in the testing cells. We also showed that TβRII could oligomerize under high expression conditions, which might prevent the observation of TβRII monomer in previous ensemble measurements with over-expressing systems. Although our results indicated that TGF-β induces the dimerization of monomeric TβRII in living cells, our results did not exclude the possibility that the ligand promotes homo- or hetero-oligemerization of TβRII homodimers.
In summary, using real-time imaging of single TβRII molecules on the living cell surface, we have uncovered the ligand-induced receptor dimerization. Our single-molecule imaging method revealed the monomer status of TGF-β receptors in resting cells. These results suggest that as for tyrosine kinase receptors, the mode of receptor activation via dimerization of monomers can be generalized to the serine/threonine kinase receptors. Therefore, single-molecule fluorescence imaging provides an approach to study the molecular interaction of TGF-β receptors and their activation process in signal transduction.
Methods
Plasmid Construction.
The DNA fragments encoding full-length TβRII and EGFR were subcloned into the HindIII and BamHI sites of pEGFP-N1 (Clontech), yielding the TβRII-GFP and EGFR-GFP expression plasmids. The plasmids were confirmed by DNA sequencing.
Cell Culture and Transfection.
HeLa or MCF7 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (HyClone) at 37 °C in 5% CO2. R1B cells were cultured in MEM (Gibco) with 10% FBS. HeLa cells were used for most of the experiments unless specified. Transfection was performed using lipofectamine2000 (Invitrogen). Cells growing in a 35-mm glass-bottom dish (Shengyou Biotechnology) were transfected with 0.2 μg/mL plasmids in the serum-free and phenol red-free DMEM or the serum-free and phenol red-free MEM (the minimal medium). To achieve a low-level protein expression, cells were incubated with the plasmid for less than 4 h, washed, and then imaged in the minimal medium under the fluorescence microscopy. To increase the protein-expression level, cells were serum-starved for the first 4 h with the plasmid, washed, changed to the completed DMEM or MEM medium with serum for another 4–12 h, and followed by fluorescence imaging in the minimal medium.
For the ligand stimulation experiments, the transfected cells which were ready for fluorescence imaging were added with 200 pM TGF-β1 (R&D) or EGF in the minimal medium for 15 min at 4 °C before fluorescence imaging. For fixed cell imaging, the transfected cells were washed with cold PBS (4 °C) twice and fixed in cold 4% paraformaldehyde/PBS solution for 10 min.
Single Molecule Fluorescence Imaging.
Single molecule fluorescence imaging was performed with objective-type total internal reflection fluorescence (TIRF) microscopy using an inverted Olympus IX71 microscope equipped with a total internal reflective fluorescence illuminator'a, 100×/1.45NA Plan Apochromat TIR objective and an intensified CCD (ICCD) camera (Pentamax EEV 512 × 512 FT, Roper Scientific) (34). The microscope was equipped with a CO2 incubation system (TOKAI HIT) and all living cell imaging was performed at 37 °C. GFP was excited at 488 nm by an argon laser (Melles Griot) with the power of 6 mW measured after the laser passing through the objective. The collected fluorescent signals were passed through two filters, BA510IF and HQ 525/50 (Chroma Technology), before directed to the ICCD camera. The gain of the ICCD camera was set at 90. As the intensity at the edge of the illumination field of TIRF microscope was about 80% of that in the center, only the central quarter of the chip (256 × 256 pixels) was used for imaging analysis to ensure homogeneous illumination. Movies of 100–500 frames were acquired for each sample at a frame rate of 10 Hz.
For the control experiment of single GFP molecule imaging on coverslips, GFP protein purified from E. coli was first dissolved in the high salt buffer (600 mM NaCl, 150 mM PBS buffer, pH 7.4) to prevent the dimer formation and then immobilized on the coverslips through biotin coupled GFP antibody (Clontech) as previously reported (34).
Image Analysis.
For analysis of single-molecule fluorescence intensity in a movie acquired from living cells, the background fluorescence was first subtracted from each frame using the rolling ball method in Image J software (National Institutes of Health). Then the first frame of each movie was used for fluorescent spot (regions of interest) selection. The image was thresholded (four times of the mean intensity of an area with no fluorescent spots), then filtered again with a user-defined program in Matlab (MathWorks Corp.) to remove discrete signals. For example, the spots covering less than three pixels were regard as noise and discarded. After the image process, the brightest pixel of each fluorescent spot within diffraction-limited size (3 × 3 pixels) was determined as the central position and a square of 3 × 3 pixels was enclosed as a region of interest to calculated integrated fluorescence intensity by MetaMorph 6.1. The spot with its peak pixel very close to another spot (<3 pixels) was excluded.
To analyze the bleaching steps, regions of interest for bleaching analysis were selected according to the method previously reported (22). Firstly, the background fluorescence was subtracted from the movie acquired from the fixed cells using the rolling ball method in Image J software. Then the first five frames of the movie were averaged. The averaged image was thresholded and filtered with the same method mentioned above for the intensity analysis. Finally, time courses of the integrated fluorescence intensity of regions of interest were extracted for bleaching analysis. Traces with erratic behavior and ambiguities (30% of traces) were discarded.
Immunoblotting.
Immunoblotting was performed as described previously to estimate the expression level of TβRII-GFP (35). Briefly, HeLa cells or those transfected with TβRII–GFP after a certain period time were lysed with the buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, and protease inhibitors), and the protein amount in the lysates was determined by a spectrophotometer. Equal amount of the lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and the immunoblotting was performed with anti-TβRII or anti-tubulin antibodies and secondary antibodies conjugated to horseradish peroxidase. Proteins were visualized by chemiluminescence.
Supplementary Material
Acknowledgments.
This work was supported by National Natural Science Foundation of China (Nos. 90713024, 20821003), the National Basic Research Program of China (2007CB935601, 2004CB720002, 2006CB910102), and Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908279106/DCSupplemental.
References
- 1.Boyd FT, Massague J. Transforming growth factor-beta inhibition of epithelial cell proliferation linked to the expression of a 53-kDa membrane receptor. J Biol Chem. 1989;264:2272–2278. [PubMed] [Google Scholar]
- 2.Massague J, Chen Y. Controlling TGF-β signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 3.Miyazono K. Positive and negative regulation of TGF-β signaling. J Cell Sci. 2000;113:1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- 4.Dijke PT, Hill CS. New insights into TGF-β-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 6.Feng X, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 7.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 8.Moustakas A, Souchelnytskyi S, Heldin C. Smad regulation in TGF-β signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 9.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen RH, Derynck R. Homomeric interactions between type II transforming growth factor-beta receptors. J Biol Chem. 1994;269:22868–22874. [PubMed] [Google Scholar]
- 11.Gilboa L, Wells RG, Lodish HF, Henis YI. Oligomeric structure of type I and type II transforming growth factor-β receptors: Homodimers form in the ER and persist at the plasma membrane. J Cell Biol. 1998;140:767–777. doi: 10.1083/jcb.140.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henis YI, Moustakas A, Lin HY, Lodish HF. The types II and III transforming growth factor-β receptors form homo-oligomers. J Cell Biol. 1994;126:139–154. doi: 10.1083/jcb.126.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldin C. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 14.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 15.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iino R, Koyama I, Kusumi A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys J. 2001;80:2667–2677. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakoshi H, et al. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 19.Xie XS, Trautman JK. Optical studies of single molecules at room temperature. Annu Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 20.Xie XS, Yu J, Yang WY. Living cells as test tubes. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 21.Haggie PM, Verkman AS. Monomeric CFTR in plasma membranes in live cells revealed by single-molecule fluorescence imaging. J Biol Chem. 2008;283:23510–23513. doi: 10.1074/jbc.C800100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji W, et al. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci USA. 2008;105:13668. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 24.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Meth. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, et al. Expression of transforming growth factor beta type II receptor leads to reduced malignancy in human breast cancer MCF-7 cells. J Biol Chem. 1994;269:26449–26455. [PubMed] [Google Scholar]
- 26.Cochet C, Kashles O, Chambaz EM, Borrello I, King CR, Schlessinger J. Demonstration of epidermal growth factor-induced receptor dimerization in living cells using a chemical covalent cross-linking agent. J Biol Chem. 1988;263:3290–3295. [PubMed] [Google Scholar]
- 27.Yu C, Hale J, Ritchie K, Prasad NK, Irudayaraj J. Receptor overexpression or inhibition alters cell surface dynamics of EGF-EGFR interaction: New insights from real-time single molecule analysis. Biochem Biophys Res Commun. 2009;378:376–382. doi: 10.1016/j.bbrc.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Bu P, Zhuang J, Feng J, Yang D, Shen X, Yan X. Visualization of CD146 dimerization and its regulation in living cells. Bba-Mol Cell Res. 2007;1773:513–520. doi: 10.1016/j.bbamcr.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Geertsma ER, Groeneveld M, Slotboom D, Poolman B. Quality control of overexpressed membrane proteins. Proc Natl Acad Sci USA. 2008;105:5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadella TW, Jovin TM. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol. 1995;129:1543–1558. doi: 10.1083/jcb.129.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P, et al. Investigation of the dimerization of proteins from the epidermal growth factor receptor family by single wavelength fluorescence cross-correlation spectroscopy. Biophys J. 2007;93:684–698. doi: 10.1529/biophysj.106.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E. Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell. 2002;13:2547–2557. doi: 10.1091/mbc.01-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letourneur O, Goetschy J, Horisberger M, Gr MG. Ligand-induced dimerization of the extracellular domain of the TGF-β receptor Type II. Biochem Biophys Res Commun. 1996;224:709–716. doi: 10.1006/bbrc.1996.1088. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Z, et al. Single-molecule study of lateral mobility of epidermal growth factor receptor 2/HER2 on activation. J Phys Chem B. 2008;112:4140–4145. doi: 10.1021/jp710302j. [DOI] [PubMed] [Google Scholar]
- 35.Zuo W, Chen Y. Specific activation of mitogen-activated protein kinase by transforming growth factor-β receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell. 2009;20:1020–1029. doi: 10.1091/mbc.E08-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.