Abstract
Promyelocytic leukemia (PML) bodies (also called ND10) are dynamic nuclear structures implicated in a wide variety of cellular processes. ALT-associated PML bodies (APBs) are specialized PML bodies found exclusively in telomerase-negative tumors in which telomeres are maintained by recombination-based alternative (ALT) mechanisms. Although it has been suggested that APBs are directly implicated in telomere metabolism of ALT cells, their precise role and structure have remained elusive. Here we show that PML bodies in ALT cells associate with chromosome ends forming small, spatially well-defined clusters, containing on average 2–5 telomeres. Using an innovative approach that gently enlarges PML bodies in living cells while retaining their overall organization, we show that this physical enlargement of APBs spatially resolves the single telomeres in the cluster, but does not perturb the potential of the APB to recruit chromosome extremities. We show that telomere clustering in PML bodies is cell-cycle regulated and that unique telomeres within a cluster associate with recombination proteins. Enlargement of APBs induced the accumulation of telomere-telomere recombination intermediates visible on metaphase spreads and connecting heterologous chromosomes. The strand composition of these recombination intermediates indicated that this recombination is constrained to a narrow time window in the cell cycle following replication. These data provide strong evidence that PML bodies are not only a marker for ALT cells but play a direct role in telomere recombination, both by bringing together chromosome ends and by promoting telomere-telomere interactions between heterologous chromosomes.
Keywords: ALT-associated PML bodies (APBs), alternative lengthening of telomeres (ALT), Herpes simplex virus ICP0, telomere clusters
PML (promyelocytic leukemia) nuclear bodies, which are present in most cells, have been extensively studied and a wealth of information concerning their composition, structure, dynamics and function is available (1). The PML protein is essential for the formation of PML bodies and provides the structural scaffold to which other proteins bind. Over 60 additional proteins have been localized to PML bodies spatially or temporally [see the information on PML bodies from the Nuclear Protein Database, (2)], and as a consequence, PML bodies have been implicated in the regulation of virtually every biological function including DNA damage responses.
In immortal cells that maintain telomeres by recombination-based alternative lengthening mechanisms (ALT), a special variety of PML bodies is found that, in addition to the proteins normally associated with PML bodies, contain telomeric DNA, telomere-specific proteins, and DNA recombination and repair proteins (3). The structure and the role of these ALT-associated PML bodies (APBs) are unknown. There is, however, a clear association between the presence of APBs and the utilization of ALT for telomere maintenance. However, in those studies disruptions of APBs have often been achieved by modulating the expression of proteins that also play important roles in normal telomere maintenance, such as shelterins or proteins of the MRE11/RAD50/NBS1 (MRN) complex (4–7). Therefore it has been difficult to determine the extent to which the observed telomere shortening is a consequence of a perturbed function of APBs as opposed to telomere dysfunction. On the other hand, it remains to be determine how much of the telomeric DNA associated with APBs belongs to true chromosome ends as opposed to extra-chromosomal telomeric repeat (ECTR) DNA also present in ALT cells (8, 9). For instance, it has been proposed that APBs allow ALT cells to prevent inappropriate damage responses by sequestering linear DNA with unrepaired ends (9). Finally, whether telomere recombination/lengthening reactions involving chromosome ends actually take place in APBs is still unknown.
We took advantage of recent improvements in imaging techniques to study the organization of APBs. Since APBs large enough to be examined this way are not very frequent in ALT cells, we used an innovative approach based on the properties of the Herpes simplex virus protein ICP0. This protein, previously called Vmw110, accumulates specifically and simultaneously in PML bodies and at centromeres, inducing the proteasomal-dependent degradation and destabilization of both structures (10, 11). Deletion of its ring finger domain prevents ICP0 from localizing and inducing damage at centromeres. Although the mutated protein still localizes to PML bodies, the ring finger deletion completely abolishes the capacity of ICP0 to induce PML degradation and nuclear body disruption, leading instead to protein accumulation and body enlargement (12). Using this approach, we were able to show that PML bodies in ALT cells are able to both recruit true chromosome ends and foster telomere-telomere recombination reactions.
Results
Telomeres Are Not Randomly Distributed but Cluster Around PML Bodies in Telomerase-Negative Tumor Cells.
Telomeres in both normal mammalian somatic cells or telomerase positive tumor cells are apparently randomly distributed in interphase nuclei, regardless of the cell type (13). The number of visible telomere spots within a nucleus typically approaches that of the expected number of chromosome extremities, indicating that telomeres are not clustered. In contrast, in telomerase-negative human cell lines the number of telomere spots detected in nuclei is surprisingly low, and can, instead, be very bright (Fig. S1 A and B). Bright telomeric spots usually correspond to APBs, i.e.: they also contain PML body components. We therefore hypothesized that PML bodies in ALT cells contain multiple telomeres. Using immunofluorescence (IF), we closely examined the organization of naturally large APB structures present in the telomerase-negative immortalized cell line VA13. Strikingly, 3D imaging followed by deconvolution analysis revealed that the shelterin protein TRF2, a telomeric marker, no longer forms a unique focus with the PML signal. Instead, multiple TRF2 foci are detected in association with the outermost layer of these APBs, whereas other components of PML bodies, for instance the BLM protein (14), are preferentially associated with the inner layers (Fig. 1A). We speculated that these multiple individual TRF2 foci normally associated with large APBs represent single telomeres that have clustered.
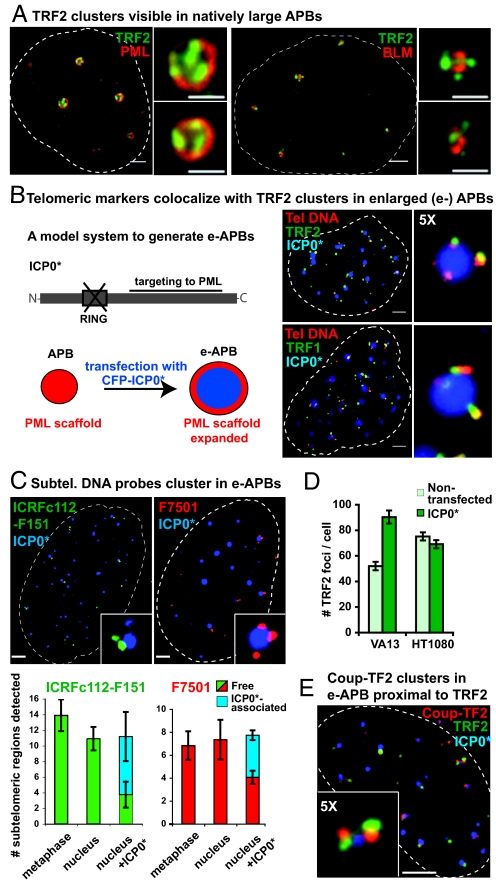
Fig. 1.
Chromosome ends associate with PML bodies to form APBs in VA13 cells. (A) Individual telomeres as detected by anti-TRF2 antibody (green) may appear associated with the surface of naturally large APBs, revealed here by either anti-PML or anti-BLM antibodies (both red). (B) Viral protein ICP0 lacking the ring finger domain fused to CFP (CFP-ICP0*, blue) was used to transiently enlarge the size of APBs (hereafter e-APBs). In e-APBs, telomeric DNA detected by PNA FISH (red) colocalizes with either TRF2 or with TRF1 protein foci (green). (C) Subtelomeric probes ICRFc112-F151 (green) and F7501 (red), cluster in e-APBs containing CFP-ICP0* (blue). These probes detect only a specific subset of chromosome arms (Fig. S3A). Histograms showing the average number of subtelomeric regions detected in 20 metaphase spreads, cell nuclei, and nuclei containing ICP0*. ICP0* associated, subtelomeric signals in proximity of ICP0*; Free, elsewhere in the nucleus. Error bars represent the standard error of the mean (SEM). (D) Histogram showing the average number of TRF2 foci expected and actually detected in interphase nuclei of native and ICP0*-transfected VA13 and HT1080 cells. Error bars, SEM. (E) Coup-TF2 is localized in close proximity of TRF2 clusters in e-APBs. (Scale bars, 2 μm and 1 μm in enlarged images.)
To test this hypothesis, we investigated whether telomeric DNA and other shelterin proteins co-localized with each individual TRF2 focus in large APBs. Since large APBs are naturally rare, we used a biological tool based on the expression of the Herpes simplex virus protein ICP0. It has previously been demonstrated that ICP0 ring finger deletion mutant (ICP0* in this paper) accumulates in PML bodies and significantly enlarges their size (12). We confirmed that a fluorescently tagged version of ICP0* also has the capacity to transiently enlarge APBs (Fig. S2 A–D). Transient transfection with ICP0* did not interfere with cell cycle progression of VA13 cells nor did it affect protein levels of major components of PML bodies or APBs (Fig. S2E). Moreover, IF co-localization studies revealed that ICP0* exclusively accumulated in the core of PML bodies or of APBs, while clusters of individual TRF2 foci were easily detected surrounding the PML shell of ICP0*-enlarged APBs (hereafter called e-APBs). Furthermore, we found that every single TRF2 focus in e-APBs colocalized with telomeric DNA and that the telomeric DNA in turn colocalized with another shelterin protein, TRF1 (Fig. 1B), indicating the presence of several individual, bona fide telomeric structures in APBs.
Chromosome Ends, Not Extra-Chromosomal Telomeric DNA, Are the Main Component of the Telomeric Material in APBs.
We wished to know whether the telomeric signals that cluster around e-APBs correspond to bona fide chromosome ends. Currently, evidence that chromosome ends associate with APBs is lacking. Telomeric markers, such as PNA telomeric probes and telomeric proteins, have been localized to APBs. However, telomerase-negative tumor cells also contain linear and circular ECTR DNA, which presumably binds shelterin proteins and may be sequestered into APBs (8, 9). To determine the nature of telomeric material present in APBs we looked for the presence of true chromosome end markers in association with APBs. We performed FISH experiments using two subtelomeric probes, ICRFc112-F151 and F7501, each specific for independently duplicated subtelomeric sequences distributed among several chromosome extremities and located at 10–20 kb from the telomeric tract (15) (Fig. S3A). Subtelomeric sequences are not found in extra-chromosomal telomeric circles (16) and are not expected to be present at detectable levels in linear low molecular-weight telomeric DNA. We frequently detected these subtelomeric regions around e-APBs (Fig. 1C), while no such association was seen with enlarged (e-) PML in HT1080 control cells (Fig. S3B). Quantification of these associations showed that while these probes detected comparable numbers of chromosome extremities on metaphase spreads and in whole nuclei, about half of signals detected in the latter were found in proximity of e-APBs. Although multiple colocalization of subtelomeric probes with the same APB is expected to be a rare event since these probes recognize only a small subset of chromosome extremities (1 out of 10 in the case of ICRFc112-F151 and 1 out of 20 for F7501), we did occasionally observe clusters of subtelomeric regions, thus confirming that multiple chromosome ends can cluster around APBs (Fig. 1C, enlarged images). In conclusion, these experiments, without ruling out the possibility that ECTR DNA is also present, strongly suggest that chromosome ends are the source of telomeric material detected in APBs.
Further supporting this interpretation is our observation that the number of individual TRF2 foci that become detectable in VA13 cells upon ICP0*-mediated enlargement of APBs is much closer to the expected number of telomeres (given the ploidy), while this number does not change upon expression of ICP0* in telomerase-positive HT1080 cells in which telomeres do not associate with PML (Fig. 1D).
In addition, orphan receptor family members have been recently found to be enriched at telomeric material purified from ALT cells (17). Independent biochemical evidence suggests that these proteins primarily bind to variant telomeric repeats typically present in juxtatelomeric and subtelomeric regions (18). We found, that one such orphan receptor, Coup-TF2, formed clusters reminiscent of TRF2 in natively large APBs and are also in close proximity to TRF2 foci in e-APBs (Fig. 1E), although they do not fully overlap, supporting the interpretation that Coup-TF2 binds to regions proximal to telomeric repeats. These and the above data, support a model in which chromosome extremities associate with the surface of the PML scaffold to form APBs.
Telomere Clusters Are a Common Characteristic of Telomerase-Negative Tumor Cells.
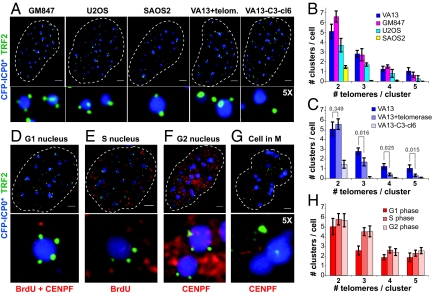
We next wanted to know whether telomere clustering is a characteristic of other ALT cells. We examined the effect of ICP0*-mediated APB enlargement in other ALT cell lines, derived either from in vitro spontaneous immortalization of SV-40 transformed cells or from telomerase-negative human tumors. Telomere clusters were found in all APB-containing ALT cell lines tested, albeit at different frequencies (Fig. 2 A and B). They were more frequent and contained more individual telomeres in the SV40-transformed fibroblast lines VA13 and GM847 than in the osteosarcoma-derived cell lines U2OS and SAOS2. In the cell lines with the most abundant telomere clusters, little less than half of all detectable telomeric signals in interphase nuclei were associated with APBs (Table 1). On average, ALT cells displayed several small clusters containing 2–3 telomeres and at least one large cluster with up to five telomeres. Consistent with the observation that telomerase expression in ALT cells most often does not disrupt APBs (19), we observed telomere clusters in VA13+telomerase cells (Fig. 2A). However, clusters bearing more than two telomeres were significantly less frequent in the presence of telomerase (Fig. 2C), suggesting that elongation of the shortest telomeres by telomerase may affect the recruitment of chromosome extremities to APBs. In an atypical ALT cell line derived from VA13 cells (VA13-C3-cl6), in which APBs are virtually absent (20), telomere clusters were rarely detected (Fig. 2 A and C), as expected. This observation indicates that ICP0* expression is not sufficient to induce telomere clustering in cells that maintain telomeres by recombination but lack APBs.
Fig. 2.
Telomere clusters are a common feature of ALT cells. (A) Telomere clusters, detected by colocalization of anti-TRF2 antibodies (green) with CFP-ICP0* (blue), are present in all ALT lines tested (GM847, U2OS, SAOS2), and in VA13 cells constitutively expressing telomerase (VA13+telom.), but are rarely observed in an atypical ALT line without APBs (VA13-C3-cl6). (Scale bars, 2 μm.) (B and C) Quantification of results from (A) based on 20 nuclei corrected for the level of random co-localization of TRF2 with ICP0* as observed in HT1080 cells (2 = 0.65 ± 0.21, 3 = 0.50 ± 0.17, 4 = 0, 5 = 0). Error bars, SEM. (D–G) Telomere clusters are present in G1, S, and G2 phase of the cell cycle but not in M. VA13 cells were transfected with CFP-ICP0* (blue) for 48 h, labeled with BrdU before fixation and stained with anti-TRF2 antibodies (green) and different markers for the cell cycle (red) as follows: G1 cells (D) were identified as negative after a double staining with markers for the S and G2 phases; S phase nuclei (E) were identified with anti-BrdU antibodies; G2 nuclei (F) were identified with anti-CENP-F antibodies and chromosome condensation allowed to distinguish CENP-F positive cells that were in M (G). (H) Quantification of results from (D–G). Error bars, SEM.
Table 1.
Quantification of the number of telomeres associated with APBs in different cell lines
| Cell line | # ICP0* foci | # TRF2 foci | % TRF2 associated with ICP0* |
|---|---|---|---|
| GM847 | 42.0 ± 3.1 | 80.8 ± 4.1 | 46.1 ± 2.1 |
| VA13 | 35.9 ± 3.0 | 90.3 ± 5.1 | 40.3 ± 2.6 |
| VA13 + telomerase | 29.1 ± 1.2 | 74.4 ± 2.4 | 38.2 ± 0.9 |
| U2OS | 38.5 ± 2.4 | 72.1 ± 4.4 | 31.3 ± 2.0 |
| SAOS2 | 29.4 ± 1.9 | 58.3 ± 3.9 | 20.0 ± 3.6 |
| VA13-C3-cl6 | 42.5 ± 2.6 | 108.3 ± 3.3 | 8.7 ± 1.5 |
Different cell lines (see Fig. 2A) were transfected with CFP-ICP0* to create e-APBs and analyzed by immunofluorescence using anti-TRF2 antibodies 24 h post transfection. Average statistics with SEM are reported based on the analysis of 20 flat projections of 3D nuclear images per cell line. The percentage of TRF2 foci associated with ICP0* was corrected for the level of random colocalization of TRF2 with ICP0* due to the flattening projections of 3D stacks as determined in HT1080 cells (7.6% ± 0.9%).
Clustering of Telomeres Around PML Bodies Is Cell Cycle Regulated.
Since it has been reported that the number of detectable APBs tends to fluctuate in a cell-cycle dependent manner (21), we examined whether telomere cluster formation also oscillated during cell cycle progression. Telomere clusters were present in VA13 cells at similar frequencies cells from G1 through G2 phases of the cell cycle (Fig. 2 D–F). However, clusters were completely absent during mitosis (Fig. 2G), suggesting that telomeres actively disengage from the PML protein scaffold during this phase, presumably to ensure proper segregation of chromosomes. This cycling of APBs and formation of telomere clusters closely mimics the natural cycling of PML nuclear bodies, which form early in G1 and are relatively stable until late in G2 when a reorganization of the scaffold occurs with persistence of PML aggregates (22). We noted that ICP0* foci also persisted throughout mitosis in large aggregates, likely associated to the PML protein, but not to telomeres. This indicates again that the presence of ICP0* aggregates is not sufficient to induce telomere recruitment in ALT cells.
Unique Telomeres in Clusters Bear Marks of Recombination.
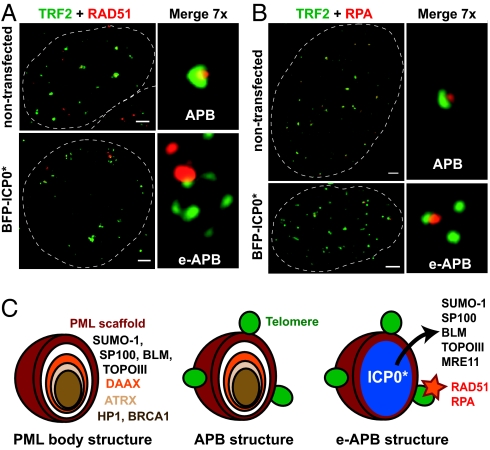
APBs contain, by definition, several DNA recombination and repair proteins (23). We therefore examined how these proteins partition in e-APBs. IF experiments using antibodies against RAD51 and replication protein A (RPA), two major proteins implicated in recombination pathways, revealed distinct and bright foci associated with the periphery of e-APBs. Co-localization studies readily indicated that a large majority of these foci are specifically associated with single telomeres (Fig. 3 A and B). Strikingly, these RAD51/RPA foci were most of the time associated with only one telomere in a cluster and therefore few such foci were usually detected per nucleus. Occasionally, both RAD51 and RPA foci lacking an associated TRF2 signal were observed on the periphery of an e-APB, suggesting the presence of recombination substrates bearing very little, or no, telomeric repeats. In contrast to RAD51 and RPA, several other components of either PML bodies or APBs (Table S1), such as the BLM helicase or MRE11, which play distinctive roles in DNA recombination (24), were not detected in association with telomeres in e-APBs but were instead found in the nucleoplasm (Fig. S4). It should be stressed that despite this induced redistribution, ICP0* expression did not affect total protein levels in these cells (Fig. S2E). Strikingly, our experiments indicate that the exclusion of MRE11, BLM, TOPOIIIα, and SP100 from e-APBs does not impact on the formation of such structures, since telomeres remained stably associated with the PML scaffold.
Fig. 3.
Recombination protein RAD51 and replication protein A (RPA) preferentially associate with particular telomeres in a cluster. (A and B) IF analyses of VA13 cells fixed 24 h posttransfection with BFP-ICP0* (not represented in the figures) revealing the co-localization of anti-RAD51 (A) or anti-RPA (B) staining (both red) with individual telomeres, as detected by anti-TRF2 antibody (green), in e-APBs. 24% (n = 20) and 35% (n = 20) of RAD51 and RPA nuclear foci, respectively, were found associated with TRF2 in e-APBs. Localization of the same proteins in unperturbed APBs in non-transfected cells is also shown. Scale bars, 2 μm. (C) Models of native PML and native and e-APB structures. Infiltration by ICP0* leads to displacement of proteins from the core of the PML scaffold but preserves the association of telomeres with the outer layer of the body.
APB-Associated Telomere Clustering Promotes Telomere-Telomere Recombination After Replication.
We finally addressed the question whether APBs play a functional role in telomere recombination. ALT cell lines are characterized by the presence of high levels of telomere exchange events between sister chromatids, T-SCEs (25). These exchanges can be revealed by the chromosome-oriented (CO-) FISH technique in which telomeric G-rich and C-rich parental strands can be distinctly visualized on metaphase chromosomes (Fig. 4A). If an exchange has occurred between sister telomeres, a mixed signal is detected on both sister chromatids. We found that in cells with e-APBs these T-SCE reactions occur at normal levels (Fig. 4B, see also Fig. S5). Therefore perturbing the structure of APBs did not interfere with postreplication recombination reactions between sister chromatids.
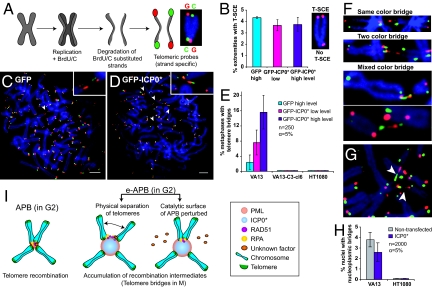
Fig. 4.
Enlargement of APBs perturbs the resolution of recombination intermediates between telomeres located on different chromosomes. (A) Chromosome-oriented (CO-) FISH procedure. (B) The level of telomere sister chromatid exchange (T-SCE) remains unperturbed in VA13 cells upon expression of ICP0* as determined by CO-FISH using a probe against the G-rich strand (red) (See also Fig. S5). n = 40–42 metaphases. Error bars, SEM. (C and D) CO-FISH analysis revealing telomere recombination intermediates or telomere bridges (arrowheads) whose incidence increases upon transfection of VA13 cells with ICP0*. (Scale bars, 5 μm.) (E) Percentage of metaphase spreads with telomere bridges in VA13, VA13-C3-cl6 and HT1080 cells. Error bars represent the confidence interval. (F) Representative images for different categories of telomere bridges based on the pattern of parental G-rich (red) and C-rich (green) telomeric DNA strands. (G) Two sister chromatids forming two independent telomere bridges (arrowheads) with two different chromosomes. (H) Increased incidence of telomere bridges is not accompanied by an increase in nucleoplasmic bridges. Error bars represent the confidence interval. (I) We propose two, non-exclusive, models of how ICP0* interferes with the recombination of telomeres in enlarged (e-)APBs: ICP0* may prematurely promote the physical separation of telomeres or perturb the catalytic surface of APBs perhaps by inducing the relocation of recombination factors from APBs into the nucleoplasm.
During our T-SCE analysis, we noticed that a significant proportion of metaphase spreads from ALT cells with e-APBs displayed thin fibers of telomeric DNA connecting two chromosome ends (Fig. 4 C–G). Extensive examination of control VA13 cells indicated that these bridges are present, albeit at much lower levels, in metaphases from ALT cells with unperturbed APBs (Fig. 4C), their frequency increasing proportionally to the expression level of ICP0* (Fig. 4E). Such telomere bridges were never detected in HT1080 cells, nor in ALT cells without APBs (VA13-C3-cl6) (Fig. 4E), suggesting that telomere-telomere interactions occur in APBs and that the enlargement perturbs such process. The CO-FISH analysis allowed us to reveal that the vast majority (over 85%) of these bridges correspond to telomeric DNA fibers in which parental C-rich and G-rich tracts alternate at least once (Fig. 4F). Such alternating signal pattern is expected on stretched telomere DNA fibers that have undergone recombination after replication. That telomere-telomere recombination occurs postreplicatively is further supported by the presence of bridges connecting sister telomeres from the same chromosome arm to two different chromosomes (Fig. 4G). Despite frequent telomere bridge formation during M phase in ALT cells highly expressing ICP0*, we did not detect any increase in the number of nucleoplasmic bridges (Fig. 4H), a biomarker for telomeric fusions (26), suggesting that these telomere-telomere interactions correspond to unstable intermediates. Together, our data provide evidence that in ALT cells telomeres on heterologous chromosomes can recombine in APBs following replication. We were unable to test the long-term consequences of enlarged APBs on telomere length maintenance, since expression of ICP0* is efficiently shut down by the cell after a few cycles.
Discussion
The structural and functional analyses of APBs presented here demonstrate that, in immortalized and tumor cells relying on alternative mechanisms for telomere maintenance, PML bodies bind bona fide chromosome ends and sequester telomeres, thus providing the required physical proximity for recombination to occur. Our observations suggest a structural model according to which APBs are organized, similarly to PML bodies (14), into concentric layers of proteins, PML constituting the outermost layer with which chromosome ends interact (Fig. 3C). It remains to be determined how telomeres interact with the PML scaffold and how they are recruited. The PML protein does not bind to naked DNA, although it may interact with chromatin (27) and thus particular chromatin marks may be required for APB formation. Alternatively, sumoylated telomeric proteins may directly interact with the SUMO binding domain of PML, consistent with the observation that sumoylation of TRF1/TRF2 by the SMC5/SMC6/MMS21 complex is required for APB formation (6). However the SMC5/SMC6/MMS21 complex accumulated in APBs only in late S/G2 phase, while we observed telomere cluster formation also in G1, suggesting that sumoylation is not involved in the initial targeting of telomeres to APBs but may be required to maintain this association. Since telomere signals can move over relatively long distances in live ALT cells (28) it is likely that the telomere clusters we observed assemble partially through this directional movement. In addition, fusions and fission of PML bodies probably contribute to the dynamics of APB formation.
Recently, lacO repeats inserted in proximity of chromosome ends in ALT cells were shown to associate with the PML protein (29). Nonetheless, these interactions display morphological characteristics that strongly resemble depicted associations of PML with foreign viral DNA (1, 30) or with hypomethylated heterochromatic DNA sequences (14). In these cases, and in contrast to telomeres clusters in APBs, the PML protein engulfs the DNA rather than the latter being associated with the surface of the PML body.
Our results also indicate that telomeres in PML bodies constitute an unexpected exception to the classic general view that telomeres show no preferential clustering in non-meiotic mammalian cells. Telomeres in somatic mammalian cells have been shown to be attached to the granular material of the nuclear matrix and randomly distributed around the nucleus (13). Here, we show that PML bodies have the capacity to recruit telomeres in some mammalian somatic cells into clusters. Although this clustering is reminiscent of the formation of telomeres bouquets during meiosis (31) or the formation of telomere clusters in vegetative budding yeast (32), one major difference is that, in the case of APBs, telomere clusters show no preference for a peripheral localization.
This report provides further and more direct evidence that telomeres on different chromosomes can directly recombine in ALT cells (33). Since the incidence of metaphase telomere bridges, which are already detectable at very low levels in native cells, increases dramatically upon ICP0* infiltration of APBs, it is reasonable to propose that such recombination occurs in APBs. Although the physical proximity of chromosome extremities in the native APB structures may favor the interaction between telomeres, proximity is clearly not sufficient, since telomeric bridges are never detected between individual telomeric structures around e-APBs in interphase nuclei. Instead, recombination events are only observed following replication, suggesting that passage of the replication fork allows telomeres in APBs to become uncapped and to interact. We propose that APBs provide both the required physical proximity and the required catalytic surface that promote telomere recombination (Fig. 4I), although they are probably not the unique place in the nucleus where telomere recombination occurs. It is also possible that recombining telomeres are recruited to APBs for resolution. It is not known how the choice of telomeres that will recombine in a given cell cycle is made. Nevertheless, the limited number of telomeres associated with RAD51 or RPA proteins and the limited number of telomere bridges that are detected in metaphase preparations of ALT cells highly expressing ICP0* both point to the existence of additional layers of regulation. Finally, our results stress the potential role of PML bodies in the formation of recombination centers involving chromosome domains in somatic cells.
Materials and Methods
Cell Culture and Plasmids.
WI38/VA13 clone 2RA (VA13) and GM847 are SV40 immortalized human lung embryonic and skin fibroblasts, respectively, while U2OS and SAOS2 are established osteosarcoma cell lines, all of which maintain their telomeres by ALT. VA13 cells that stably express hTERT and hTR (VA13+telomerase) maintain telomeres by both mechanisms, ALT and telomerase. VA13-C3-cl6 is a clonal cell line derived from VA13 that maintains telomeres by an atypical ALT pathway in the absence of APBs (20). HT1080 is a telomerase-positive human lung sarcoma cell line. Cells were maintained in culture as described (25). The plasmid carrying an N-terminal enhanced green fluorescent protein fusion to the ICP0 ring finger deletion (pAL30) has been described (10). The control plasmid pAL39 expressing the enhanced GFP alone was obtained by removing the ICP0* fragment from pAL30 using BglII and SmaI restriction sites. Color variants of the GFP-ICP0* fusion were generated by exchanging an internal fragment of GFP with the corresponding fragment from either the enhanced blue fluorescence protein to generate the BFP-ICP0* fusion (pAL118) or the enhanced cyan fluorescence protein to obtain the CFP-ICP0* fusion (pAL119).
Indirect Immunofluorescence (IF).
Fifteen thousand cells were seeded in 4-well cell culture cover slips (VWR) and transfected with plasmid DNA (pAL118 or pAL119) using Lipofectamine 2000 (Invitrogen). Alternatively, 1 million cells were transfected with 2 μg plasmid using AMAXA nucleofection technology. Twenty-four h or 48 h post-transfection, cells were fixed using the following protocol: 5 min in Solution T (0.5% Triton-X-100, 50 mM Tris, pH 8, 150 mM NaCl, 5 mM MgCl2, and 300 mM sucrose), 15 min in Solution F (3% formaldehyde, 1× PBS, and 300 mM sucrose), three washes with PBS, 10 min in Solution T. Fixed slides were stored in PBS at 4 °C. For IF, fixed cells were blocked with 10% goat or horse serum in PBS, incubated sequentially in different primary antibodies, followed by fluorescently labeled secondary antibodies. All incubation steps were done in a humid incubator at 37 °C for 30 min. Slides were mounted in Vectashield with 0.2 μg/mL 4′,6-diamidino-2-phenylindole (DAPI). Images were taken with a 3D deconvolution microscope (Leica DM6000 B or Nikon) using the MetaMorph software. Final images are composed of arithmetic stacks of 20–30 deconvolved images, each 0.2 μm in depth.
Antibodies.
Primary antibodies used were mouse anti-TRF1 (ab10579) 1/500, anti-TRF2 (IMG-124A) 1/500, anti-PML 5E10 1/50, anti-actin-HRP (sc-47778) 1/10,000, rabbit anti-RAD51 (Calbiochem, PC130) 1/500, anti-RPA-32 (GeneTex, RB-RPA32-UP100) 1/1,000, anti-WRN (sc-5629) 1/250, anti-SP100 (Chemicon, AB1380) 1/1,000, anti-PML (Chemicon, AB1370) 1/1,000, anti-BLM (ab2179) 1/100, anti-MRE11 (ab397) 1/1,000, anti-Coup-TF2 (ab50487) 1/100, anti-BRCA1 (ab2956) 1/100, anti-CENP-F (ab5) 1/400, goat anti-GFP (ab6673), and rat anti-BrdU (ab6326) 1/50. Secondary antibodies coupled to various Alexa fluorophores were used for detection (Invitrogen).
PNA FISH, and Subtelomeric FISH.
Fixed cells were co-denatured at 80 °C for 3 min in the presence of a Telomeric C-rich (CCCTAA)3 PNA probe (labeled with either Alexa 488 or Cy3, Applied Biosystems) dissolved at 5 μg/mL in a hybridization mix containing 70% formaldehyde, 10 mM Tris (pH = 7.2), 5% Mg-buffer (25 mM MgCl2, 9 mM citric acid, and 82 mM Na2HPO4), and 0.5% Boehringer blocking powder. Following 1 h incubation in the dark, slides were washed twice for 15 min in 50% formamide, 10 mM Tris (pH = 7.2), and 0.1% BSA, then three times for 5 min in 100 mM Tris (pH = 7.5), 100 mM NaCl, and 0.08% Tween-20. For detection of subtelomeric DNA in cell nuclei cosmids carrying subtelomeric regions, F7501 (GenBank accession number L78442) and ICRFc112-F151 (GenBank accession number Y13543), were obtained from Barbara Trask (Fred Hutchinson Cancer Research Center, Seattle, WA) and Gilles Vergnaud (IGM, Orsay, France), respectively. The FISH procedure was as described (15).
Chromosome-Oriented (CO-) FISH and T-SCE Analysis of Cells Expressing ICP0*.
Twenty million cells were transfected with 40 μg GFP-ICP0* (pAL30) or GFP (pAL39) using AMAXA, collected by trypsinization after 24 h and subjected to FACS. Cells expressing high or low levels of GFP were collected and maintained in culture for another 24 h in the presence of 10 μM BrdU/3.3 μM BrdC. Before harvesting, cells were treated with colcemide (0.5 μg/mL, 4 h) and following a hypotonic shock and fixation (ethanol:acetic acid, 3:1) metaphase spreads were prepared. The CO-FISH procedure was as described (25) with the following modifications: Consecutive hybridizations were carried out with a C-rich Cy3-(CCCTAA)3 PNA probe (Applied Biosystems) and a G-rich (Fam)GGGT+TAGGG+T+TAG+GGTTAGGG+T+TAG+GG+T+TAGGG+ TTA(Fam) LNA probe (Proligo) The latter was applied at 5 μg/mL (25–30 μL per slide) in a hybridization mix containing 50% formamide, 2× SSC, and 0.5% Boehringer blocking powder.
Supplementary Material
Acknowledgments.
We thank Zofia Maciorowski and Annick Viguier for help with cell sorting, Stéphane Koundrioukoff for help with immunofluorescence assays, and all lab members for comments on the manuscript. We thank the Nikon Imaging Centre at Institut Curie-CNRS and the PICT-IBiSa Imaging Facility. This work was supported by grants from Agence Nationale pour la Recheche, La Ligue Contre le Cancer, and La Association pour la Recherche contre le Cancer, (to A.L.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907689106/DCSupplemental.
References
- 1.Bernardi R, Pandolfi PP. Structure, dynamics, and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 2.Dellaire G, Farrall R, Bickmore WA. The Nuclear Protein Database (NPD): Sub-nuclear localization and functional annotation of the nuclear proteome. Nucleic Acids Res. 2003;31:328–330. doi: 10.1093/nar/gkg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeager TR, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 4.Jiang WQ, et al. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol Cell Biol. 2005;25:2708–2721. doi: 10.1128/MCB.25.7.2708-2721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- 6.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 7.Zhong ZH, et al. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 8.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasching CL, Neumann AA, Muntoni A, Yeager TR, Reddel RR. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- 10.Everett RD, Earnshaw WC, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett RD, et al. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luderus ME, et al. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luciani JJ, et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci. 2006;119:2518–2531. doi: 10.1242/jcs.02965. [DOI] [PubMed] [Google Scholar]
- 15.Der-Sarkissian H, Vergnaud G, Borde YM, Thomas G, Londono-Vallejo JA. Segmental polymorphisms in the proterminal regions of a subset of human chromosomes. Genome Res. 2002;12:1673–1678. doi: 10.1101/gr.322802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooney AJ, Tsai SY, O'Malley BW, Tsai MJ. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- 20.Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–7901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- 21.Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci. 2000;113:4577–4585. doi: 10.1242/jcs.113.24.4577. [DOI] [PubMed] [Google Scholar]
- 22.Chen YC, Kappel C, Beaudouin J, Eils R, Spector DL. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol Biol Cell. 2008;19:3147–3162. doi: 10.1091/mbc.E08-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 24.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 26.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 27.Torok D, Ching RW, Bazett-Jones DP. PML nuclear bodies as sites of epigenetic regulation. Front Biosci. 2009;14:1325–1336. doi: 10.2741/3311. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar C, et al. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jegou T, et al. Dynamics of telomeres and PML nuclear bodies in a telomerase negative human cell line. Mol Biol Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett RD, Murray J, Orr A, Preston CM. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol. 2007;81:10991–11004. doi: 10.1128/JVI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de La Roche Saint-Andre C. Alternative ends: Telomeres and meiosis. Biochimie. 2008;90:181–189. doi: 10.1016/j.biochi.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.